With an aim of identifying interventions that could reduce or attenuate drug-induced hepatocyte toxicity, hepatocytes were generated from a renewable resource and exposed to varying levels of paracetamol. A novel RNA that reduced paracetamol-induced hepatocyte toxicity was identified, representing an important advance for the field.
Keywords: Drug-induced liver injury, MicroRNA, Hepatocyte, Apoptosis, Necrosis, Paracetamol
Abstract
The liver performs multiple functions within the human body. It is composed of numerous cell types, which play important roles in organ physiology. Our study centers on the major metabolic cell type of the liver, the hepatocyte, and its susceptibility to damage during drug overdose. In these studies, hepatocytes were generated from a renewable and genetically defined resource. In vitro-derived hepatocytes were extensively profiled and exposed to varying levels of paracetamol and plasma isolated from liver-failure patients, with a view to identifying noncoding microRNAs that could reduce drug- or serum-induced hepatotoxicity. We identified a novel anti-microRNA, which reduced paracetamol-induced hepatotoxicity and glutathione depletion. Additionally, we identified a prosurvival role for anti-microRNA-324 following exposure to plasma collected from liver failure patients. We believe that these studies represent an important advance for the field, demonstrating the power of stem cell-derived systems to model human biology “in a dish” and identify novel noncoding microRNAs, which could be translated to the clinic in the future.
Significance
The liver performs vital functions within the human body and is composed of numerous cell types. The major metabolic cell type of the liver, the hepatocyte, is susceptible to damage during drug overdose. In these studies, hepatocytes were generated from a renewable resource and exposed to varying levels of paracetamol, with a view to identifying interventions that could reduce or attenuate drug-induced liver toxicity. A novel noncoding RNA that reduced paracetamol-induced hepatocyte toxicity was identified. These findings may represent an important advance for the field.
Introduction
The liver is a multifunctional and highly regenerative organ. In both the acute and chronic settings, liver damage has dire consequences for health. A common cause of liver damage is adverse reactions to prescription drugs, which can lead to drug-induced liver injury (DILI). This creates major problems for patients, clinicians, the pharmaceutical industry and regulatory authorities [1]. It has been reported that in the U.K., approximately 15% of the hospital inpatients suffer from liver toxicity in response to medications during admissions, with 20% of these patients readmitted again after 1 year and a 2% mortality rate [2, 3]. The annual cost to the National Health Service in the U.K. has been estimated at £450 million, with the costs growing year by year [4].
In the context of drug overdose or serious adverse reactions, liver failure can be acute and life-threatening, and in serious cases requires orthotopic liver transplantation. Although transplantation is highly successful, such an approach has many limitations [5], justifying basic science attempts to develop better human models of liver injury and novel intervention strategies. With this in mind, we have studied the importance of microRNAs (miRs) in regulating human drug metabolism and their potential to reduce liver toxicity in response to toxic levels of paracetamol.
miRs are small noncoding RNAs that are approximately 20–24 nucleotides long, and their major function is to fine-tune gene expression. Enzyme processing of primary and precursor microRNAs generates a mature microRNA that is incorporated into the RNA-induced silencing complex (RISC). The complex recognizes the target mRNA through perfect and imperfect base pairing with the target miRs [6]. It is known that microRNAs play roles in regulating the first and third phase of drug metabolism (7–12]. However the second phase of drug metabolism, drug conjugation, has not been studied in detail. Drug conjugation is crucial to human drug processing, and perturbation of this process can lead to life-threatening liver injury. To test the importance of miRs in regulating phase II drug metabolism, we opted to study the metabolism of a commonly used analgesic, paracetamol (acetaminophen). When taken in the appropriate amounts, paracetamol is modified by sulfuryl transferases and UDP glucuronosyl transferases and removed from the body without organ damage [13]. However, when paracetamol is taken above the recommended dose, it is metabolized by phase I enzymes to generate a toxic intermediate N-acetyl-p-benzoquinone imine (NAPQI), which, if untreated, can lead to hepatocyte cell death and liver failure, placing the patient in a life-threatening situation. To promote nontoxic paracetamol metabolism, in the context of drug overdose, we identified and used candidate miRs to regulate its metabolism.
In summary, we have identified a novel microRNA that regulates phase II drug metabolism, promoting nontoxic paracetamol drug metabolism. Moreover, we demonstrate a supportive role for microRNAs in managing the toxic nature of human liver failure plasma. We believe our findings are novel and provide proof of concept. These studies exemplify the power of pluripotent stem cell-derived models to identify new approaches to treating human liver damage.
Materials and Methods
Cell Culture and Differentiation
Human embryonic stem cells (hESCs) (H9) were cultured and differentiated as described [5, 14–16].
Primary Human Hepatocyte Culture
Cryopreserved human primary hepatocytes were purchased from Thermo Fisher Scientific Life Sciences (Waltham, MA, http://www.thermofisher.com). In this study, female line (Hu8119; HMCPIS) and male line (Hu8182; HMCPIS) were chosen. Both lines were donated from Caucasian patients of 21–27 years of age. The cryoplateable hepatocytes were plated and maintained as per vendor’s instruction. Briefly, hepatocytes were resuspended in thawing medium (CM3000) and plated onto Matrigel in a 48-well plates. Subsequently, cells were placed in the incubator at 37°C (5% CO2) for 24 hours. At 24 hours after replating, the medium was changed to incubation medium (CM4000). At 48 hours after replating, hepatocyte metabolic activity (CYP3A and CYP1A2) were measured by using Promega pGlo luciferase-based assays (Promega, Madison, WI, http://www.promega.com).
