Abstract
The transcription factors Mesp1 and Mesp2 have essential roles in the migration and specification of multipotent progenitor cells at the onset of cardiogenesis. Chiapparo et al. (2016. J. Cell Biol. http://dx.doi.org/10.1083/jcb.201505082) identify common Mesp functions in fate specification and Mesp1-specific targets controlling the speed and direction of progenitor cell migration.
The vertebrate heart forms from multipotent cardiovascular progenitor cells that are specified at gastrulation as they migrate from the primitive streak in the posterior region of the embryo to the anterior lateral mesoderm (Fig. 1). Once they reach this region, cardiac progenitor cells differentiate in the cardiac crescent, rapidly followed by the formation of a linear heart tube. Addition of late-differentiating progenitor cells from the second heart field drives subsequent heart tube elongation, accompanied by rightward looping and the onset of chamber morphogenesis.
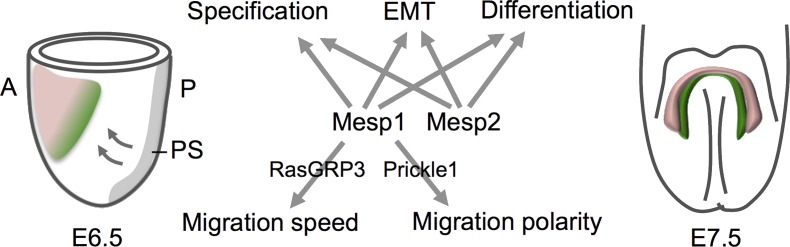
Figure 1.
Differential regulation of Rasgrp3 and Prickle1 by Mesp1 and Mesp2 controls migration speed and directionality of nascent cardiac mesoderm. A lateral view of an embryonic day (E) 6.5 mouse embryo (left), showing how cells that transiently express Mesp1 in the primitive streak (PS) migrate (arrows) to form cranial and cardiogenic mesoderm (denoted by the pink and green shaded area). A ventral view of an E7.5 embryo (right) shows differentiated cardiomyocytes in the cardiac crescent (pink), whereas second heart field cells (green), derived from later Mesp1-expressing progenitor cells, retain progenitor cell status and progressively contribute to the elongating heart tube. Mesp1 and Mesp2 are both required for cardiovascular progenitor cell specification, EMT, and differentiation. Chiapparo et al. (2016) now identify Mesp1-specific targets (Prickle and RasGRP3) that regulate the speed and polarity of cardiovascular progenitor cell migration in differentiating ES cells. A, anterior; P, posterior.
The earliest sign of cardiovascular development in the developing mouse embryo is transient expression of the bHLH transcription factor Mesp1 in the primitive streak (Saga et al., 2000). Although cardiac specification and differentiation occur in the absence of Mesp1, the migration of cardiovascular progenitor cells is delayed, impairing the formation of a single linear heart tube. Transcriptional up-regulation of the neighboring and paralogous gene Mesp2, coexpressed with Mesp1 during gastrulation, suggests there is functional redundancy between these factors in cardiac specification and differentiation. This was confirmed by the absence of cranial and cardiac mesoderm in Mesp1 Mesp2 double mutant embryos (Kitajima et al., 2000). Further studies in chimeric embryos revealed that Mesp1 Mesp2 double mutant cells do not contribute to the developing heart, indicating a cell autonomous role in cardiac specification. Genetic lineage tracing using Cre recombinase has shown that Mesp1-expressing progenitor cells give rise to all cardiac cell types, including myocardium, the endocardial lining of the heart, and the outer epicardial layer. In addition, the Mesp1 lineage contributes to endothelial cells, smooth muscle, and cranial and extraembryonic mesoderm (Saga et al., 2000). Recently, clonal analysis has revealed that these diverse fates reflect Mesp1 activation in temporally and molecularly distinct progenitor cell populations. These cell populations contribute sequentially to different cranial and cardiovascular derivatives, including endocardial or myocardial cells of the linear heart tube and, later, multipotent cardiopharyngeal mesoderm, which gives rise to both second heart field and head muscle progenitor cells (Lescroart et al., 2014). The upstream role of Mesp1 in cardiogenesis is conserved across chordates. The single ascidian Mesp gene is essential for specification of the cardiopharyngeal lineage, which gives rise to the heart and pharyngeal muscles in Ciona, and regulates the genetic program controlling cardiopharyngeal progenitor cell migration (Satou et al., 2004; Christiaen et al., 2008).
