Abstract
Background:
Survivin is an oncoprotein silenced in normal mature tissues but reactivated in serous ovarian cancer (SOC). Although transcriptional activation is assumed for its overexpression, the long 3'-untranslated region (3'-UTR) in survivin gene, which contains many alternate polyadenylation (APA) sites, implies a propensity for posttranscriptional control and therefore was the aim of our study.
Methods:
The abundance of the coding region, the proximal and the distal region of survivin mRNA 3'-UTR, was evaluated by real-time polymerase chain reaction (PCR) in SOC samples, cell lines, and normal fallopian tube (NFT) tissues. The APA sites were confirmed by rapid amplification of cDNA 3' ends and DNA sequencing. Real-time PCR were used to screen survivin-targeting microRNAs (miRNAs) that were inversely correlated with survivin. The expression of an inversely correlated miRNA was restored by pre-miRNA transfection or induction with a genotoxic agent to test its inhibitory effect on survivin overexpression.
Results:
Varying degrees of APA were observed in SOC by comparing the abundance of the proximal and the distal region of survivin 3'-UTR, and changes of 3'-UTR correlated significantly with survivin expression (r = 0.708, P < 0.01). The main APA sites are proved at 1197 and 1673 of survivin 3'-UTR by DNA sequencing. Higher level of 3'-UTR proximal region than coding region was observed in NFT, as well as in SOC and cell lines. Among the survivin-targeting miRNAs, only a few highly expressed miRNAs were inversely correlated with survivin levels, and they mainly targeted the distal part of the 3'-UTR. However, in ovarian cancer cells, restoration of an inversely correlated miRNA (miR-34c) showed little effect on survivin expression.
Conclusions:
In NFT tissues, survivin is not transcriptionally silenced but regulate posttranscriptionally. In SOC, aberrant APA leads to the shortening of survivin 3'-UTR which enables it to escape the negative regulation of miRNAs and is responsible for survivin up-regulation.
Keywords: 3’-Untranslated Region Shortening, Alternative Polyadenylation, MicroRNA, Serous Ovarian Cancer, Survivin
INTRODUCTION
Posttranscriptional control is an important mechanism in the regulation of gene expression.[1] It is typically mediated by microRNAs (miRNAs) and RNA-binding proteins (RBPs) that recognize and bind to elements in the 3'-untranslated region (3'-UTR) of mRNAs. However, more than 50% of human genes contain alternative polyadenylation (APA) sites in their 3'-UTRs,[2] resulting in mRNA isoforms with the same coding sequences but different 3'-UTR lengths. As shorter 3'-UTR lengths may lack miRNA and RBP-binding sites, these genes may escape the negative regulation of miRNAs and RBPs.
Although genes with shorter 3'-UTRs have been reported in many cancers, much more remains to be investigated. First, the knowledge about APA of these genes is mainly from high-throughput genome-wide sequencing and bioinformatic analysis,[3,4,5] only a few genes are verified to have different 3'-UTR lengths in tissues by other methods. Second, the notion that 3'-UTR shortening results in higher gene expression come mainly from a few studies using luciferase reporter assays.[6] No correlation was found between 3'-UTR shortening and gene expression levels on transcriptome scale.[3,4,5] Finally, miRNA dysregulation has been identified in many cancers, including ovarian cancer.[7,8] Aberrant APA and miRNA dysregulation are interconnected aspects of posttranscriptional regulation in cancer, but the coordinating mechanism between APA and miRNAs is underexplored.
Although comprehensive shortening of 3'-UTR in ovarian cancer has never been reported, we previously found high expression of HMGA2, an oncofetal protein, to be significantly associated with its 3'-UTR shortening in ovarian cancer.[9] We reason that other oncogenes with long 3'-UTRs could also be regulated by APA.
Survivin (official gene name: BIRC5, NM_001168.2) is a member of the inhibitor of apoptosis family and affects multiple cellular functions, including proliferation, invasion, and cell survival control. Like HMGA2, survivin is widely expressed in fetal tissues and restrictively regulated during development. In most normal adult tissues, survivin expression appears to be negligible, but it is strongly expressed in various cancers, including ovarian cancer.[10] Elevated survivin expression is associated with enhanced proliferation, reduced apoptosis, resistant to chemotherapy, metastatic dissemination, tumor recurrence, and poor prognosis.[11,12,13] Like HMGA2, survivin mRNA has a long 3'-UTR of 2100 nt with nine putative APA sites as predicted by PolyA_SVM software (http://exon.umdnj.edu/polya_svm_server/index.html) and nearly 40 miRNA target sites as predicted by TargetScan software (www.targetscan.org). Although transcriptional reactivation is assumed for “tumor-specific” survivin overexpression,[14] posttranscriptional regulation is highly possible. Therefore, in this study, we focused on the critical factors concerning posttranscriptional regulation of survivin, including APA and survivin-targeting miRNAs, using serous ovarian cancer (SOC) samples.