RNA Isolation and Quantitative Polymerase Chain Reaction
RNA was isolated, reverse-transcribed, and used for TaqMan polymerase chain reaction as described in Szkolnicka et al. [5]. The reference numbers of particular primers can be found in supplemental online Table 4.
RT2 Profiler PCR Array
Total RNA from hESC-derived hepatocytes (day 18) and primary human hepatocytes (purchased from 3H Biomedical AB, Uppsala, Sweden, http://www.3hbiomedical.com) was reverse-transcribed using RT2 First Strand Kit (Qiagen, Hilden, Germany, http://www.qiagen.com) according to the manufacturer’s instructions. The quantitative polymerase chain reaction (qPCR) was performed using the RT2 profiler PCR Array system (Qiagen) in accordance with the manufacturer's instructions.
MicroRNA Profiling
Total RNA from hESC-derived hepatocytes (day 18; n = 3) and primary human hepatocytes (purchased from 3H Biomedical AB) were analyzed on Agilent miRNA platform (using Agilent’s SurePrint G3 Human v16 microRNA 8 × 60K microarray slides; miRBase Version 16.0; Agilent, Santa Clara, CA, http://www.agilent.com) following Sistemic proprietary standard operating procedures. Then, 100 ng of total RNA, from a working solution at 50 ng/μl in nuclease-free water, was used as input for each microarray experiment. Each slide contained eight individual arrays, and each array was identified by a unique barcode and contained capture probes for 1,349 microRNAs (1205 human; 144 viral). The microarray data were normalized by using Sistemic’s in-house preprocessing and data quality-control (QC) methods (Sistemic, Glasgow, U.K., http://www.sistemic.co.uk). Detection calls (present or absent) for individual miRNAs were compared across the samples. The detection calls were calculated by using the Agilent Feature Extraction software (Version 10.7.3.1). A detailed description of how these calls are made is available in the “Feature Extraction Reference Guide” on the Agilent website (http://www.genomics.agilent.com).
miR-mRNA Binding Analysis
TargetScanHuman 6.2 is an online tool that predicts microRNA binding sites at the 3′ untranslated region of the biological target. The program focuses on the presence of conserved and nonconserved sites that match the seed region of each microRNA [17].
Immunofluorescence
Cell cultures at day 18 of cellular differentiation were fixed in 100% ice-cold methanol at −20°C for 30 minutes. After fixation, cell cultures were washed twice with phosphate-buffered saline (PBS) at room temperature. Cells were blocked with 0.1% PBS-Tween containing 10% bovine-specific antigen (BSA) for 1 hour and subsequently incubated with primary antibodies diluted in PBS-0.1% Tween/1% BSA (the GSTT1 antibody was diluted in PBS-0.1% Tween/2% BSA) at 4°C overnight. The next day, the primary antibody was removed, and the fixed monolayers were washed three times with PBS-0.1% Tween/1% BSA (the GSTT1 antibody was washed in PBS-0.1% Tween/2% BSA). After this, the cells were incubated with the appropriate secondary antibody diluted in PBS for 1 hour at room temperature and washed three times with PBS. After washing, the Hoechst 33342 (NucBlue Live Cell Stain ReadyProbes; Thermo Fisher Scientific Life Sciences) diluted in PBS (according to the manufacturer’s instructions) was added to the fixed monolayer and incubated for 20 minutes at room temperature. After removal, cell cultures were mounted with Mountant Perma-Fluor (Thermo Fisher Scientific Life Sciences). Stained monolayers were analyzed by an Olympus TH4-200 microscope (Olympus Corp., Tokyo, Japan, http://www.olympus.co.jp) and Volocity 4 software (PerkinElmer, Waltham, MA, http://www.perkinelmer.com). The percentage of positive cells and standard deviation were estimated from at least four random fields of view. The list of antibodies and dilutions are provided in supplemental online Table 4.
Paracetamol Dosing and Toxicity Studies
Paracetamol (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com) was diluted in ethanol (Sigma-Aldrich) and prepared at 0.5 M stock concentration. Different paracetamol concentrations (0, 1, 2, 5, 10, 20, and 50 mM) were prepared in HepatoZYME supplemented with factors (10 ng/ml hepatocyte growth factor [HGF]; 20 ng/ml oncostatin M [OSM]. At day 17, cells were treated with specific drug concentration and left for 24 hours in the incubator at 37°C. At day 18, adenosine triphosphate (ATP) production was measured by the CellTiter-Glo Luminescent Cell Viability Assay (Promega). For primary human hepatocytes, the cells were treated with the same range of paracetamol concentrations as described above at 48 hours after replating. The concentrations were prepared by diluting the stock solution in specific volumes of incubation medium (CM4000) and 2% bovine serum albumin (Sigma-Aldrich). Cells were incubated with the drug for 24 hours at 37°C. Subsequently, ATP production was measured by the CellTiter-Glo Luminescent Cell Viability Assay (Promega).
