Abstract
Natural modifications of peptidoglycan modulate the innate immune response. Peptidoglycan derivatives activate this response via the intracellular innate immune receptor, Nod2. To probe how these modifications alter the response, a novel and eficient carbohydrate synthesis was developed to allow for late-stage modification of the amine at the 2-position. Modification of the carbohydrate was found to be important for stabilizing Nod2 and generating the proper response. The native Nod2 ligands demonstrate a significant increase in the cellular stability of Nod2. Moreover, changing the identity of the natural ligands at the carbohydrate 2-position allows for the Nod2-dependent immune response to be either up-regulated or down-regulated. The ligand structure can be adjusted to tune the Nod2 response, suggesting that other innate immune receptors and their ligands could use a similar strategy.
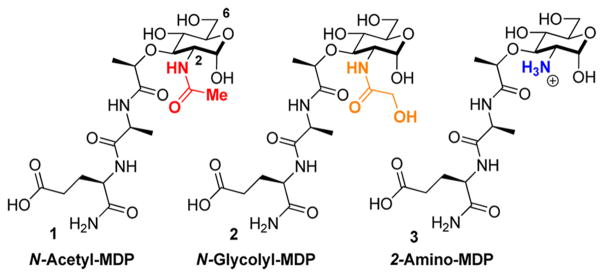
The innate immune system must recognize and destroy pathogenic bacteria while maintaining the proper balance of the trillion commensal bacteria. One method by which the innate immune system detects bacteria is via the peptidoglycan, a strong polymer of carbohydrates and peptides that provides protection to the vulnerable bacterial cell. Peptidoglycan fragments such as N-acetyl-muramyl dipeptide (MDP) (1, Figure 1) and N-glycolyl-MDP (2, Figure 1) activate an immune response.1 Compound 1 is a highly conserved component of peptidoglycan and is found in both Gram-positive and Gram-negative bacteria, while 2 is specific to the Gram-positive bacteria Mycobacterium avium, the bacterium commonly found in patients suffering from Crohn’s disease, a debilitating, noncurable, inflammatory bowel disease. In order to evade an innate immune response, pathogenic bacteria such as Staphylococcus aureus, Listeria monocytogenes, and Bacillus cereus utilize acetylation and deacetylation strategies to avoid detection,3 generating fragments such as 2-amino-MDP (3, Figure 1). However, it is not known how specific immune receptors respond to these peptidoglycan modifications (Figure 1).
Figure 1.
Naturally occurring muramyl dipeptides (MDPs) with modifications at the 2-position.
Recently, 1 was shown to directly interact with the human innate immune receptor protein Nod2,4 ultimately resulting in the activation of a signaling cascade known as the inflammatory response via the NF-κB and MAP kinase pathways,5 while 2 has been demonstrated to elicit a more robust Nod2-dependent inflammatory response via the same pathways.6 The differential effects of both the presence and identity of acyl substituents could have broad implications in immune recognition of bacteria. Here we investigate how acetylation/deacetylation of these peptidoglycan fragments modulates molecular recognition by Nod2.
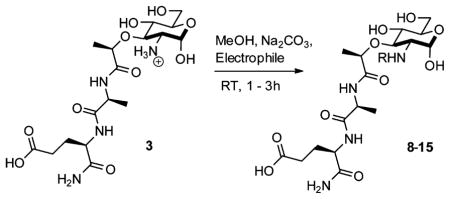
To further study the interaction of N-substituted MDPs (Figure 1) and Nod2, we required a synthetic route for MDP that offers selective functionalization of the 2-position. Existing synthetic strategies require extensive carbohydrate manipulation,7 troublesome dipeptide couplings,8 or expensive carbohydrate starting materials,9 and to date, no existing syntheses allow for a facile, late-stage modification at the 2-position.10 Additionally, no syntheses exist for the deacetylated peptidoglycan derivatives. We thus examined a rapid and modular synthesis that would allow for alterations at the 2-position and provide a divergent route for a variety of peptidoglycan and peptidoglycan-like derivatives. We present here a synthetic route that (1) allows general exploration of function via acylation at the carbohydrate 2-position and (2) utilizes an azide, a sterically unobtrusive acid-and base-stable protecting group,11 to mask the amine in the synthetic approach.