METHODS
Tissue specimens and ovarian cancer cell lines
Forty specimens of high-grade SOC tissues were obtained from the Gynecology Tissue Bank of the Peking University People's Hospital, and five normal fallopian tubes (NFTs) from women who underwent salpingectomy for reasons other than adnexal malignancy. The NFTs were used as normal controls in this study, as they are proposed to be primary origins of high-grade SOC.[15,16] All specimens were obtained with informed consent, using protocols and procedures approved by the Ethics Committee at Peking University People's Hospital. Tissues samples were snap-frozen immediately after resection and stored at −80°C until use. Ovarian cancer cell lines SKOV3, SKOV3.ipl, CAOV3, OVCA429, and A2780 were maintained in standard DMEM, supplemented with 10% fetal calf serum.
RNA extraction
We used the miRNeasy Mini Kit (Qiagen, Germany) to extract high molecular weight (HMW) RNA (>200 nt) and low molecular weight (LMW) RNA (<200 nt), following manufacturer's instructions. The integrity of HMW RNAs was verified by 1% agarose gel electrophoresis. LMW RNAs were examined by electrophoresis using 3.5% NuSieve agarose gels (Cambrex, Rockland, USA). RNA concentration was measured by absorbance at 260 nm.
Real-time polymerase chain reaction analysis for mRNA expression
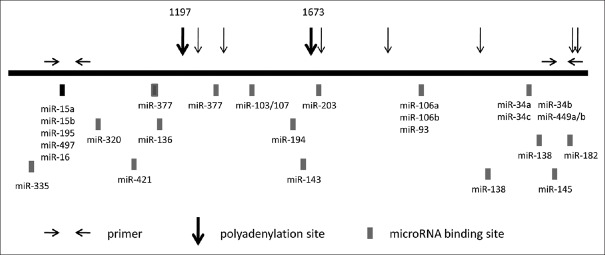
HMW RNAs were reverse-transcribed using random primers and SuperScript III reverse transcriptase (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's protocol. Three sets of primers aimed at coding region, proximal and distal region of 3'-UTR were designed to evaluate the expression of different parts of the survivin gene by real-time polymerase chain reaction (PCR). The sequence of the primer pairs were coding region: GGA CCA CCG CAT CTC TAC AT and CCA AGT CTG GCT CGT TCT CA; proximal region of 3'-UTR: TTG TCT TGA AAG TGG CAC CAG and AAG CCC GGG AAT CAA AAC AGC; and distal region of 3'-UTR: CAG AAG AGA CCA GCA AGC C and AAA CCA CAT GAG ACT TTA TTG GC. The position of the primers is shown in Figure 1.
Figure 1.
Schematic illustration of suvivin 3’-UTR. Horizontal arrows: primer locations for the real-time PCR reaction; solid boxes: miRNA-binding sites; vertical arrows: predicted polyadenylation sites; miRNA: MicroRNA; 3'-UTR: 3'-untranslated region; PCR: Polymerase chain reaction.
Real-time PCR was conducted with Opticon 2 (BioRad, USA) using the SYBR Green Real-time PCR Master Mix Kit (Toyobo, Osaka, Japan). The specificity of amplification was verified by the presence of a single peak on the dissociation curve. Relative expressions of different regions in each sample were normalized to glyceraldehyde-3-phosphate dehydrogenase as an internal reference.
The microRNA expression analysis by real-time polymerase chain reaction
LMW RNAs (1.5 µg) were polyadenylated by poly (A) polymerase and reverse transcribed using an anchored oligo-dT RT primer with a 36-nt extension at its 5'-ends (5'-GCT GTC AAC GAT ACG CTA CGT AAC GGC ATG ACA GTG [T] 24VN-3' [where V = A, G, or C; N = A, G, C, or T]). Reverse transcription was performed at 50°C without RNA denaturation using SuperScript III reverse transcriptase (Life Technologies, Carlsbad, CA, USA).
The miRNA-derived cDNAs were diluted 200–500 folds and amplified by real-time PCR using the SYBR Green Real-time PCR Master Mix Kit (Toyobo, Osaka, Japan). The first 20 nucleotides at the 5' end of the RT primer were defined as the common reverse primer (5'-GCTGTCAACGATACGCTACG-3'). Forward primers were carefully designed to ensure better discrimination of the miRNA isoforms and to avoid inaccurate quantification caused by length heterogeneity, as previously described.[17] Annealing conditions were optimized for each miRNA primer, using the gradient function of the Opticon 2 (BioRad, Hercules, CA, USA). Different miRNA primers were arrayed on plates to allow simultaneous amplification of different miRNAs at different annealing temperatures. The specificity of each reaction was determined by a single peak on dissociation curve, which was also confirmed by an 80-bp band on a 3.5% NuSieve GTG agarose gel (miRNAs: about 22 nt in length; extended oligo-dT RT primers: 62 nt). Pre-miRNAs were not amplified using 200-fold diluted miRNA-derived cDNA, possibly because pre-miRNA hairpin structures are less efficiently reverse-transcribed.