In Vitro Modulation of Paracetamol Toxicity Using MicroRNAs
At day 17, cells were transfected with scrambled controls, miRs, or antagomirs to miR-24, miR-324 for 24 hours at 37°C. At day 18, hESC-derived hepatocytes were exposed to the concentration of paracetamol resulting in a 50% depletion in ATP (IC50 − 12.85 mM). Twenty-four hours later (day 19), the toxic effect of the drug was measured using the CellTiter – Glo Luminescent Cell Viability Assay (Promega) and the GSH/GSSG-Glo Assay (Promega).
In Vitro Modulation of Paracetamol Toxicity Using N-Acetylcysteine
N-Acetylcysteine (NAC; Sigma-Aldrich) was prepared at 1 M stock concentration. At day 17 of differentiation, hESC-derived hepatocytes were treated with 1 mM NAC diluted in HepatoZYME supplemented with 10 ng/ml HGF and 20 ng/ml OSM. At day 18 of differentiation, hESC-derived hepatocytes were exposed to the concentration of paracetamol resulting in 50% death (IC50 = 12.85 mM) for another 24 hours. At day 19, ATP levels were measured using CellTiter – Glo Luminescent Cell Viability Assay (Promega).
Cell Viability Assay
Cellular ATP levels were measured by using CellTiter-Glo Luminescent Cell Viability Assay (Promega) according to the manufacturer’s instructions, and the luminescence signal was detected by the luminometer (Promega). The IC50 of paracetamol (acetaminophen [APAP]) was estimated from the function f(x) = ax + b.
Reduced Glutathione Assay
The amount of reduced glutathione produced in the hESC-derived hepatocytes was measured by the GSH/GSSG-Glo Assay from Promega and carried out according to the manufacturer’s instructions.
Patient Information, Sample Collection, and Processing
Ethical approval for the study was provided by the Scotland “A” Research and Ethics Committee, and written informed consent was obtained. Three female donors or their nominated next of kin consented to blood sampling. Paracetamol hepatotoxicity was prospectively defined as previously described [18]. Peripheral blood samples were obtained on the day of admission to the Scottish Liver Transplantation Unit. Serum was collected after centrifuging blood samples at 1,000g for 15 minutes at 4°C within 1 hour after collection. Serum was then immediately aliquotted, and stored and −80°C until thawing for the experiments. Importantly, no paracetamol was detectable in the serum samples used in the study. Patient blood biochemistry and normal ranges are provided in supplemental online Table 5.
Results
Defining Metabolic Gene Expression and Function in Stem Cell-Derived Hepatocytes
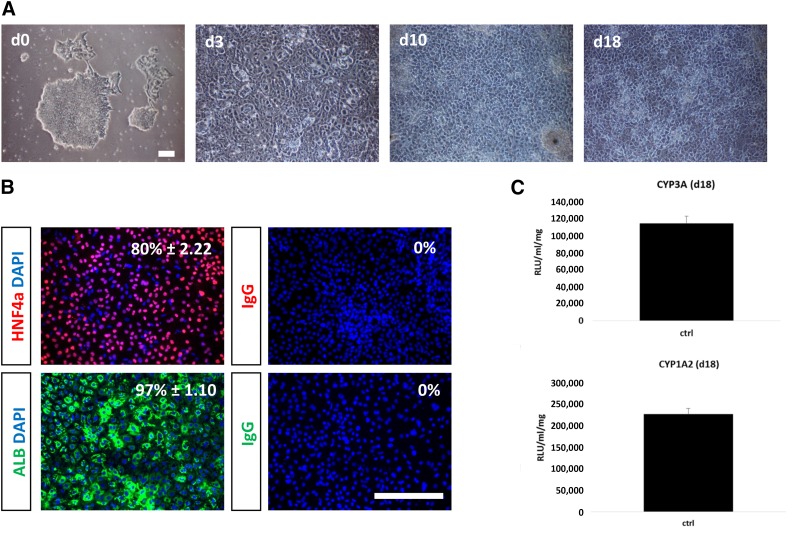
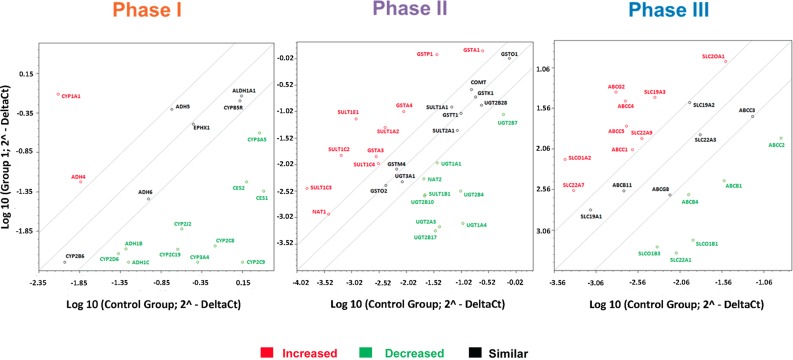
hESC-derived hepatocytes were produced in vitro by using established methodology [5, 16] and demonstrated appropriate cell morphology, gene expression, and appreciable levels of metabolic function (Fig. 1A–1C). Cyp3A activity in stem cell-derived hepatocytes was estimated at 8.4% of the female and 22% of male adult hepatocytes (Fig. 1C; supplemental online Fig. 1), whereas Cyp1A2 activity was estimated to be 0.009% and 0.06% of female and male hepatocytes, respectively (Fig. 1C; supplemental online Fig. 1). After validation, stem cell-derived hepatocytes were characterized for drug-metabolizing gene expression using microarray technology. Throughout these studies, stem cell-derived hepatocytes were compared with adult human hepatocytes. From these studies, we demonstrated that stem cell-derived hepatocytes expressed transcripts for phase I, II, and III drug metabolism, some of which were at reduced levels in comparison with primary hepatocytes (Fig. 2; supplemental online Table 1).