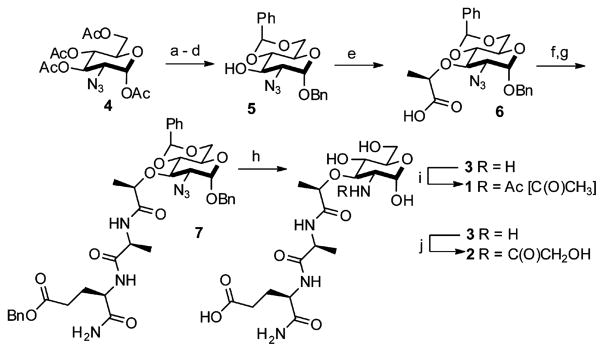
The synthesis begins with the inexpensive and readily obtainable starting material glucosamine hydrochloride, which is easily converted to 5 in six high-yielding steps using notably a diazo transfer to install the azide 4 and a Koenigs-Knorr-type reaction to protect the anomeric position (Scheme 1).12 The coupling reactions of a dipeptide fragment to modularly protected muramic acid derivatives were low yielding; thus, we opted to perform the amino acid couplings sequentially. Global deprotection was performed using Pd/C and H2 to expose the 2-amino functionality and yield 3, an N-deacetylated naturally occurring MDP. Acetylation of the 2-amino position under mild conditions furnished the N-acetyl-MDP target 113 and the N-glycolyl-MDP target 210 in 10 linear steps from glucosamine hydrochloride with overall yields of 33% and 28%, respectively. The synthesis provides a simplified route to the minimal peptidoglycan fragments known to activate Nod2.6,14
Scheme 1.
Synthetic Route for the Preparation of N-Acetyl-and N-Glycolyl-Muramyl Dipeptidesa
aReagents and conditions: (a) hydrazine acetate, DMF, 80%; (b) 1. oxalyl chloride, cat. DMF, DCM; 2. benzyl alcohol, cat. Ag2CO3, cat. AgOTf, DCM, 85%; (c) cat. NaOMe, MeOH, quantitative; (d) cat. pTSA, benzaldehyde dimethyl acetal, DMF, 95%; (e) NaH, (S)-2-chloropropionic acid, DMF, 76%; (f) 1. HBTU, N-methylmorpholine, DMF, 2. L-alanine methyl ester hydrochloride, 81%; (g) 1. KOH, MeOH, 2. HOBt, EDC, DMF, 2,4,6-collidine, 3. D-isoglutamine benzyl ester perchlorate, 89%; (h) Pd/C, H2O, MeOH, AcOH, quantitative; (i) H2O, NaHCO3, Ac2O, quantitative (j) MeOH, Na2CO3, acetoxyacetic acid NHS ester, 85%.
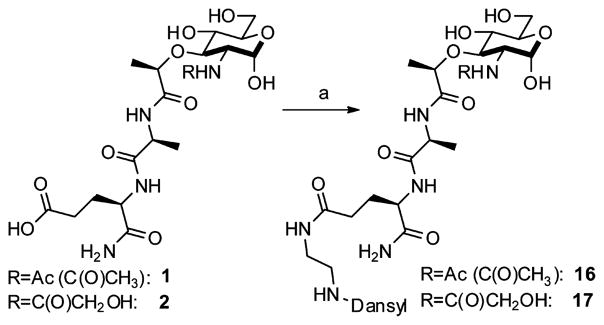
Having established a synthesis that allowed for the late-stage modification of the 2-amino position and the potential to yield a variety of new, 2-amino-functionalized MDPs via the naturally occurring deacetylated MDP 3, we set out to produce other peptidoglycan-like derivatives (Table 1) to explore their effect on Nod2 signaling. Functionality was chosen for coupling that would produce MDP derivatives applicable for use as chemical tools to study biological systems from peptidoglycan biosynthesis to peptidoglycan-induced innate immune system activation. We synthesized 8 and 9, the acetylated and methyl ether versions of 2, to probe how altering the glycolyl substitution affects Nod2 recognition. Compound 10 inverses the electronics from the parent compound 1. Compounds 11 and 12 represent extended acetylations reminiscent of 1 and 2. Finally, 13–15 represent MDP derivatives that are applicable to click chemistry, fluorophore-based assays, and anchoring to avidin/streptavidin, respectively. Additionally, this strategy allows for installation of orthogonal modification, permitting the synthesis of peptidoglycan derivatives with chemical tools at locations other than the 2-position (Scheme 2). Finally, one can envision the installation of a 13C- or 14C-labeled acetate to yield the isotopes of 1, tools that are important in studying innate immunity, for NMR analysis and other biological studies.
