Abstract
Morphological abnormalities observed typically in the brains of adults with Down syndrome are identical with those present in patients with Alzheimer’s disease. However, only some adults with Down syndrome suffer from early dementia, whereas others remain unaffected. We aimed to identify the genomic background responsible for this observation. We performed cognitive assessment and genome expression analysis of blood mononuclear cells in seniors with Down syndrome. Unaffected elderly patients and younger patients with severe cognitive disability or cognitive deterioration differed significantly with regard to the MTRNR2L12 gene. Our findings suggest the potential value of this gene as a blood marker of early dementia in individuals with Down syndrome.
Keywords: Alzheimer’s disease, blood marker, cognitive assessment, Down syndrome, early dementia, microarrays
INTRODUCTION
Histological changes typical of Alzheimer’s disease (AD) are constantly reported in autopsied brain tissue of deceased individuals with Down syndrome (trisomy 21) older than 30–40 years [1–3]. This fact suggests the important role that the overexpression of genes encoded on the 21st chromosome plays in the development of AD. In particular, the APP gene encoding amyloid-β protein precursor is an obvious candidate due to its role in the development of amyloid plaques. Nevertheless, genome-wide association studies suggest the importance of several other genes [4].
Interestingly, significant variability of cognitive profiles can be observed in adult persons with Down syndrome. On the one hand, symptoms of AD-like dementia develop in these individuals frequently in the fourth or fifth decade of life. On the other hand, some seniors with Down syndrome remain unaffected by AD-like dementia. This phenomenon suggests thepresence of genetic variability that influences meta-bolic pathways crucial for the development of AD-like dementia in individuals with Down syndrome.
Although genome expression is tissue specific, it has been suggested that several genes play central roles in ubiquitous metabolic pathways following similar expression patterns in the brain and the blood [5, 6]. Consequently, the alteration of genome expression in the brain in the case of AD development might be partially detectable in the blood.
In light of these facts, we aimed to find potential genomic blood markers of AD-like dementia development in adults with Down syndrome.
METHODS
We focused on a cohort of 48 adults with Down syndrome older than 35 years who were institutionalized or attended day-care centers; these individuals were recruited from the five largest cities in Poland. All of these persons or their legal representatives signed informed consent forms to participate in the study and the protocol was accepted by the Jagiellonian University Ethics Committee.
To enable the detection of potential markers of early dementia, the participants were divided into two age groups: younger persons (younger than 55) and older persons (older than 55). The cognitive function of each study participant was assessed using the short form of the Prudhoe Cognitive Function Test (PCFT) [7]. Cognitive status was defined as good/acceptable for PCFT scores above 25% ; a severe cognitive disability was inferred from PCFT scores below 25% . Furthermore, disability was presumed if administration of the PCFT was not possible due to the person’s inability to cooperate. History of cognitive deterioration within last three years was also recorded in persons younger than 55 if reported by a caregiver or appropriate medical records. In all assessed persons, hypothyroidism was excluded as well as poorly controlled epilepsy and other serious medical conditions, which could result in cognitive decline.
We next conducted the genomic study. Subgroups of patients with good/acceptable cognitive statuses and individuals with severe cognitive disabilities from both age categories were cross compared. In addition, subgroups of younger persons with a history of cognitive deterioration and older participants with good/acceptable cognitive statuses were compared.
Blood samples (2.5 mL) were obtained from the study participants and used to extract total RNA from blood white cells (PAXgene system; PreAnalytiX). The whole genome expression was next assessed using SurePrint Human Gene Expression 8×60K v2 Microarrays (Agilent, Santa Clara, USA) that were scanned on an Agilent SureScan G2600D scanner according to the manufacturer’s protocol. SurePrint G3 Human Gene Expression Microarrays provide a comprehensive coverage of the entire human genome (50.599 genes and transcripts) with a high specificity of target detection and a very wide dynamic range. This analysis allows for detection of biological features encompassing both very high and very lowexpression.
We performed microarray data analysis using GeneSpring Software (http://www.genespring.com). We also used single-color expression technology dedicated to Agilent microarrays. For detecting statistically significant differences between the subgroups of study participants, we used a moderated t-test. We also applied a Benjamini-Hochberg false discovery rate (FDR) correction for multiple testing. A corrected value of p < 0.05 was considered significant with fold change (FC) greater than 2.
The primary microarray data were added to the GEO public repository and are accessible via GEO Series accession number GSE63870 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63870).