Figure 1.
Stagewise human embryonic stem cell (hESC) differentiation to the hepatocyte lineage. (A): Phase-contrast imaging demonstrated that cells underwent sequential morphological changes during transit from stem cell (day 0), to definitive endoderm (day 3), to hepatoblast (day 10), to hepatocyte (day 18). (B): Immunocytochemistry demonstrating upregulation of HNF4a and albumin during hepatic specification at day 18. Negative controls performed with corresponding immunoglobulin G. The percentage of positive cells is provided in the top right of each panel. This was calculated from four random fields of view and is shown as ± SD. (C), Cyp3A and Cyp1A2 metabolism were assessed in day 18 hepatocyte-like cells using the Promega pGlo system. Experiments were performed in triplicate and measured on a luminometer. The units of activity quoted are relative light units per milliliter of supernatant per milligram of protein. Scale bars = 100 μm. Abbreviations: ALB, albumin; d, day; DAPI, 4′,6-diamidino-2-phenylindole; HNF4a, hepatocyte nuclear factor 4a; IgG, immunoglobulin G RLU, relative light units.
Figure 2.
Stem cell-derived (day 18) and primary human hepatocyte gene expression. The scatter plots represent expression of the major metabolic genes involved in phase I, II, and III drug metabolism. Gene expression was performed by using the Human Drug Metabolism RT2 Profiler PCR Array (Qiagen) according to the manufacturer’s instructions. The scatter plot represents the changes in gene expression. The graph plots the log10 of normalized gene expression levels in a control condition, primary human hepatocytes (x axis) versus an experimental condition, human embryonic stem cell (hESC)-derived hepatocytes (y axis). Symbols outside the boundary area indicate fold differences larger than threefold.
Detailed Study of Paracetamol Metabolism in Stem Cell-Derived Hepatocytes
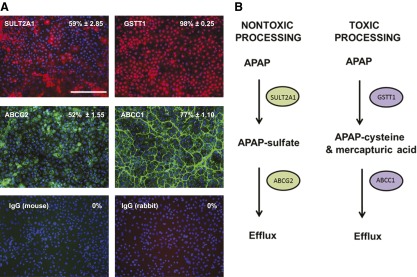
In order to ascertain which metabolic pathways were intact, in stem cell-derived hepatocytes, the data from the array experiments were analyzed using PharmGKB software (Stanford University, Stanford, CA, http://www.pharmgkb.org). From the analysis, we determined that a number of metabolic pathways were intact in our stem cell-derived system, including paracetamol metabolism (supplemental online Fig. 2). When paracetamol is taken within the therapeutic range, it is metabolized and excreted normally by phase II and III enzymes (supplemental online Fig. 2). However, if the drug is taken at higher doses than recommended, phase I enzymes generate NAPQI, which is toxic and leads to glutathione depletion and, ultimately, cell death (supplemental online Fig. 2). The results generated from the array experiments were validated by immunostaining, focusing on key phase II and III proteins. Phase II enzymes from the nontoxic pathway, SULT2A1, and toxic pathway, GSTT1, were expressed in 59% and ∼98% cells, respectively (Fig. 3A; supplemental online Table 1). Importantly, stem cell-derived hepatocytes also expressed phase III drug transporters important in each pathway. Approximately 52% and 77% of cells expressed ABCG2 and ABCC1, respectively (Fig. 3A; supplemental online Table 1). We therefore hypothesized that stem cell-derived hepatocytes had the correct machinery to process paracetamol in a nontoxic and toxic manner (Fig. 3B). To test this, we exposed stem cell-derived hepatocytes to a range of concentrations of paracetamol, ranging from 0 to 50 mM. After exposure, cell viability was monitored by ATP production. From these studies, we demonstrated that stem cell-derived hepatocyte cell death increased in a dose-dependent fashion with an IC50 value of 12.85 mM. Although differences in cytochrome P450 metabolic capacity were identified between stem cell-derived hepatocytes and primary hepatocytes, the paracetamol IC50 values obtained in vitro were comparable to both female and male hepatocytes, at 10.51 and 12.6 mM, respectively (supplemental online Fig. 3).
Figure 3.
The expression of enzymes and transporters involved in paracetamol (APAP) metabolism. (A): Representation of protein expression of phase II enzymes (GSTT1 and SULT2A1) and phase III transporters (ABCC1 and ABCG2) in stem cell-derived hepatocytes. The percentage of positive cells is provided in the top right of each panel. This was calculated from four random fields of view and is shown as ± SD. The images were taken at ×20 magnification. Scale bar = 100 μm. (B): Phase II enzymes and phase III transporters play major role in a toxic and nontoxic pathways of paracetamol metabolism. In the nontoxic pathway, paracetamol is metabolized by SULT2A1 enzymes to produce APAP sulfate metabolite that is effluxed from the cell by ABCG2 transporter. In a toxic pathway, paracetamol is metabolized by GSTT1 enzyme to produce APAP cysteine (mercapturic acid) metabolite that is effluxed from the cell by ABCC1 transporter. Abbreviations: ABCC1, adenosine triphosphate-Binding Cassette Transporter Subfamily C member 1; ABCG2, adenosine triphosphate-Binding Cassette Transporter Subfamily G member 2; APAP, acetaminophen; GSTT1, glutathione-S-transferase θ 1; SULT2A1, sulfotransferase 2A1.