Table 1.
Generation of Peptidoglycan Derivatives from 2-Amino-MDP (3)
| |||
|---|---|---|---|
| Entry | Electrophile | Product | Yield |
| 1a,b | Acetoxyacetic acid NHS ester | 8,R=C(O)CH2OAc | 48% |
| 2 | Methoxyacetic anhydride | 9,R = C(O)CH2OMe | 78% |
| 3 | Ethyl triflouroacetate | 10,R=C(O)CF3 | 92% |
| 4c | Succinic anhydride | 11,R = C(O)(CH2)2CO2H | 66% |
| 5d | Levulinic acid NHS ester | 12, R = C(O)(CH2)2C(O)Me | 77% |
| 6d | 2-Azidoacetic acid NHS ester | 13,R=C(O)CH2N3 | 88% |
| 7c,e | Dansyl chloride | 14, R= Dansyl | 48% |
| 8 | Biotin NHS ester | 15, R = Biotin | 79% |
Electrophile prepared according to literature precedent.15
Per-formed in H2O with NaHCO3.
Stirred 24 h.
Electrophile prepared with EDC/NHS.
Performed in H2O/DMF.
Scheme 2.
Orthogonal Reactions for Chemical Toolsa
aReagents and conditions: (a) HOBt, EDC, 2,4,6-collidine, MeOH, 5-dimethylaminonaphthalene-1-(N-(2-aminoethyl))sulfonamide, quantitative.
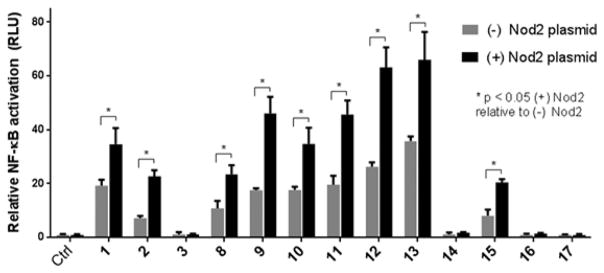
To directly determine how modification at the 2-position affects innate immune system activation, the MDP derivatives 1–3, 8–17 were assayed for activity (Figure 2, Table 2) using a Nod2-dependent NF-κB reporter assay.16 All compounds were tested at 20 μM, as we have shown in an in vitro binding assay that Nod2 binds to 1 with a KD of 51 nM.4a In addition, select compounds (1–3, 12, 13) were tested at a range of concentrations (0.2, 2, 20, and 200 μM) to ensure NF-κB activation studies were done at an effective concentration (Figure S1). As expected, 1 and 2 activate NF-κB in a Nod2-dependent manner (Figure 2, Table 2). Compound 3, the N-deacetylated-MDP, did not activate NF-κB, thus suggesting that N-acylation is an important feature for recognition by the Nod2 signaling cascade. Most modifications are tolerated at the 2-position, with 9–13 activating Nod2 ~2-fold greater than the untreated control. The fluorescent derivatives 14, 16, and 17 do not activate, regardless of fluorophore position. In contrast, placement of a biotin molecule, containing four methylenes (separating the amide from the heterocycle), at the 2-position (15) still allows activation of NF-κB, albeit to a lesser extent than the other acylated derivatives. These data suggest that the installation of a fluorophore is possible with increased linker length. The data also show that there is significant NF-κB activation in HEK293T cells, which follows the same trend as the Nod2-induced effect. This could be from the endogenous Nod2 and/or other targets of the bacterial cell wall ligands. Overall these data demonstrate that modification of the 2-position can be used for optimization/modulation of the Nod2 response.