Reference miRNAs were selected by comparing expression variances of 5S RNA and nine abundant miRNAs representing different miRNA families (including miR-26a, let-7b, miR-30a-5p, miR-195, miR-98, miR-125a, miR-15b, miR-29a, and miR-34a) using NormFinder,[18] a model-based comparison approach. The miR-26 was identified as the most stable miRNA and used as the reference miRNA for normalization.
Rapid amplification of cDNA 3' ends
The GeneRacer kit (Life Technologies, USA) was used for rapid amplification of cDNA 3' ends (3' RACE) PCR with modifications. Briefly, RNAs were revered-transcribed at 60°C using ThermoScript reverse transcriptase (Life Technologies, USA) and an anchored oligo(d) T primer containing an adaptor sequence, (GCT GTC AAC GAT ACG CTA CGT AAC GGC ATG ACA GTG [T] 24VN). RNase H was added after reverse transcription. The cDNA was then amplified as follows: the outer 3' RACE adaptor primer (GCT GTC AAC GAT ACG CTA CG) and the outer gene-specific primer (GCA CCA CTT CCA GGG TTT AT) were used for the first amplification, followed by a nested PCR reaction with the inner 3' RACE adaptor primer (CGC TAC GTA ACG GCA TGA CAG TG) and the inner gene-specific primer (TTG TCT TGA AAG TGG CAC CAG). The PCR products were separated on agarose gels. DNA fragments were cloned into pCR4-TOPO plasmid and sequenced to identify the 3' ends of the targeted mRNAs.
Transfection of pre-microRNA-34c precursor
SKOV3.ipl cells were transfected with 30 nmol/L pre-miR-34c or pre-miR-neg precursor (Ambion, USA) using siPORT NeoFX (Ambion, USA) according to the manufacturer's instruction. Cells were harvested at 48 h for RNA extraction.
Induction of miR-34c expression in cancer cells
We treated the p53-wild-type ovarian cancer cell line A2780 with 0.5 µg/ml of the genotoxic agent adriamycin and harvested the cells for RNA extraction at 8 h, 16 h, and 24 h. Induction of miR-34c expression was verified by real-time PCR.
Statistical analysis
Data analysis was performed using SPSS software version 17.0 (SPSS Inc., Chicago, IL, USA) and Graphpad Prism 5.0 (Graphpad Software Inc., La Jolla, USA). Nonparametric method Mann–Whitney U-test was used to test the statistical difference of expression levels between two groups that were not normally distributed. Student's t-test was used to examine data with normal distribution. The Spearman correlation coefficient was used to test the association between data of two groups that were not normally distributed. A value of P < 0.05 was considered statistically significant.
RESULTS
Survivin coding region expression level
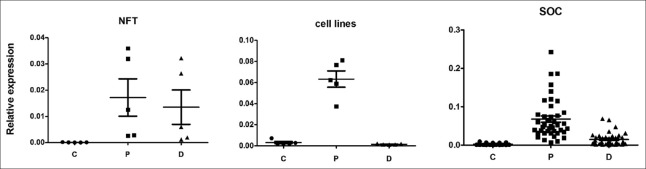
Coding region expression was evaluated by real-time PCR using a primer pair that spanned exon 1 to exon 2 of the survivin gene, which enabled detection of all mRNA isoforms. Survivin was detected in all SOC samples, but negative in NFT tissues, when cDNAs were diluted at 1:10 (the dilution we usually use). When we changed the dilution to 1:2, survivin was detected in NFT samples, with cycle threshold >30. Coding region expression levels significantly differed between SOC and NFT tissues [Figure 2, P < 0.01], whereas its expression in the five ovarian cancer cell lines was similar to those of most SOC samples [Figure 1].
Figure 2.
Expression of survivin coding region in NFT, ovarian cancer cell lines and SOC samples. *P < 0.01. NFT: Normal fallopian tube; SOC: Serous ovarian cancer.
Expression of survivin 3'-untranslated region proximal region is higher than that of the coding region
Expression levels of the 3'-UTR proximal and distal regions were evaluated by real-time PCR using two sets of primers for up- and down-stream 3'-UTR, respectively [Figure 2]. In all SOC samples, 3'-UTR proximal region expression was much higher than that of the coding region (the ratio of the proximal/coding region 8.5–123.0 times); the five ovarian cancer cell lines showed results similar to those of the SOC samples (8.6–45.0 times). Interestingly, although very low survivin coding-region mRNA was expressed in the NFT tissues, survivin 3'-UTR expression was 127–417 times that of the coding region [Figure 3], which suggests that transcription was not silenced. The ratio of the proximal region of 3'-UTR to coding region in NFT was significantly higher than that in SOC and ovarian cancer cell lines (P < 0.01 for both).
Figure 3.