MicroRNA Profiling of Stem Cell-Derived and Primary Hepatocytes
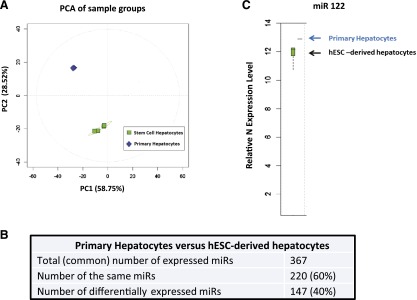
Once we had established that stem cell-derived hepatocytes responded to paracetamol appropriately, we wished to study microRNA expression within stem cell-derived hepatocytes. miRs are known to be potent regulators of gene expression, and our hypothesis was that miRs could play an important role in modulating paracetamol metabolism and, therefore, drug overdose in hepatocytes. To screen for miRs that regulate this process, day-18 hESC-derived hepatocytes were harvested and compared to adult hepatocytes using the Agilent miRNA platform. From these studies, we determined that stem cell-derived hepatocytes and primary human hepatocytes expressed 367 miRs in common, with 220 miRs being expressed at similar levels and 147 miRs differentially expressed (Fig. 4A, 4B; supplemental online Table 2). Of note, the major miR expressed in the liver, miR-122, was expressed at similar levels between primary and hESC-derived hepatocytes (Fig. 4C). Next, we evaluated similarly expressed miRs using TargetScanHuman6.2 (Whitehead Institute for Biomedical Research, Cambridge, MA, http://www.targetscan.org) to predict novel miRs that may regulate phase II enzymes important in paracetamol metabolism. The “hits” from our screen were ranked by their context scores [19, 20]. Our analysis predicted that miR-24 and -324 could potentially regulate GSTT1 and SULT2A1, respectively, and those miRs served as the focus of further experimentation (supplemental online Table 3).
Figure 4.
hESC-derived hepatocytes (hESC-heps) and primary human hepatocytes (PHH) microRNA expression profile. (A): Principal component analysis overview plot demonstrates strong clustering by cell type. (B): Statistical analysis of the microRNA array demonstrates 367 reliably detected microRNAs in both hESC-heps and PHH; 220 microRNAs have a similar expression in both systems and 147 microRNAs are differentially expressed. (C): microRNA 122 is expressed in hESC-heps at the same level as in PHH. The microRNA array was carried out by Sistemic Limited. The RNA samples (four replicates of PHH and four experimental samples of hESC-derived hepatocytes) were run on the Agilent miRNA platform. Abbreviations: hESC, human embryonic stem cell; miR, microRNA; PC1, principal component 1; PC2, principal component 2; PCA, principal component analysis.
MicroRNA 324 Regulation of Paracetamol-Induced Toxicity in Stem Cell-Derived Hepatocytes
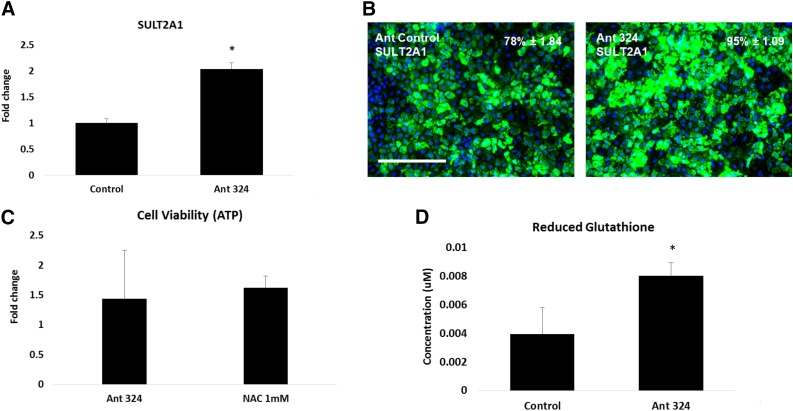
In order to test our hypothesis that stem cell-derived hepatocyte susceptibility to paracetamol overdose could be regulated by miRs, we transfected synthetic noncoding RNAs. Stem cell-derived hepatocyte transfection was optimized, with synthetic RNAs used at a concentration of 50 nM for 24 hours before incubation with a toxic dose of paracetamol. To examine the effects of antagomir transfection on phase II enzyme expression, we performed qPCR and immunostaining. hESC-derived hepatocytes transfected with the scrambled control served as our baseline throughout. In response to transfection with the antagomir for miR-324, stem cell-derived hepatocytes expressed greater levels of SULT2A1 (Fig. 5A, 5B). In contrast, transfection with miR-324 or the noncoding RNAs for miR-24 did not result in gene expression changes for SULT2A1 or GSTT1 (supplemental online Figs. 4, 5). Once we had established that SULT2A1 expression could be modulated successfully, we measured cell viability in response to a toxic dose of paracetamol. Cell viability was improved following antagomir 324 transfection in hESC-derived hepatocytes. The improvement in cell viability was comparable to that of cells treated with the current clinical practice, NAC administration (Fig. 5C). Notably, increased cell viability was paralleled by a twofold increase in reduced glutathione levels (Fig. 5D).