Figure 2.
NF-κB activation with standard deviation for peptidoglycan derivatives. HEK293T cells were transfected using lipofectamine with (±) Nod2 plasmid, NF-κB reporter, and a renilla control for 16 h. The cells were treated with stimuli for 12 h, harvested, and tested for luciferase activity (* = P < 0.05, activates in a Nod2-dependent manner). All compounds were tested at 20 μM. Procedures and tabulated data (Table S1) can be found in the Supporting Information. Two effects are seen: (1) the native effect of the MDP derivatives (gray bar vs control) on NF-κB signaling, which could be due to native Nod2 or other targets activated by MDP; and (2) the induced effect of the MDP derivatives (gray bars vs black bars for a given compound) on overexpressed Nod2.16,17
Table 2.
Cellular Effects of Peptidoglycan Derivatives on Nod2 Signaling and Stabilitya
| Compound | NF-κB Fold Activation ± SD | Half-life (h)± SD |
|---|---|---|
| Control | No activation | 7.1 ±1.4 |
| 1 | 1.8 ±0.3 | 15.9 ±4.1* |
| 2 | 3.1 ±0.3 | 23.1 ±2.2* |
| 3 | No activation | 7.7 ±0.7 |
| 8 | 2.2 ±0.3 | Not tested |
| 9 | 2.6 ±0.4 | 10.5 ±2.6* |
| 10 | 1.9 ±0.3 | 15.0 ±1.8* |
| 11 | 2.3 ±0.3 | Not tested |
| 12 | 2.4 ±0.3 | 11.7 ±2.7* |
| 13 | 1.8 ±0.3 | 8.9 ±0.9* |
| 15 | 2.5 ±0.3 | Not tested |
| 16 | No activation | 8.0 ±0.7 |
 18 |
2.8±0.119 | 19.0 ±1.8* |
(+) Nod2 plasmid NF-κB activation normalized to the (−) Nod2 plasmid. Nod2 band intensities were plotted against time assuming first-order decay (ln(Ir) vs time). The rate constant was calculated using the negative slope of the line (k = −slope), and the corresponding half-life was calculated (T1/2 = ln(2)/k).21 SD = standard deviation.
= P < 0.05, stabilizes compared to the untreated control. The control, 3, and 16 did not significantly activate.
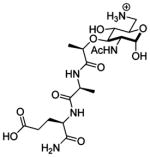
Nod2 is a transiently stable protein.18 It is well-known that ligand binding can affect the stability of a receptor.19 We considered that the interaction of peptidoglycan derivatives with Nod2 could alter the stability of the protein. To determine if ligand interaction changes the half-life of Nod2, we analyzed the naturally occurring MDPs 1–3 and a subset of the peptidoglycan derivatives with increased activity (12, 13) or structural similarity (9, 10) to 1 or 2. We made use of a tetracycline-induced Nod2-expressing cell line,18 an excellent match for endogenous Nod2 cell lines, and cycloheximide inhibition of protein synthesis to determine the half-life of Nod2 in the presence of the MDP derivatives.20 Half-life was determined by first-order decay according to literature precedent.21 Treatment with either 1 or 2 significantly increases the Nod2 half-life (Figure 3, Table 2). When Nod2 is not treated with a peptidoglycan derivative, the half-life is 7.1 ± 1.4 h. However, when cells are treated with 1, the half-life is 15.9 ± 4.1 h, and if cells are treated with 2, the half-life is 23.1 ± 2.2 h. The N-deacetylated derivative 3 provided no stabilization of Nod2 (Table 2). Notably, the N-glycolyl derivative 2 promotes greater stabilization of Nod2 than the N-acetyl derivative 1, suggesting that Nod2-dependent NF-κB activation and Nod2 stabilization are correlated for the naturally occurring compounds 1–3 (Table 2). As 3 is the only compound tested that contains an amine, which will be positively charged at cellular pH, we were concerned that cell permeability may be a factor. We analyzed the ability of 6-amino-MDP (18, Table 2), a MDP derivative that replaces the 6-primary-hydroxy group with an amine. This compound is known to activate NF-κB in a Nod2-dependent manner.11a Despite the presence of the charged ammonium ion, 18 increased the half-life of Nod2 by nearly 3-fold (Table 2), indicating that the presence of a free amine does not prevent cellular access.