Expression of survivin coding region, and 3'-UTR proximal and distal regions in different samples. C: Coding region; P: Proximal region; D: Distal region; NFT: Normal fallopian tube; SOC: Serous ovarian cancer; 3'-UTR: 3'-untranslated region.
Ratios of proximal/distal 3'-untranslated region expression correlated significantly with coding region expression
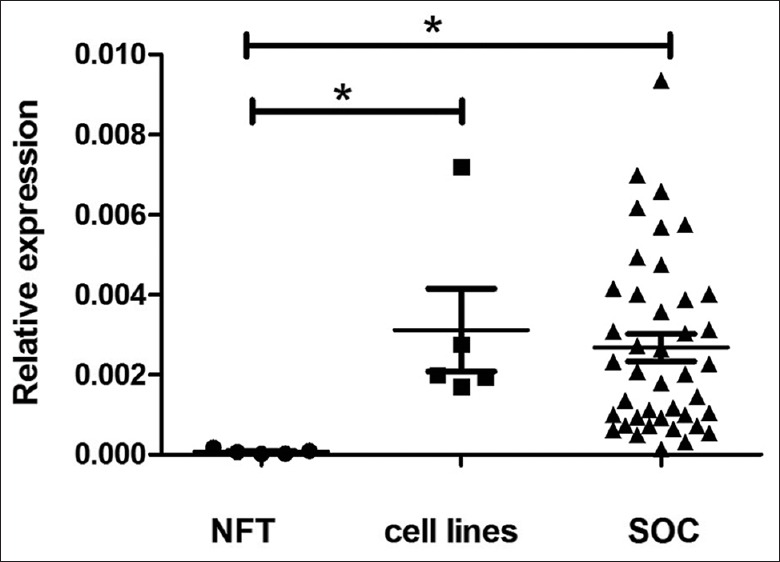
In all SOC samples, expression of the 3'-UTR proximal region was higher than that of the distal region, as evaluated by real-time PCR, which was also true for the 5 ovarian cancer cell lines [Figure 3]. The ratio of the proximal/distal region of the 3'-UTR varied in different SOC samples [from 1.5 to 96.0, Figure 4], and correlated significantly with the expression level of the coding region [Figure 5, Spearman's correlation coefficient, r = 0.708, P < 0.01]. In general, the higher the ratio of the proximal/distal region of the 3'-UTR, the higher the expression level of the coding region. In contrast, in NFT specimen, the copy numbers of the distal region was almost equal to the copy numbers of the proximal region of survivin 3'-UTR [the proximal/distal ratio range from 1.1 to 2.2, Figures 3 and 4]. The ratio of the proximal/distal region of the 3'-UTR was lower in NFT than in SOC and ovarian cancer cell lines [Figure 4, P < 0.01 for both].
Figure 4.
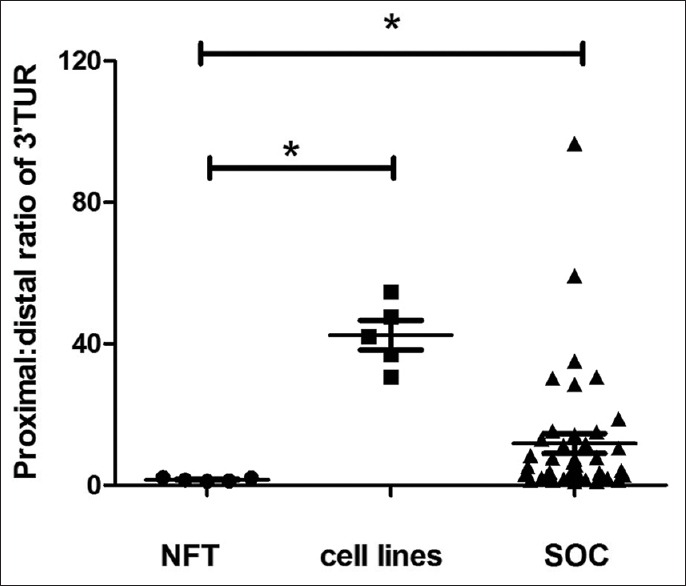
Ratio of proximal/distal regions of 3'-UTR expression in different samples. NFT: Normal fallopian tube; SOC: Serous ovarian cancer; 3'-UTR: 3'-untranslated region.
Figure 5.