The validation of the microarray data was performed using Real-Time Quantitative PCR (RQ-PCR). A total of 120 ng of RNA was used for the reverse transcription of each sample by means of Superscript II reverse transcriptase (Life Technologies, Carlsbad, USA). Next, SYBR Green chemistry was employed for measuring the gene expression (measurements in duplication), with the beta-2-microglobulin gene (B2M) serving as the control gene (the sequences of the starters are 5’- GAGTATGCCTGCCGTGTG-3’ and 5’- AATCCAAATGCGGCATCT-3’). ABI PRISM 7300 Real-time PCR System (Life Technologies, Carlsbad, USA) with a universal instrument setting was used. We performed the data analysis using the comparative cycle threshold (Ct) method of relative quantification (ΔCt = Ct (gene of interest) -Ct (control gene) ; ΔΔCt = ΔCt (tested group) -ΔCt (control group) ; FC = 2 - ΔΔCt).
RESULTS
A group of 23 women (aged 35–67 years; mean age: 49 years) and 25 men (aged 35–67 years; mean age: 46 years) entered the study. Among them, 36 persons (aged 37–67 years; mean age: 50 years) were institutionalized and 12 persons (aged 35–57 years; mean age: 40 years) lived with their families and attended day-care centers. The “younger” and the “older” subgroups of the study participants consisted of 35 persons and 13 persons, respectively. Severe cognitive disability was not recorded in patients younger than 40. Good/acceptable cognitive function and severe cognitive disability were observed in 33 and 15 participants, respectively. The frequency of severe disability was similar for women (8/23) and for men (7/25). Only one severely disabled person attended a day-care center.
The PCFT scores in the subgroups of persons with good/acceptable cognitive function ranged from 15/30–30/30 points (mean: 26/30) in the younger subgroup and 17/30–29/30 points (mean: 20/30) in the older subgroup. In the group with severe cognitive disability, the PCFT could be administered only in three cases (the scores ranged from 4/30–6/30 points).
Cognitive deterioration was recorded in five persons younger than 55. In two cases with documented deterioration, administration of the PCFT was possible (these participants scored 15/30 and 23/30 points, respectively).
Details of the studied population are summarized in Table 1.
Table 1.
The participants of the study
| Patient’s Code |
Age | Gender | Day-care center/ Institutionalized |
Age group | Cognitive status | Cognitive deterioration <55 y |
PCFT score (Short form of the test, maximal score = 30 points) |
| 1 | 53 | M | Institutionalized | Younger | Severe disability | Yes | Inability to cooperate |
| 2 | 50 | F | Institutionalized | Younger | Severe disability | Inability to cooperate | |
| 3 | 48 | M | Institutionalized | Younger | Severe disability | 6/30 | |
| 4 | 47 | F | Institutionalized | Younger | Severe disability | Yes | Inability to cooperate |
| 5 | 45 | M | Institutionalized | Younger | Severe disability | Inability to cooperate | |
| 6 | 40 | M | Day-care center | Younger | Severe disability | 6/30 | |
| 7 | 40 | F | Institutionalized | Younger | Severe disability | Yes | Inability to cooperate |
| 8 | 62 | M | Institutionalized | Older | Good/acceptable | 17/30 | |
| 9 | 62 | M | Institutionalized | Older | Good/acceptable | 14/16* | |
| 10 | 60 | F | Institutionalized | Older | Good/acceptable | 25/30 | |
| 11 | 59 | F | Institutionalized | Older | Good/acceptable | 29/30 | |
| 12 | 57 | F | Day-care center | Older | Good/acceptable | 25/30 | |
| 13 | 52 | F | Institutionalized | Younger | Good/acceptable | Yes | 15/30 |
| 14 | 50 | M | Institutionalized | Younger | Good/acceptable | Yes | 23/30 |
| 15 | 47 | F | Institutionalized | Younger | Good/acceptable | 30/30 | |
| 16 | 47 | M | Institutionalized | Younger | Good/acceptable | 24/30 | |
| 17 | 47 | F | Institutionalized | Younger | Good/acceptable | 20/30 | |
| 18 | 45 | F | Day-care center | Younger | Good/acceptable | 19/30 | |
| 19 | 45 | F | Institutionalized | Younger | Good/acceptable | 18/30 | |
| 20 | 44 | M | Institutionalized | Younger | Good/acceptable | 28/30 | |