Differentiating pluripotent embryonic stem (ES) cells provide a powerful system in which to dissect the mechanisms that underlie early fate decisions and have been used extensively to study the onset of cardiac differentiation. The analysis of Mesp1 function and targets during ES cell cardiac differentiation has provided molecular insights into how Mesp1 rules the cardiovascular hierarchy. Induced overexpression experiments and use of fluorescent reporter alleles have shown that Mesp1 irreversibly promotes cardiovascular progenitor cell fate by directly and/or indirectly regulating the expression of genes involved in epithelial–mesenchymal transition (EMT), as well as the expression of early mesodermal and core cardiac transcription factors (Bondue et al., 2008, 2011; Lindsley et al., 2008; David et al., 2009; Chan et al., 2013; Soibam et al., 2015; den Hartogh et al., 2016). Bipotent cardiopharyngeal progenitor cells with dual cardiac and skeletal muscle potential have recently been identified using this system (Chan et al., 2016). Together, these studies suggest that transient Mesp1 activity leads to the cell-autonomous activation of a series of context-dependent targets and primes genes for activation after Mesp1 expression has been down-regulated. This priming by Mesp1 may occur via the activation of pro-differentiation signaling pathway components, as well as by the direct regulation of target gene chromatin structure (Bondue et al., 2008; Soibam et al., 2015). One Mesp1 target is Mesp1 itself; other upstream regulators of Mesp1 expression in the primitive streak have been identified, including the T-box transcription factors Brachyury and Eomesodermin, Oct4, and components of the canonical Wnt signaling pathway (Liu and Schwartz, 2013). Other genes, including Mesp2, are down-regulated on Mesp1 expression, ensuring unidirectional lineage specification (Bondue et al., 2008). However, the relative roles of Mesp1 and Mesp2 in the regulation of cell migratory activity and cell fate specification have remained unclear.
In this issue Chiapparo et al. address this question by comparing the function and targets of Mesp2 with those of Mesp1 in differentiating ES cells using an inducible gain-of-function approach. A detailed characterization of the resulting phenotypes revealed that Mesp1 and Mesp2 have indistinguishable roles in cardiovascular progenitor cell specification, in the onset of EMT, and in myocardial and endothelial cell differentiation, consistent with previous evidence that Mesp2 can induce progenitor cell and EMT markers (Lindsley et al., 2008). In contrast Mesp1, but not Mesp2, is required cell autonomously for rapid progenitor cell migration, monitored using time-lapse microscopy in a monolayer scratch assay. Similarly, Mesp1 specifically drives the polarized migration of overexpressing cells, accompanied by reorganization of the actin cytoskeleton and the appearance of oriented stress fibers. Importantly, these observations provide a molecular explanation for the phenotypes of Mesp1 and Mesp1 Mesp2 double mutant mouse embryos in which progenitor cell migration is impaired and cardiovascular specification is ablated, respectively.
Chiapparo et al. (2016) provide additional molecular insights into Mesp protein subfunctionalization by microarray analysis. Numerous commonly regulated genes were identified, with roles in cardiovascular specification and differentiation, as well as in EMT (Fig. 1). The authors focused on two genes, Rasgrp3 and Prickle1, that are differentially regulated by Mesp1 and Mesp2 and that might mediate Mesp1-specific functions. Rasgrp3 encodes a guanine nucleotide exchange factor that switches on Ras GTPase activity and downstream extracellular signal-regulated kinase (ERK) signaling. Chiapparo et al. (2016) present evidence that a Mesp1-RasGRP3-ERK cascade drives the fast migration of Mesp1-expressing cells. In support of this, Rasgrp3 is expressed in mesoderm emerging from the primitive streak, and Rasgrp3 expression and phospho-ERK levels are reduced in Mesp1 mutant embryos. Both Mesp proteins can bind to target sites in the first intron of Rasgrp3, although only Mesp1 appears to up-regulate Rasgrp3 transcription, suggesting the existence of yet to be identified Mesp1-specific coactivators. RasGRP3 overexpression in the ES cell differentiation system promotes ERK activation and ERK-dependent migration, although less so than on Mesp1 expression, indicating that additional targets are required for Mesp1-regulated ERK activity. Indeed, RasGRP3 levels may act together with other inputs, such as FGF-driven ERK activation, to accelerate Mesp1-expressing progenitor cell migration. In Ciona, Mesp-dependent FGF signaling drives cardiopharyngeal progenitor migration by up-regulating the gene that encodes the GTPase RhoDF, promoting actin filament growth and membrane protrusions (Christiaen et al., 2008). Using gene editing in ES cells, Chiapparo et al. (2016) show that RasGRP3 is required for fast migration after Mesp1 overexpression. Rasgrp3 null embryos, however, survive gastrulation, potentially through the activity of other guanine nucleotide exchange factors including additional members of the Rasgrp family. Ras activation by RasGRP3 accompanies angiogenesis and a Rasgrp3 lacZ gene trap allele has been shown to be expressed at sites of blood vessel formation (Roberts et al., 2004). Furthermore, RasGRP3 is required for Endothelin stimulated endothelial cell migration, although overexpression of RasGRP3 in endothelial cells, in contrast to ES cells, affects the direction but not the speed of migration (Randhawa et al., 2011). Future experiments will define whether RasGRP3 specifically controls migration speed in Mesp1-expressing progenitor populations with endothelial fates.