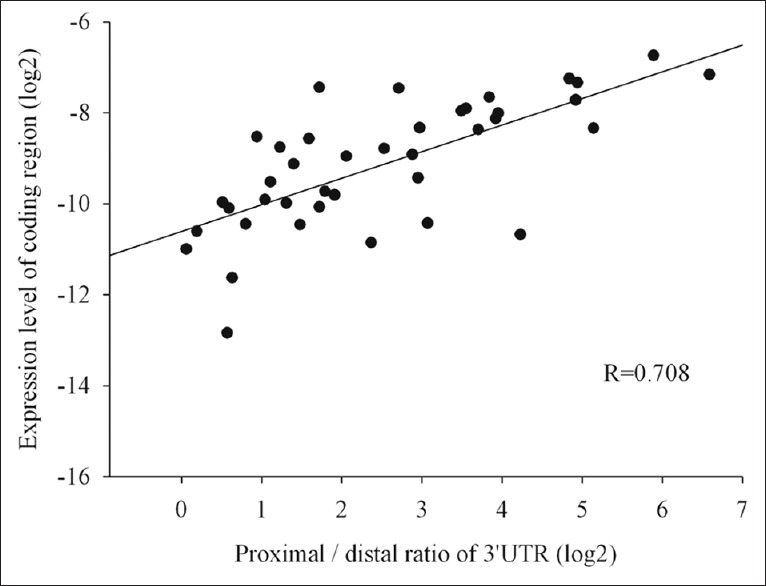
Ratio of proximal/distal regions of survivin 3'-UTR significantly correlated with expression of survivin coding region in SOC samples (n = 40; r = 0.708; P < 0.01). SOC: Serous ovarian cancer; 3'-UTR: 3'-untranslated region.
Validation of alternate polyadenylation by rapid amplification of cDNA 3' ends and sequencing
Representative SOC and NFT specimens were selected for 3' RACE-PCR. The 3' RACE nested PCR products of SOC samples showed four bands on agarose gel at 2000 bp, 1100 bp, 900 bp, and 600 bp [Figure 6]. However, no specific bands were detected in NFT sample. When the four bands were recovered and cloned for sequencing, the 600-bp and 1100-bp PCR products confirmed the presence of two APA sites at 1197 and 1673 [Figure 1]. However, even after we repeated the experiment several times, the clones from the two weak bands (2000 bp and 900 bp) were found to be the same as the 600-bp PCR products. When the 2000-bp and 900-bp DNA fragments were recovered and reloaded in agarose gel, we saw all four binds in the gel. Then, the reason was discovered. As the 3' RACE product was a mixture of four DNA fragments which had the same sequence in the first 600 bp of the 5' ends and the last 40 bp at the 3' ends. Single-strand DNA fragments of different lengths would therefore hybridize together, especially when much fewer 2000-bp and 900-bp fragments were available than other fragments.
Figure 6.
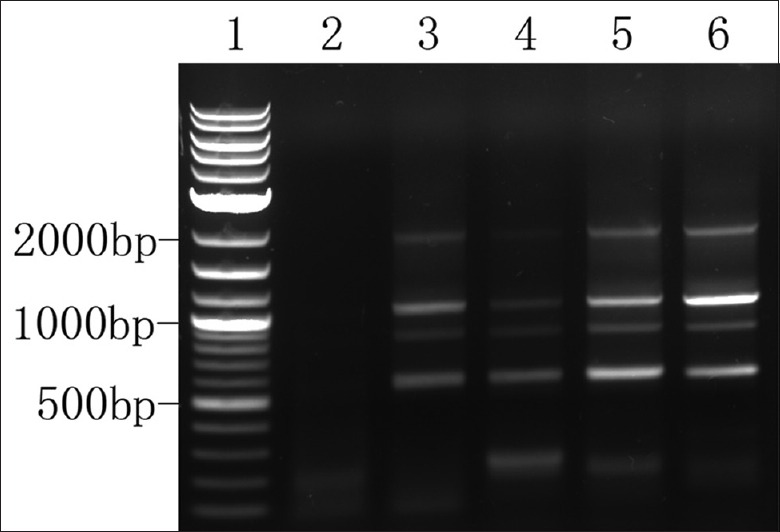
Validation of the shortening of survivin 3'-UTR by 3' RACE. Lane 1: 2-log DNA ladder. Lane 2: 3' RACE nested PCR product of NFT specimen. Lanes 3–6: 3' RACE nested PCR product of SOC samples with varying coding region expression levels and proximal/distal ratios; 3'-UTR: 3'-untranslated region; 3' RACE: Rapid amplification of cDNA 3' ends. SOC: Serous ovarian cancer; PCR: Polymerase chain reaction; NFT: Normal fallopian tube.
Expression of microRNAs that target survivin 3'-untranslated region
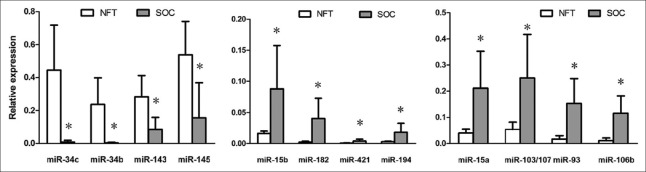
The miRNAs that target the 3'-UTRs are major posttranscriptional regulators of gene expression. Consequently, we inspected expression levels of 24 miRNAs (miR-34b, miR-34c, miR-449a/b, miR-34a, miR-15b, miR-15a, miR-136, miR-138, miR-143, miR-182, miR-203, miR-320, miR-377, miR-421, miR-497, miR-194, miR-103/107, miR-145, miR-106b, miR-106a, miR-93, miR-195, miR-16, and miR-335), which are predicted to target different parts of survivin 3'-UTR [Figure 1] by TargetScan online software. In NFT tissues, miR-145, miR-34c, miR-143, miR-34b, miR-195, miR-106a, and miR-16 (sorted in descending order of expression level in NFT) were high expression miRNAs; the others were medium- or low-expression miRNAs. In SOC, eight miRNAs (miR-15b, miR-15a, miR-182, miR-93, miR-106b, miR-103/107, miR-421, and miR-194) were upregulated and four miRNAs (miR-34b, miR-34c, miR-143, and miR-145) down-regulated compared with NFT [Figure 7, P < 0.01 for all]. Notably, the four down-regulated miRNAs are highly expressed in NFT, and three of them target the distal part of the 3'-UTR [Figure 2].