| 21 | 44 | M | Institutionalized | Younger | Good/acceptable | 22/30 | |
| 22 | 44 | M | Institutionalized | Younger | Good/acceptable | 16/30 | |
| 23 | 43 | F | Institutionalized | Younger | Good/acceptable | 27/30 | |
| 24 | 43 | M | Institutionalized | Younger | Good/acceptable | 20/30 | |
| 25 | 42 | M | Day-care center | Younger | Good/acceptable | 30/30 | |
| 26 | 40 | F | Day-care center | Younger | Good/acceptable | 30/30 | |
| 27 | 40 | M | Institutionalized | Younger | Good/acceptable | 29/30 | |
| 28 | 40 | M | Day-care center | Younger | Good/acceptable | 24/30 | |
| 29 | 40 | M | Institutionalized | Younger | Good/acceptable | 20/30 | |
| 30 | 39 | F | Institutionalized | Younger | Good/acceptable | 30/30 | |
| 31 | 39 | M | Institutionalized | Younger | Good/acceptable | 27/30 | |
| 32 | 38 | F | Day-care center | Younger | Good/acceptable | 30/30 | |
| 33 | 38 | F | Institutionalized | Younger | Good/acceptable | 28/30 | |
| 34 | 38 | M | Day-care center | Younger | Good/acceptable | 21/30 | |
| 35 | 37 | M | Institutionalized | Younger | Good/acceptable | 29/30 | |
| 36 | 37 | M | Institutionalized | Younger | Good/acceptable | 28/30 | |
| 37 | 36 | M | Day-care center | Younger | Good/acceptable | 30/30 | |
| 38 | 36 | M | Day-care center | Younger | Good/acceptable | 30/30 | |
| 39 | 36 | F | Day-care center | Younger | Good/acceptable | 16/30 | |
| 40 | 35 | F | Day-care center | Younger | Good/acceptable | 28/30 | |
| 41 | 67 | F | Institutionalized | Older | Severe disability | ** | Inability to cooperate |
| 42 | 67 | M | Institutionalized | Older | Severe disability | ** | Inability to cooperate |
| 43 | 64 | F | Institutionalized | Older | Severe disability | 4/30 | |
| 44 | 63 | M | Institutionalized | Older | Severe disability | ** | Inability to cooperate |
| 45 | 63 | F | Institutionalized | Older | Severe disability | ** | Inability to cooperate |
| 46 | 62 | F | Institutionalized | Older | Severe disability | ** | Inability to cooperate |
| 47 | 60 | M | Institutionalized | Older | Severe disability | Inability to cooperate | |
| 48 | 58 | F | Institutionalized | Older | Severe disability | Inability to cooperate |
*Only partial application of the test possible because of serious visual impairment in the patient (questions requiring visual skills were omitted). **Various degree of cognitive decline observed in the last years.
An initial comparison of the younger and older subgroups by means of microarrays revealed significant differences with regard to three transcripts (ESPNL, XLOC_007536 and USP27X-AS1). However, these transcripts do not seem to be involved in the pathogenesis of dementia. No significant differences were detected when comparing the group with good/acceptable cognitive statuses and the group with severe cognitive disabilities. On the other hand, subgroups of younger patients with severe cognitive disabilities (n = 7) and patients with cognitive deterioration (n = 5) differed significantly when compared with older patients with good/acceptable cognitive statuses (n = 5; corrected p = 0.00026 and 0.00024; FC = 17.76 and 18.66, respectively) with regard to the expression of a single gene: MTRNR2L12, an isoform of humanin, considered to be a protective factor in familial AD [8].
Since no commercial RQ-PCR assays for expression measurement of the MTRNR2L12 gene are available on the market, specific starters were designed, based on the sequence of the transcript ENST00000600213 (http://www.ensembl.org) and the sequence of the respective microarray probe (A_21_P0003289) located on the SurePrint Human Gene Expression 8×60K v2 Microarray. Starters with the following sequences were used: 5’- GCTGTCTCTTACTTTCAACCAGTG-3’ (forward) and 5’- CATAGTCTTGCTATGTTATGCCCG-3’ (reverse). A validation procedure with SYBR Green chemistry confirmed the expression difference discussed above for younger patients with severe cognitive disabilities and individuals with cognitive deterioration, compared with older patients with good/acceptable cognitive statuses (ΔΔCt = −5.36; FC = 41; p < 0.0001 and ΔΔCt = −5.16; FC = 35.7; p < 0.0001, respectively).