Figure 5.
Antagomir of microRNA 324 upregulated SULT2A1 gene and protein expression and increased cell survival after exposure to paracetamol. At day 17, human embryonic stem cell-derived hepatocytes (hESC-heps) were transfected with the antagomir of the corresponding microRNA at 50 nM for 24 hours. (A, B): Transfection with the Ant-324-5p upregulated SULT2A1 gene expression by twofold (A) and protein expression by 20% (B) in comparison with the scrambled control. At day 17, hESC-hepatocytes were either transfected with antagomirs or treated with 1 mM N-acetylcysteine (NAC) concentration for 24 hours. At day 18, the antagomir-transfected or NAC pre-exposed hESC-hepatocytes were exposed to paracetamol concentration that causes 50% of the cell death (IC50 = 12.85 mM) for another 24 hours. At day 19, the cell viability was measured using CellTiter Assay (Promega) and Glutathione Depletion Assay (Promega). (C, D): The antagomir of microRNA 324 significantly increased ATP to the levels comparable with NAC (fold increase was calculated in comparison with control; the values for Ant ctrl = 2.17 × 108; Ant 324= 3.12 × 108; H2O (vehicle) = 2.38 × 108; NAC = 3.86 × 108 (C) and enhanced reduced glutathione levels (D). Levels of significance were measured by Student’s t test. ∗, p < .05. The percentage of positive cells is provided in the top right of each panel. This was calculated from four random fields of view and is quoted as ± SE. Scale bar = 100 μm. Abbreviations: Ant 324, antagomir 324-5p; Ant ctrl; scrambled antagomir control; ATP, adenosine triphosphate; SULT2A1, sulfotransferase 2A1.
Inhibition of MicroRNA-324 Reduces Cell Necrosis in Response to Plasma From Patients With Fulminant Liver Failure
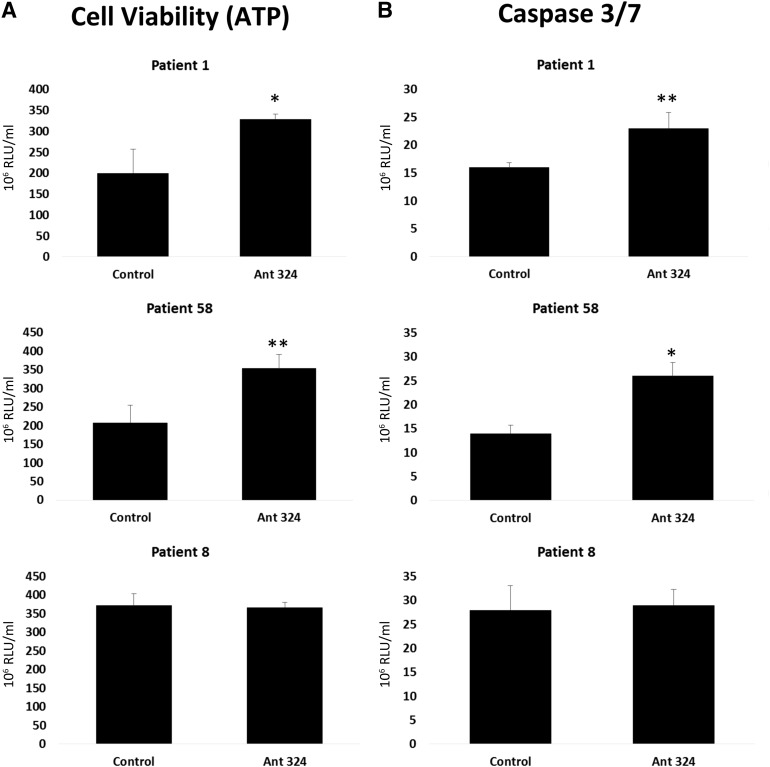
Following on from acute paracetamol-induced hepatocyte injury, we studied the toxic nature of patient’s derived plasma on stem cell-derived hepatocytes. As described before, stem cell-derived hepatocytes were differentiated and transfected with either the scrambled control or the antagomir for miR-324. At 24 hours post-transfection, stem cell-derived hepatocytes were incubated with human plasma from three female donors (anonymized patient details and blood biochemistry are supplied in supplemental online Table 5). After exposure to plasma, stem cell-derived hepatocyte ATP levels were measured. Notably, stem cell-derived hepatocytes transfected with antagomir 324 displayed greater levels of ATP, which was significantly increased over scrambled controls in two of the three patients (Fig. 6A). In parallel, we measured caspase 3 and 7 activity in stem cell-derived hepatocytes. As for ATP, we also observed a significant increase in caspase activity after transfection with antagomir 324 in two of the three patients (Fig. 6B). Taken together, these data suggest that increasing the levels of SULT2A1 gene expression in stem cell-derived hepatocytes redirects cell necrosis to apoptosis, following challenge with plasma from liver failure patients.
Figure 6.