Figure 7.
Dysregulated miRNAs in SOC compared with NFT. *P < 0.01. miRNA: MicroRNA; NFT: Normal fallopian tube; SOC: Serous ovarian cancer.
Effects of restoring miR-34c on survivin expression in cancer cells
Survivin up-regulation in ovarian cancer coincided with the down-regulation of miR-34b and miR-34c. To assess the effect of miR-34 overexpression on survivin expression, we followed survivin mRNA levels by real-time PCR at 48 h after pre-miR-34c transfection in SKOV3.ipl cells. The transfection efficiency was estimated to be nearly 100% according to our observations on cells transfected by a fluorescence-labeled control RNA fragment. However, although exotic miR-34c overexpression results in G1/S cell-cycle arrest,[19] it did not change the survivin expression level [Figure 8; P > 0.05], compared with mock transfected cells.
Figure 8.
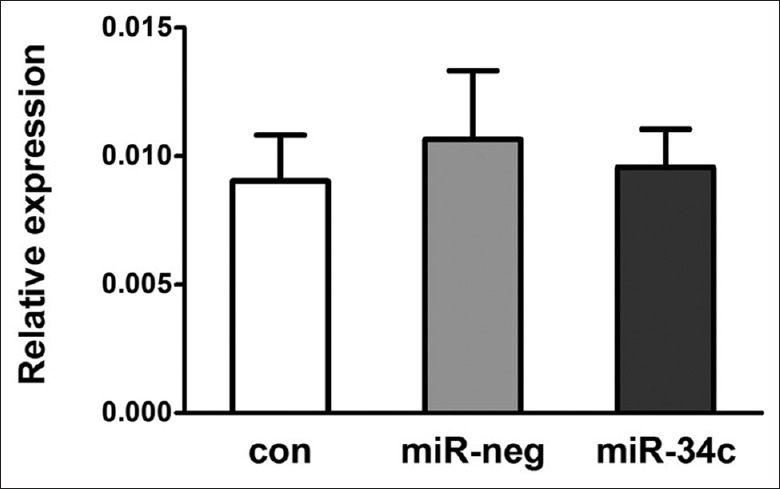
Survivin coding region expression at 48 h after pre-miR-34c transfection in SKOV3.ipl cells. con: Control; neg: Negative; miRNA: MicroRNA.
When A2780 cells were treated with adriamycin, p53 expression was induced, miR-34b and miR-34c were upregulated about 20-fold (data published previously).[19] Expression levels of the 3'-UTR proximal, as well as survivin coding region, decreased gradually [Figure 9]. At 24 h after treatment, coding region expression decreased 90%, while the 3'-UTR proximal region decreased 80%. The ratio of the proximal/distal region also decreased gradually.
Figure 9.
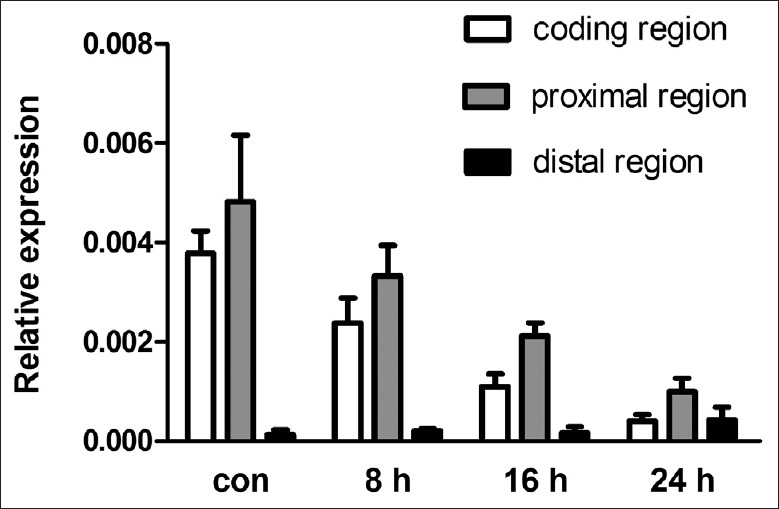
Expression of survivin coding region, and 3'-UTR proximal and distal regions in A2780 cells treated with adriamycin. con: Control; 3'-UTR: 3'-untranslated region.