Figure 3.
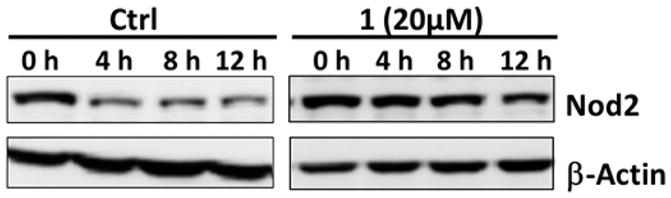
Peptidoglycan derivatives stabilize Nod2. HEK 293T-Nod2myc/Tet-op cells were incubated with MDP derivatives or water (control), and lysates were collected after cycloheximide treatment during the indicated time intervals. Equal amounts of lysates were subjected to Western blot and probed using rabbit anti-myc antibody. β-actin was used as a loading control. Procedures and all Western blots for half-lives shown in Table 2 are in the Supporting Information (Figures S2–S8).
Other N-substituted derivatives tested significantly stabilized Nod2 compared to the untreated control (Table 2), indicating that these modifications likely do not alter the ability of the ligand to bind and/or stabilize Nod2. The correlation between Nod2-dependent NF-κB activation and Nod2 stabilization was not as strong for these compounds as for the natural compounds 1–3, indicating that these compounds may have differential permeabilites and/or cell degradation pathways. It is important to note that 3 and 16, which did not activate, also did not stabilize (Table 2). The half-life data demonstrate that peptidoglycan derivatives are capable of altering the stability of the receptor, implying that ligand recognition is important for the signaling processes by initiating and maintaining the response. We note earlier studies using mouse Nod2 showed that treatment with very high concentrations (200 μM) of 1 led to the degradation of Nod222 when protein synthesis is not inhibited, suggesting there are other feedback mechanisms to control Nod2 levels in the cell. The data presented here demonstrate the ability to modulate Nod2 stability via natural or unnatural variation at the site of acetylation.
In their natural environments, virulent bacteria modify their peptidoglycan to evade detection by the innate immune system.3,23 Acetylation of the 6-position of the carbohydrate and deacetylation of the 2-postion prevents cell wall degradation by lysozyme.3c,d Interestingly, these data demonstrate that deacetylation of the 2-postion also eliminates the ability of the peptidoglycan fragment to signal through the Nod2-dependent pathway. Acetylation of the 6-position does not affect the ability to stimulate the Nod2-dependent immune response.11a These data suggests that the deacetylation strategy used by bacteria is two-pronged in that the modification (3) yields lysozyme-resistant peptidoglycan that does not elicit an immune response via the Nod2-dependent pathway.
In addition to its role in recognizing pathogenic bacteria, Nod2 is critical for maintaining the proper balance of commensal bacteria.24 Interestingly, Nod2 mutations that correlate with development of the inflammatory bowel disease, Crohn’s disease, are unable to activate Nod2.5a Previous experiments have shown that these mutations are unstable compared to the wild-type protein, and signaling can be restored via stability of Nod2 by the chaperone protein, Hsp70.18 This report shows that simple peptidoglycan derivatives stabilize Nod2 and modifications at the 2-position alter the stability of Nod2. Thus, peptidoglycan derivatives produced by these methods may stabilize Crohn’s-associated Nod2 mutations, providing therapeutic leads.