Whereas migration speed is enhanced in Rasgrp3-expressing ES cells, Chiapparo et al. (2016) found that the direction of migration is unpolarized in these cells and that the inactivation of Rasgrp3 does not affect cell polarity after Mesp1 overexpression. They went on to identify Prickle1 as a Mesp1-specific target that controls the directionality of migration. Prickle1 is a core component of the planar cell polarity pathway that orients cells within epithelia. Like Rasgrp3, Prickle1 is expressed in cells ingressing through the primitive streak and is decreased in Mesp1 null embryos. Mesp1, but not Mesp2, directly binds to target sites in the first intron of Prickle1, suggesting that differential DNA binding, as well as different cofactor interactions, may distinguish the two Mesp proteins. Overexpression of Prickle1 in ES cells increased the polarity but not the speed of cardiovascular progenitor cell migration. Loss of Prickle1 has revealed that it is required before gastrulation for apicobasal polarity of epiblast cells and that its loss is associated with defects in extracellular matrix deposition and spindle orientation (Tao et al., 2009). Interestingly, Prickle1 has recently been shown to play a later role in heart development. The Beetlejuice mutation was identified in an N-ethyl-N-nitrosourea mutagenesis screen as a novel Prickle1 allele that causes defective development of late-differentiating second heart field cells (Gibbs et al., 2016). In particular, polarized cell orientation and epithelial tissue architecture are lost in the transition zone where differentiation takes place, resulting in a failure of heart tube elongation and congenital heart defects. Consistent with the Mesp1 study, Prickle1 was found to specifically regulate directionality of embryonic fibroblast migration in a wound-closure assay. Prickle1 thus plays iterative roles in the control of cell polarity and migration during heart development.
The dissection of events upstream and downstream of Mesp1 is proving to be a powerful approach to probe the earliest steps in cranial mesoderm and cardiovascular lineage decisions in vitro and in vivo. In the study by Chiapparo et al. (2016), new insights are provided into cardiovascular progenitor cell biology. These define how intrinsic and extrinsic inputs are integrated to coordinate the specification and migration of nascent cardiogenic mesoderm in differentiating ES cells. Further work is needed to resolve the sequential sets of direct Mesp targets in vivo, as well as the identification of Mesp primed genes. This is particularly challenging given the transient expression of endogenous Mesp1 in different progenitor cell populations. For example, do the Mesp1 targets identified by Chiapparo et al. (2016) regulate the speed and direction of migration of all, or subsets of, Mesp1-expressing cells as they leave the primitive streak? The mechanisms underlying the differential binding and transactivation properties of the two Mesp proteins also remain to be identified. The work of Chiapparo et al. (2016) confirms that progenitor cell migration and specification are separable processes yet coordinated by common transcriptional regulators, consistent with earlier analysis of the single Mesp homologue in Ciona (Christiaen et al., 2008). Finally, although focused on the earliest events in cardiovascular progenitor cell biology, the study of Mesp1 function can provide insights that are relevant to regenerative medicine and the etiology of human congenital defects, as pathological variants of MESP1 have been identified in patients with congenital heart defects (Werner et al., 2016).
Acknowledgments
Work in R.G. Kelly's laboratory is supported by grants from the Fondation pour la Recherche Médicale, Association Française contre les Myopathies, and Fondation Leducq.
The author declares no competing financial interests.