DISCUSSION
Although transcriptional reactivation is assumed, we demonstrate that APA leads to the shortening of 3'-UTR plays a significant role in survivin overexpression. We not only observed varying degrees of APA in different samples, but also verified the main APA sites at 1197 and 1673 of 3'-UTR. Moreover, we found the correlation between the ratios of proximal/distal region and survivin coding region expression level in the SOC samples. While high-throughput methods did not reveal the correlation between the extent of 3'-UTR changes and gene expression levels, possibly because of their inferiority to real-time PCR in sensitivity and linear range of quantification. In SOC, shorter 3'-UTR enabled survivin gene to escape suppression by miRNAs and other negative regulators. While in NFT tissues, the almost equal expression levels of 3'-UTR proximal region and distal region enabled miRNAs and other negative regulators more effectively bind to its 3'-UTR. Therefore, higher ratio of survivin 3'-UTR proximal region to coding region was observed. Although expression of the coding region was negligible in NFT, high level of its 3'-UTR suggested that survivin transcription was not suppressed but constitutively transcribed.
Two mechanisms have been proposed for the negative regulation by miRNAs of their target genes: repression of protein translation or degradation of target mRNA.[20,21] Our results strongly support the latter mechanism, in which miRNAs mainly act by triggering deadenylation and decay of target mRNAs from the 5'-end through base pairing to the 3'-UTR of mRNAs. First, much higher expression of survivin 3'-UTR proximal region than that of the coding region indicated that its coding region was decayed. Second, the negative results in the 3' RACE assay (wherein an extended oligo [dT] primer was used in reverse transcription) for NFT samples supported the deadenylation of survivin mRNA in normal tissue.
By analyzing 24 miRNAs that target different parts of survivin 3'-UTR, we found only four miRNAs (miR-145, miR-34c, miR-143, and miR-34b) were significantly down-regulated in SOC, and their expression levels were inversely correlated to survivin expression. In NFT, these four miRNAs showed very high-level expression, and three of them targeted the distal part of 3'-UTR. The miR-34a, another member of miR-34 family, targeting the same site in survivin mRNA, has been reported to negatively regulate survivin expression in gastric cancer.[22] However, miR-34a showed a relatively low expression level in NFT and did not correlate with the expression of survivin. These data suggest that both expression level and location of targeting site are important factors in the effects of miRNAs. The eight up-regulated miRNAs which target scattered parts of the 3'-UTR, showed medium or low expression levels. Although their roles cannot be explained here, they are components of the molecular network and may regulate other genes in ovarian cancer.
To further understand the relationship of APA and miRNAs, we tested the effects of restoring miR-34 expression in ovarian cancer cells. We found exotic over-expression of miR-34c did not change survivin expression level significantly. When p53 wild-type cell line A2780 was treated with a genotoxic agent, miR-34c was induced and survivin expression was repressed. However, while the coding region of survivin decreased 90%, the 3'-UTR proximal region also decreased 80%, which implies that transcriptional suppression by p53 (a transcriptional repressor of survivin),[23] not miR-34b/c up-regulation, is the major contributor to survivin down-regulation. As no NFT-derived cell lines were available, we were unable to test the effect of miR-34c knock-down on survivin expression.
Our data suggest that the four highly expressed miRNAs could effectively suppress the expression of survivin in NFT. But in SOC, although down-regulation of the four miRNAs coincides with the shortening of 3'-UTR, APA is more important for survivin up-regulation, as APA controls whether expression should be regulated and to what extent. On the other hand, multi-targeting miRNAs could selectively regulate a specific gene with the cooperation of APA. However, further investigation is needed to understand the coordinating mechanism between APA and miRNAs and how they are regulated.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Footnotes
Edited by: Yuan-Yuan Ji and Qiang Shi
REFERENCES
- 1.An J, Zhu X, Wang H, Jin X. A dynamic interplay between alternative polyadenylation and microRNA regulation: Implications for cancer (Review) Int J Oncol. 2013;43:995–1001. doi: 10.3892/ijo.2013.2047. doi: 10.3892/ijo.2013.2047. [DOI] [PubMed] [Google Scholar]