In conclusion, acylation of peptidoglycan derivatives at the 2-position allows for the tuning of Nod2 stability and NF-κB response. This new, rapid, tunable, high-yielding synthesis of peptidoglycan derivatives allowed the production of analogues to probe the substrate requirements for activation and stabilization effects on the protein. Nature produces ligands (1, 2) that are capable of stabilizing Nod2 and other ligands (3) that produce no stabilizing effect. Moreover, synthetic, novel peptidoglycan-like derivatives can activate and stabilize Nod2, informing on requirements for modulating the innate immune signaling cascade in response to bacterial cell wall fragments.
Supplementary Material
Acknowledgments
This publication was made possible by the Delaware COBRE program, supported by a grant from the National Institute of General Medical Sciences (NIGMS 1 P30 GM110758 and 1 P20 GM104316-01A1) from the National Institutes of Health. C.L.G. is a Pew Scholar in the Biomedical Sciences, supported by the Pew Charitable Trusts. We thank Neal Zondlo, John Koh, and Thomas Hanson for critical reading of this manuscript.
Footnotes
Notes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Experimental procedures, spectral data, NF-κB assay conditions, detailed half-life protocols, Western blot analysis, and complete refs 3c, 5a, 6, and 12b. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.5b01607.
References
- 1.(a) Kindt T, Goldsby R, Osborne B. Kuby Immunology. 6. W.H. Freeman and Co; New York: 2007. [Google Scholar]; (b) Ellouz F, Adam A, Ciorbaru R, Lederer E. Biochem Biophys Res Commun. 1974;59:1317. doi: 10.1016/0006-291x(74)90458-6. [DOI] [PubMed] [Google Scholar]; (c) Rubino SJ, Magalhaes JG, Philpott D, Bahr GM, Blanot D, Girardin SE. Innate Immun. 2013;19:493. doi: 10.1177/1753425912471691. [DOI] [PubMed] [Google Scholar]
- 2.Raymond JB, Mahapatra S, Crick DC, Pavelka MS., Jr J Biol Chem. 2005;280:326. doi: 10.1074/jbc.M411006200. [DOI] [PubMed] [Google Scholar]
- 3.(a) Sansonetti PJ, Di Santo JP. Immunity. 2007;26:149. doi: 10.1016/j.immuni.2007.02.004. [DOI] [PubMed] [Google Scholar]; (b) Bera A, Herbert S, Jakob A, Vollmer W, Gotz F. Mol Microbiol. 2005;55:778. doi: 10.1111/j.1365-2958.2004.04446.x. [DOI] [PubMed] [Google Scholar]; (c) Boneca IG, et al. Proc Natl Acad Sci USA. 2007;104:997. [Google Scholar]; (d) Vollmer W. FEMS Microbiol Rev. 2008;32:287. doi: 10.1111/j.1574-6976.2007.00088.x. [DOI] [PubMed] [Google Scholar]
- 4.(a) Grimes CL, de Ariyananda LZ, Melnyk JE, O’Shea EK. J Am Chem Soc. 2012;134:13535. doi: 10.1021/ja303883c. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mo J, Boyle JP, Howard CB, Monie TP, Davis BK, Duncan JA. J Biol Chem. 2012;287:23057. doi: 10.1074/jbc.M112.344283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Inohara N, et al. J Biol Chem. 2003;278:5509. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]; (b) Moreira LO, Zamboni DS. Front Immunol. 2012;3:328. doi: 10.3389/fimmu.2012.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coulombe F, et al. J Exp Med. 2009;206:1709. doi: 10.1084/jem.20081779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Kiso M, Kaneda Y, Okumura H, Hasegawa A, Azuma I, Yamamura Y. Carbohydr Res. 1980;79:C17. doi: 10.1016/s0008-6215(00)85145-9. [DOI] [PubMed] [Google Scholar]; (b) Hasegawa A, Okumura H, Nishibori K, Kaneda Y, Kiso M, Azuma I. Carbohydr Res. 1981;97:337. doi: 10.1016/s0008-6215(00)80713-2. [DOI] [PubMed] [Google Scholar]