References
- Bondue A., Lapouge G., Paulissen C., Semeraro C., Iacovino M., Kyba M., and Blanpain C.. 2008. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 3:69–84. 10.1016/j.stem.2008.06.009 [DOI] [PubMed] [Google Scholar]
- Bondue A., Tännler S., Chiapparo G., Chabab S., Ramialison M., Paulissen C., Beck B., Harvey R., and Blanpain C.. 2011. Defining the earliest step of cardiovascular progenitor specification during embryonic stem cell differentiation. J. Cell Biol. 192:751–765. 10.1083/jcb.201007063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.S., Shi X., Toyama A., Arpke R.W., Dandapat A., Iacovino M., Kang J., Le G., Hagen H.R., Garry D.J., and Kyba M.. 2013. Mesp1 patterns mesoderm into cardiac, hematopoietic, or skeletal myogenic progenitors in a context-dependent manner. Cell Stem Cell. 12:587–601. 10.1016/j.stem.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.S., Hagen H.R., Swanson S.A., Stewart R., Boll K.A., Aho J., Thomson J.A., and Kyba M.. 2016. Development of bipotent cardiac/skeletal myogenic progenitors from MESP1+ mesoderm. Stem Cell Rep. 6:26–34. 10.1016/j.stemcr.2015.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiapparo G., Lin X., Lescroart F., Chabab S., Paulissen C., Pitisci L., Bondue A., and Blanpain C.. 2016. Mesp1 controls the speed, polarity, and directionality of cardiovascular progenitor migration. J. Cell Biol. 10.1083/jcb.201505082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiaen L., Davidson B., Kawashima T., Powell W., Nolla H., Vranizan K., and Levine M.. 2008. The transcription/migration interface in heart precursors of Ciona intestinalis. Science. 320:1349–1352. 10.1126/science.1158170 [DOI] [PubMed] [Google Scholar]
- David R., Stieber J., Fischer E., Brunner S., Brenner C., Pfeiler S., Schwarz F., and Franz W.M.. 2009. Forward programming of pluripotent stem cells towards distinct cardiovascular cell types. Cardiovasc. Res. 84:263–272. 10.1093/cvr/cvp211 [DOI] [PubMed] [Google Scholar]
- den Hartogh S.C., Wolstencroft K., Mummery C.L., and Passier R.. 2016. A comprehensive gene expression analysis at sequential stages of in vitro cardiac differentiation from isolated MESP1-expressing-mesoderm progenitors. Sci. Rep. 6:19386 10.1038/srep19386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs B.C., Damerla R.R., Vladar E.K., Chatterjee B., Wan Y., Liu X., Cui C., Gabriel G.C., Zahid M., Yagi H., et al. 2016. Prickle1 mutation causes planar cell polarity and directional cell migration defects associated with cardiac outflow tract anomalies and other structural birth defects. Biol. Open. 5:323–335. 10.1242/bio.015750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima S., Takagi A., Inoue T., and Saga Y.. 2000. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development. 127:3215–3226. [DOI] [PubMed] [Google Scholar]
- Lescroart F., Chabab S., Lin X., Rulands S., Paulissen C., Rodolosse A., Auer H., Achouri Y., Dubois C., Bondue A., et al. 2014. Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat. Cell Biol. 16:829–840. 10.1038/ncb3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley R.C., Gill J.G., Murphy T.L., Langer E.M., Cai M., Mashayekhi M., Wang W., Niwa N., Nerbonne J.M., Kyba M., and Murphy K.M.. 2008. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell. 3:55–68. 10.1016/j.stem.2008.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., and Schwartz R.J.. 2013. Transient Mesp1 expression: a driver of cardiac cell fate determination. Transcription. 4:92–96. 10.4161/trns.24588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randhawa P.K., Rylova S., Heinz J.Y., Kiser S., Fried J.H., Dunworth W.P., Anderson A.L., Barber A.T., Chappell J.C., Roberts D.M., and Bautch V.L.. 2011. The Ras activator RasGRP3 mediates diabetes-induced embryonic defects and affects endothelial cell migration. Circ. Res. 108:1199–1208. 10.1161/CIRCRESAHA.110.230888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D.M., Anderson A.L., Hidaka M., Swetenburg R.L., Patterson C., Stanford W.L., and Bautch V.L.. 2004. A vascular gene trap screen defines RasGRP3 as an angiogenesis-regulated gene required for the endothelial response to phorbol esters. Mol. Cell. Biol. 24:10515–10528. 10.1128/MCB.24.24.10515-10528.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga Y., Kitajima S., and Miyagawa-Tomita S.. 2000. Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc. Med. 10:345–352. 10.1016/S1050-1738(01)00069-X [DOI] [PubMed] [Google Scholar]
- Satou Y., Imai K.S., and Satoh N.. 2004. The ascidian Mesp gene specifies heart precursor cells. Development. 131:2533–2541. 10.1242/dev.01145 [DOI] [PubMed] [Google Scholar]
- Soibam B., Benham A., Kim J., Weng K.C., Yang L., Xu X., Robertson M., Azares A., Cooney A.J., Schwartz R.J., and Liu Y.. 2015. Genome-wide identification of MESP1 targets demonstrates primary regulation over mesendoderm gene activity. Stem Cells. 33:3254–3265. 10.1002/stem.2111 [DOI] [PubMed] [Google Scholar]
- Tao H., Suzuki M., Kiyonari H., Abe T., Sasaoka T., and Ueno N.. 2009. Mouse prickle1, the homolog of a PCP gene, is essential for epiblast apical-basal polarity. Proc. Natl. Acad. Sci. USA. 106:14426–14431. 10.1073/pnas.0901332106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner P., Latney B., Deardorff M.A., and Goldmuntz E.. 2016. MESP1 mutations in patients with congenital heart defects. Hum. Mutat. 37:308–314. 10.1002/humu.22947 [DOI] [PMC free article] [PubMed] [Google Scholar]