Human embryonic stem cell (hESC)-derived hepatocytes transfected with the antagomir 324-5p. At 24 hours post-transfection, transfection, hESC-hepatocytes were exposed to 20% plasma from patients with fulminant hepatic failure for a further 24 hours (patients 1, 8, and 58). ATP levels and caspase 3/7 activation were determined using Promega Glo technology. Units of activity are expressed as RLU per milliliter (n = 4). Levels of significance are quoted and measured by Student’s t test. ∗, p < .05; ∗∗, p < .01. Abbreviations: Ant 324;, antagomir 324-5p; ATP, adenosine triphosphate; RLU, relative light units.
Discussion
Despite major progress in the knowledge and management of human liver injury, there are approximately 2,000 cases per year of acute liver failure (ALF) in the U.S. [21–23]. Paracetamol overdose is a major cause of ALF, with critical damage done to the hepatocyte compartment of the liver, and accounts for approximately 50% of cases [24, 25]. Although hepatocyte cell death occurs in large numbers, the manner by which the cells die after overdose remains complicated [26].
The hepatotoxic dose of paracetamol is considered to be greater than 75 mg/kg. This translates into toxic blood concentrations that range between 25 and 150 mg/l [27, 28]. Currently, treatment with N-acetylcysteine (NAC) is the most effective strategy in treating paracetamol-induced liver injury. Patients who have ingested 75–150 mg/kg in 24 hours can be considered for NAC treatment, with NAC usually prescribed if more than 150 mg/kg was ingested within 24 hours (based on the guidelines issued by the National Poisons Information Service U.K.; http://www.npis.org). Although successful, oral and intravenous NAC treatment may elicit serious side effects [29–32], and therefore new approaches to treat acute intoxication are necessary. In order to study the important nature of microRNAs in human drug toxicity, reliable and renewable liver models are required. In this vein, we have used pluripotent stem cells to generate human hepatocyte-like cells. The in vitro model used in these studies is serum-free and has already demonstrated promise in modeling human drug metabolism and toxicity [5, 16, 33, 34]. Moreover, in these studies, we support those observations because stem cell-derived hepatocytes display similar IC50 values to male and female primary hepatocytes (supplemental online Fig. 3).
microRNAs are potent noncoding RNAs that can alter mammalian gene expression and therefore represent promising candidates for modulating enzymatic pathways [7–12, 35] and treating human disease. Several studies have shown that regulation of different microRNAs may potentially serve as effective therapeutics [36–38]. In recent years, there has been a focus on miR regulation of phase I enzymes involved in human drug metabolism. Studies have demonstrated that miR-27b regulates CYP3A4 and CYP1B1, and miR-126* controls CYP2A3 expression [7, 8, 39]. Other studies have also focused on microRNA regulation of drug transporters such as P-glycoprotein and breast cancer-resistant protein [11, 12, 40–42]. Although phase I and III of drug metabolism have been studied, in detail, there is still little known about regulation of phase II enzymes by microRNAs. Phase II enzymes, such as glutathione-S-transferases (GSTs) and sulfotransferases (SULTs), are essential to detoxify multiple prescription medicines. Therefore, these enzymes serve as important clinical targets. To test whether microRNAs could regulate phase II enzymes (SULT2A1 and GSTT1) that play an important role in paracetamol metabolism, we used TargetScan Human software to identify novel noncoding RNAs. Inhibition of miR-324-5p in hESC-derived hepatocytes resulted in increased SULT2A1 expression (Fig. 5A, 5B). This led to improved cell survival following exposure to toxic levels of paracetamol and was comparable with current clinical practice (Fig. 5C). Notably, the inhibition of miR-324-5p was paralleled by an increase in reduced glutathione production (Fig. 5D). In contrast, modulation of miR-24-3p did not have any effect on GSTT1 expression, cell viability, and reduced glutathione production in the paracetamol studies. Although TargetScan is considered to be the most effective tool for predicting miR-target binding sites [43, 44], algorithm-prone errors may have led to this false prediction.
Given the promising effects of the antagomir 324 with paracetamol, we were keen to assess its efficacy after exposure to liver failure serum from paracetamol-poisoned female patients. Importantly, paracetamol was not present in patient sera, and, therefore, we were studying the supportive effects of antagomir 324 in the context of patient recovery. The inhibition of miR-324-5p by antagomir 324 resulted in increased cell viability and caspase activity in two patients (Fig. 6), suggesting a potential switch from cell necrosis to apoptosis. Contrary to these two patients, antagomir 324 did not have any rescue effect after exposure to the third patient’s serum. The exact reason for this is unknown; however, this patient possessed the lowest concentration of serum alanine transaminase (ALT), which may have reflected a lesser liver injury and therefore lower toxic load to the cells (supplemental online Table 5).
Conclusion
We demonstrate that a novel miR inhibitor, antagomir 324, plays a major role in the regulation of SULT2A1, improving cell survival in the context of acute injury and patient recovery after paracetamol overdose. We believe that these studies are novel and offer serious promise to reduce the toxic effects of paracetamol overdose.
Supplementary Material
Acknowledgments
We thank Sistemic for performing the microRNA profiling experiments. The microRNA profiling experiments were supported by the Scottish Stem Cell Network. D.S. and B.L.-V. were supported by Medical Research Council Ph.D. studentships. S.J.F. and D.C.H were supported by U.K. Regenerative Medicine Platform Awards MR/K026666/1 and MR/L022974/1. K.J.S. was supported by Chief Scientist Office, Scotland, Grant ETM/191.