- 8.Okumura H, Tokushima Y, Saiki I, Azuma I, Kiso M, Hasegawa A. Carbohydr Res. 1983;122:87. doi: 10.1016/0008-6215(83)88409-2. [DOI] [PubMed] [Google Scholar]
- 9.Heibert CK, Kopp WC, Richerson HB, Barfknecht CF. J Med Chem. 1983;26:1729. doi: 10.1021/jm00366a014. [DOI] [PubMed] [Google Scholar]
- 10.Xing S, Gleason JL. Org Biomol Chem. 2014;13:1515. doi: 10.1039/c4ob02147a. [DOI] [PubMed] [Google Scholar]
- 11.(a) Grimes CL, Podolsky DK, O’Shea EK. Bioorg Med Chem Lett. 2010;20:6061. doi: 10.1016/j.bmcl.2010.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Masuko S, Bera S, Green DE, Weiwer M, Liu J, DeAngelis PL, Linhardt RJ. J Org Chem. 2012;77:1449. doi: 10.1021/jo202322k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Li Y, Zhou Y, Ma Y, Li X. Carbohydr Res. 2011;346:1714. doi: 10.1016/j.carres.2011.05.024. [DOI] [PubMed] [Google Scholar]; (d) Crich D, Dudkin V. J Am Chem Soc. 2001;123:6819. doi: 10.1021/ja010086b. [DOI] [PubMed] [Google Scholar]
- 12.(a) Goddard-Borger ED, Stick RV. Org Lett. 2007;9:3797. doi: 10.1021/ol701581g. [DOI] [PubMed] [Google Scholar]; (b) Ye H, et al. Org Lett. 2013;15:18. doi: 10.1021/ol4021095. [DOI] [PubMed] [Google Scholar]; (c) Shing TKM, Perlin AS. Carbohydr Res. 1984;130:65. [Google Scholar]
- 13.McFarlane EF, Martinic C. Aust J Chem. 1983;36:1087. [Google Scholar]
- 14.Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ. J Biol Chem. 2003;278:8869. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 15.Mitsos C, Zografos A, Igglessi-Markopoulou O. J Hetercycl Chem. 2002;39:1201. [Google Scholar]
- 16.Barnich N, Aguirre JE, Reinecker HC, Xavier R, Podolsky DK. J Cell Biol. 2005;170:21. doi: 10.1083/jcb.200502153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao L, Kwon MJ, Huang S, Lee JY, Fukase K, Inohara N, Hwang DH. J Biol Chem. 2007;282:11618. doi: 10.1074/jbc.M608644200. [DOI] [PubMed] [Google Scholar]
- 18.Mohanan V, Grimes CL. J Biol Chem. 2014;289:18987. doi: 10.1074/jbc.M114.557686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.(a) Pissios P, Tzameli I, Kushner P, Moore DD. Mol Cell. 2000;6:245. doi: 10.1016/s1097-2765(00)00026-5. [DOI] [PubMed] [Google Scholar]; (b) Zhang X, Stevens RC, Xu F. Trends Biochem Sci. 2015;40:79. doi: 10.1016/j.tibs.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. Nat Chem Biol. 2010;6:209. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belle A, Tanay A, Bitincka L, Shamir R, O’Shea Erin K. Proc Natl Acad Sci USA. 2006;103:13004. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee KH, Biswas A, Liu YJ, Kobayashi KS. J Biol Chem. 2012;287:39800. doi: 10.1074/jbc.M112.410027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bishop JL, Boyle EC, Finlay BB. Proc Natl Acad Sci USA. 2007;104:691. doi: 10.1073/pnas.0611133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, Tlaskalova-Hogenova H, Kobayashi KS. Proc Natl Acad Sci USA. 2009;106:15813. doi: 10.1073/pnas.0907722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.