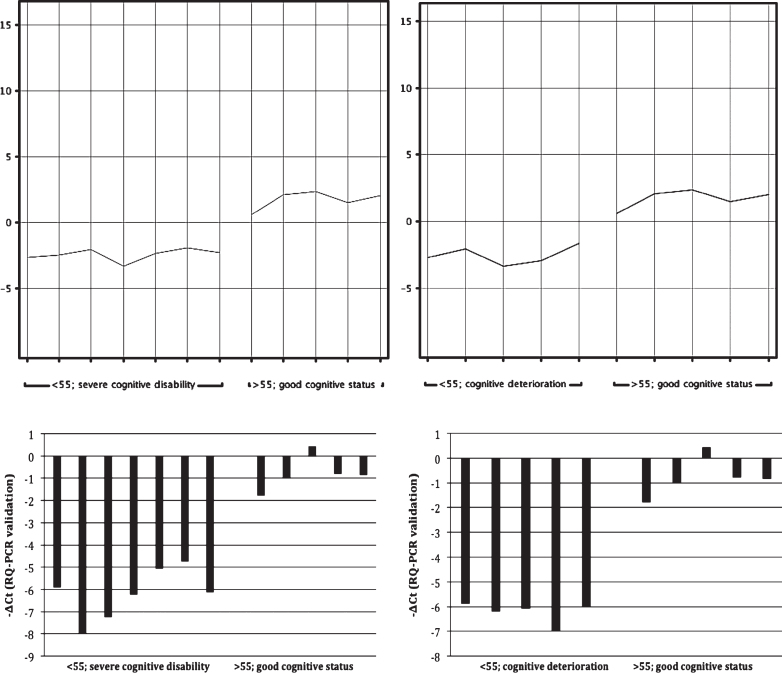
The results of the genomic study are shown in Fig. 1 and in Table 2.
Fig.1.
Expression of the MTRNR2L12 gene. Comparison of the groups of older (>55 y) patients with good/acceptable cognitive status and of younger (<55 y) patients with severe cognitive disability is presented on the left. Comparison of the same group of older patients with the group of younger (<55 y) patients with documented dementia is depicted on the right. Normalized intensity values representing expression of the MTRNR2L12 gene (microarray analysis) are visible above whereas the corresponding -ΔCt values (the RQ-PCR confirmatory analysis) are shown below.
Table 2.
Details of the statistical analysis
| Compared subgroups of patients | Number of upregulated genes/downregulated genes (FC >2, uncorrected p) |
Number of statistically significant genes (corrected p) |
Names of the statistically significant genes |
RQ-PCR validation |
| Older (>55 y) versus younger (<55 y) |
379 / 316 | 3 |
ESPNL (FC = 2.2; p = 0.047) XLOC_007536 (FC = 2.08; p = 0.047) USP27X-AS1 (FC = 2.03; p = 0.047) |
Not analyzed |
| Good/acceptable cognitive status versus severe cognitive disability |
31 / 59 | 0 | – | Not analyzed |
| Institutionalized versus day-care center |
34 / 82 | 0 | – | Not analyzed |
| Older (>55 y) with good/acceptable cognitive status versus younger (<55 y) with severe intellectual disability |
1376 / 794 | 1 |
MTRNR2L12 (FC = 17.76; p = 0.00026) |
ΔΔCt = −5.36 FC = 41, p < 0.0001 |
| Older (>55 y) with good/acceptable cognitive status versus younger (<55 y) with cognitive deterioration |
971 / 1301 | 1 |
MTRNR2L12 (FC = 18.66; p = 0.00024) |
ΔΔCt = −5.16 FC = 35.7 p < 0.0001 |
DISCUSSION
The PCFT is easy to administer, which was validated for the detection of cognitive decline and oncoming dementia. The degree of intellectual disability strongly correlates with PCFT score. However, it should be noted that potential dynamics of the symptoms of cognitive disability were not assessed in the present study, since each participant was examined only once. Therefore, a conclusive differentiation between rapid cognitive deterioration (typical of early-onset AD) and baseline severe intellectual disability related to Down syndrome itself was possible only in a few patients based on symptoms reported by the caregivers or medical records, if available. Nevertheless, the genomic analysis revealed very similar results for younger patients with severe cognitive disability and younger patients with cognitivedeterioration.
Although the findings of the present study are limited by the relatively small sample size, the results of the genome expression analysis suggest a possible role of MTRNR2L12, a humanin isoform, in the early development of dementia in Down syndrome. Humanin is a polypeptide that is highly conserved across species [8, 9]. It is a 24 amino acid-long molecule encoded by a 75bp open reading frame sequence of the mitochondrial 16S rRNA (MT-RNR2 gene). Humanin was shown to restore cellular ATP levels and to protect against several AD-related toxicities in neuronal cells by means of modulation of oxidative stress and apoptosis [8]. It is secreted from cells and can be found in plasma.