- 2.Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, et al. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–83. doi: 10.1101/gr.132563.111. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu Y, Sun Y, Li Y, Li J, Rao X, Chen C, et al. Differential genome-wide profiling of tandem 3' UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 2011;21:741–7. doi: 10.1101/gr.115295.110. doi: 10.1101/gr.115295.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris AR, Bos A, Diosdado B, Rooijers K, Elkon R, Bolijn AS, et al. Alternative cleavage and polyadenylation during colorectal cancer development. Clin Cancer Res. 2012;18:5256–66. doi: 10.1158/1078-0432.CCR-12-0543. doi: 10.1158/1078-0432.CCR-12-0543. [DOI] [PubMed] [Google Scholar]
- 5.Lai DP, Tan S, Kang YN, Wu J, Ooi HS, Chen J, et al. Genome-wide profiling of polyadenylation sites reveals a link between selective polyadenylation and cancer metastasis. Hum Mol Genet. 2015;24:3410–7. doi: 10.1093/hmg/ddv089. doi: 10.1093/hmg/ddv089. [DOI] [PubMed] [Google Scholar]
- 6.Mayr C, Bartel DP. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–84. doi: 10.1016/j.cell.2009.06.016. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miles GD, Seiler M, Rodriguez L, Rajagopal G, Bhanot G. Identifying microRNA/mRNA dysregulations in ovarian cancer. BMC Res Notes. 2012;5:164. doi: 10.1186/1756-0500-5-164. doi: 10.1186/1756-0500-5-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Iterson M, Bervoets S, de Meijer EJ, Buermans HP, 't Hoen PA, Menezes RX, et al. Integrated analysis of microRNA and mRNA expression: Adding biological significance to microRNA target predictions. Nucleic Acids Res. 2013;41:e146. doi: 10.1093/nar/gkt525. doi: 10.1093/nar/gkt525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He X, Yang J, Zhang Q, Cui H, Zhang Y. Shortening of the 3' untranslated region: An important mechanism leading to overexpression of HMGA2 in serous ovarian cancer. Chin Med J. 2014;127:494–9. doi: 10.3760/cmaj.issn.0366-699920130843. [PubMed] [Google Scholar]
- 10.Turan G, Usta CS, Usta A, Kanter M, Tavli L, Karacan M, et al. The expression of HER-2/neu (c-erbB2), survivin and cycline D1 in serous ovarian neoplasms: Their correlation with clinicopathological variables. J Mol Histol. 2014;45:679–87. doi: 10.1007/s10735-014-9591-2. doi: 10.1007/s10735-014-9591-2. [DOI] [PubMed] [Google Scholar]
- 11.Guha M, Altieri DC. Survivin as a global target of intrinsic tumor suppression networks. Cell Cycle. 2009;8:2708–10. doi: 10.4161/cc.8.17.9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altieri DC. Targeting survivin in cancer. Cancer Lett. 2013;332:225–8. doi: 10.1016/j.canlet.2012.03.005. doi: 10.1016/j.canlet.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mobahat M, Narendran A, Riabowol K. Survivin as a preferential target for cancer therapy. Int J Mol Sci. 2014;15:2494–516. doi: 10.3390/ijms15022494. doi: 10.3390/ijms15022494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boidot R, Végran F, Lizard-Nacol S. Transcriptional regulation of the survivin gene. Mol Biol Rep. 2014;41:233–40. doi: 10.1007/s11033-013-2856-0. doi: 10.1007/s11033-013-2856-0. [DOI] [PubMed] [Google Scholar]
- 15.Reade CJ, McVey RM, Tone AA, Finlayson SJ, McAlpine JN, Fung-Kee-Fung M, et al. The fallopian tube as the origin of high grade serous ovarian cancer: Review of a paradigm shift. J Obstet Gynaecol Can. 2014;36:133–40. doi: 10.1016/S1701-2163(15)30659-9. [DOI] [PubMed] [Google Scholar]
- 16.Perets R, Wyant GA, Muto KW, Bijron JG, Poole BB, Chin KT, et al. Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in Brca; Tp53;Pten models. Cancer Cell. 2013;24:751–65. doi: 10.1016/j.ccr.2013.10.013. doi: 10.1016/j.ccr.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He XJ, Zhang Q, Liu YJ, Pan XY. Increasing specificity of real time PCR to detect microRNA through primer design and annealing temperature increase (in Chinese) Beijing Da Xue Xue Bao. 2009;41:691–8. doi: 10.3969/j.issn.167X.2009.06.016. [PubMed] [Google Scholar]
- 18.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50. doi: 10.1158/0008-5472.CAN-04-0496. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, He XJ, Ma LP, Li N, Yang J, Cheng YX, et al. Expression and significance of microRNAs in the p53 pathway in ovarian cancer cells and serous ovarian cancer tissues (in Chinese) Chin J Oncol. 2011;33:885–90. doi: 10.3760/cma.j.issn.0253-3766.2011.12.002. [PubMed] [Google Scholar]
- 20.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 22.Cao W, Fan R, Wang L, Cheng S, Li H, Jiang J, et al. Expression and regulatory function of miRNA-34a in targeting survivin in gastric cancer cells. Tumour Biol. 2013;34:963–71. doi: 10.1007/s13277-012-0632-8. doi: 10.1007/s13277-012-0632-8. [DOI] [PubMed] [Google Scholar]
- 23.Mirza A, McGuirk M, Hockenberry TN, Wu Q, Ashar H, Black S, et al. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 2002;21:2613–22. doi: 10.1038/sj.onc.1205353. doi: 10.1038/sj/onc/1205353. [DOI] [PubMed] [Google Scholar]