Bioinformatic studies revealed several MT-RNR2-like nuclear loci encoding 15 humanin-like peptides [10]. The MTRNR2L12 gene located on chromosome 3 (3q11.2) is nearly identical to the mitochondrial gene MT-RNR2 encoding humanin. Interestingly, there is a polymorphic site (rs6484338) within MTRNR2L12, predicted to cause the Ser12Leu amino acid exchange, which in turn results in the production of a peptide sequence identical to mitochondrial humanin. Since the presence of the MTRNR2L12 peptide has been confirmed in brain tissue, it can be assumed that MTRNR2L12 exerts effects similar to those reported for humanin.
In conclusion, the expression of MTRNR2L12 might be a new, easy-to-measure blood marker of severe cognitive disability and, possibly, of early dementia in patients with Down syndrome. Further studies should be performed in order to evaluate the potential usefulness of the measurement of MTRNR2L12 transcript in patientswith AD.
AUTHORS’ CONTRIBUTION
MBM conceived the study, carried out the cognitive assessment and molecular genetic studies, performed the statistical analysis and drafted the manuscript. JJP participated in the design of the study, reviewed the stratification of the entire group of patients with regard to the cognitive status and the results of micro array analysis and helped to draft the manuscript. AM reviewed the medical records of the study participants (the author was then blinded to the results of genomic expression data statistical analysis). All authors have read and approve the final version of the manuscript.
ACKNOWLEDGMENTS
The Polish National Science Centre (DEC-2011/03/B/NZ5/01328) sponsored the study.
Authors’ disclosures available online (http://j-alz.com/manuscript-disclosures/14-3030r3).
REFERENCES
- 1.Dalton AJ, Crapper-McLachlan DR. Clinical expression of Alzheimer’s disease in Down syndrome. Psychiatr Clin North Am. 1986;9:959–970. [PubMed] [Google Scholar]
- 2.Epstein CJ. Scriver CE, Beaudet AL, Sly WS. The Metabolic Basis of Inherited Diseases. New York: McGraw-Hill; 1989. Down syndrome (Trisomy 21) pp. 291–327. [Google Scholar]
- 3.Margallo-Lana ML, Moore PB, Kay DWK, Perry RH, Reid BE, Berney TP, Tyrer SP. Fifteen-year follow-up of 92 hospitalized adults with Down’s syndrome: Incidence of cognitive decline, its relationship to age and neuropathology. J Intellect Disabil Res. 2007;51:463–477. doi: 10.1111/j.1365-2788.2006.00902.x. [DOI] [PubMed] [Google Scholar]
- 4.Tosto G, Reitz C. Genome-wide association studies in Alzheimer’s disease: A review. Curr Neurol Neurosci Rep. 2013;13:381. doi: 10.1007/s11910-013-0381-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liew CC, Ma J, Tang HC, Zheng R, Dempsey AA. The peripheral blood transcriptome dynamically reflects system wide biology: A potential diagnostic tool. J Lab Clin Med. 2006;147:126–132. doi: 10.1016/j.lab.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan PF, Fan C, Perou CM. Evaluating the comparability of gene expression in blood and brain. Am J Med Genet. 2006;141B:261–268. doi: 10.1002/ajmg.b.30272. [DOI] [PubMed] [Google Scholar]
- 7.Kay DW, Tyrer SP, Margallo-Lana ML, Moore PB, Fletcher R, Berney TP, Vithayathil E. Preliminary evaluation of a scale to assess cognitive function in adults with Down’s syndrome: The Prudhoe Cognitive Function Test. J Intellect Disabil Res. 2003;47:155–168. doi: 10.1046/j.1365-2788.2003.00451.x. [DOI] [PubMed] [Google Scholar]
- 8.Hashimoto Y, Niikura T, Tajima H, Yasukawa T, Sudo H, Ito Y, Kita Y, Kawasumi M, Kouyama K, Doyu M, Sobue G, Koide T, Tsuji S, Lang J, Kurokawa K, Nishimoto I. A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer’s disease genes and Aβ. Proc Natl Acad Sci U S A. 2001;98:6336–6341. doi: 10.1073/pnas.101133498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo B, Zhai D, Cabezas E, Welsh K, Nouraini S, Satterthwait AC, Reed JC. Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature. 2003;423:456–461. doi: 10.1038/nature01627. [DOI] [PubMed] [Google Scholar]
- 10.Bodzioch M, Lapicka-Bodzioch K, Zapala B, Kamysz W, Kiec-Wilk B, Dembinska-Kiec A. Evidence for potential functionality of nuclearly-encoded humanin isoforms. Genomics. 2009;94:247–256. doi: 10.1016/j.ygeno.2009.05.006. [DOI] [PubMed] [Google Scholar]