Author Contributions
D.S.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, manuscript proofreading; B.L.-V.: collection and/or assembly of data, data analysis and interpretation; J.K.M.: provision of patient samples, data analysis and interpretation; K.J.S.: provision of patient samples, data analysis and interpretation, manuscript proofreading; S.J.F.: supervision, data analysis and interpretation, manuscript proofreading; D.C.H.: conception and design, supervision, data analysis and interpretation, manuscript writing, financial support, final approval of manuscript, manuscript proofreading.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
References
- 1.Olsen AK, Whalen MD. Public perceptions of the pharmaceutical industry and drug safety: Implications for the pharmacovigilance professional and the culture of safety. Drug Saf. 2009;32:805–810. doi: 10.2165/11316620-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 2.Davies EC, Green CF, Taylor S, et al. Adverse drug reactions in hospital in-patients: A prospective analysis of 3695 patient-episodes. PLoS One. 2009;4:e4439. doi: 10.1371/journal.pone.0004439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies EC, Green CF, Mottram DR, et al. Emergency re-admissions to hospital due to adverse drug reactions within 1 year of the index admission. Br J Clin Pharmacol. 2010;70:749–755. doi: 10.1111/j.1365-2125.2010.03751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pirmohamed M, James S, Meakin S, et al. Adverse drug reactions as cause of admission to hospital: Prospective analysis of 18 820 patients. BMJ. 2004;329:15–19. doi: 10.1136/bmj.329.7456.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szkolnicka D, Farnworth SL, Lucendo-Villarin B, et al. Accurate prediction of drug-induced liver injury using stem cell-derived populations. Stem Cells Translational Medicine. 2014;3:141–148. doi: 10.5966/sctm.2013-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan YZ, Gao W, Yu AM. MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab Dispos. 2009;37:2112–2117. doi: 10.1124/dmd.109.027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsuchiya Y, Nakajima M, Takagi S, et al. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 2006;66:9090–9098. doi: 10.1158/0008-5472.CAN-06-1403. [DOI] [PubMed] [Google Scholar]
- 9.Yu AM. Role of microRNAs in the regulation of drug metabolism and disposition. Expert Opin Drug Metab Toxicol. 2009;5:1513–1528. doi: 10.1517/17425250903307448. [DOI] [PubMed] [Google Scholar]
- 10.Yu A-M, Pan Y-Z. Noncoding microRNAs: Small RNAs play a big role in regulation of ADME? Acta Pharm Sin B. 2012;2:93–101. doi: 10.1016/j.apsb.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kovalchuk O, Filkowski J, Meservy J, et al. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther. 2008;7:2152–2159. doi: 10.1158/1535-7163.MCT-08-0021. [DOI] [PubMed] [Google Scholar]
- 12.Zhu H, Wu H, Liu X, et al. Role of MicroRNA miR-27a and miR-451 in the regulation of MDR1/P-glycoprotein expression in human cancer cells. Biochem Pharmacol. 2008;76:582–588. doi: 10.1016/j.bcp.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chun LJ, Tong MJ, Busuttil RW, et al. Acetaminophen hepatotoxicity and acute liver failure. J Clin Gastroenterol. 2009;43:342–349. doi: 10.1097/MCG.0b013e31818a3854. [DOI] [PubMed] [Google Scholar]
- 14.Hay DC, Fletcher J, Payne C, et al. Highly efficient differentiation of hESCs to functional hepatic endoderm requires ActivinA and Wnt3a signaling. Proc Natl Acad Sci USA. 2008;105:12301–12306. doi: 10.1073/pnas.0806522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hay DC, Pernagallo S, Diaz-Mochon JJ, et al. Unbiased screening of polymer libraries to define novel substrates for functional hepatocytes with inducible drug metabolism. Stem Cell Res (Amst) 2011;6:92–102. doi: 10.1016/j.scr.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Szkolnicka D, Farnoworth SL, Lucendo-Villarin B, et al. Deriving functional hepatocytes from pluripotent stem cells. Curr Protoc Stem Cell Biol. 2014;30:1G.5.1–1G.5.12. doi: 10.1002/9780470151808.sc01g05s30. [DOI] [PubMed] [Google Scholar]
- 17.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 18.Craig DG, Lee P, Pryde EA, et al. Serum neopterin and soluble CD163 as markers of macrophage activation in paracetamol (acetaminophen)-induced human acute liver injury. Aliment Pharmacol Ther. 2013;38:1395–1404. doi: 10.1111/apt.12530. [DOI] [PubMed] [Google Scholar]
- 19.Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimson A, Farh KK, Johnston WK, et al. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoofnagle JH, Carithers RL, Jr, Shapiro C, et al. Fulminant hepatic failure: Summary of a workshop. Hepatology. 1995;21:240–252. [PubMed] [Google Scholar]
- 22.Polson J, Lee WM, American Association for the Study of Liver Disease AASLD position paper: The management of acute liver failure. Hepatology. 2005;41:1179–1197. doi: 10.1002/hep.20703. [DOI] [PubMed] [Google Scholar]
- 23.Fontana RJ. Acute liver failure including acetaminophen overdose. Med Clin North Am. 2008;92:761–794, viii. doi: 10.1016/j.mcna.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nourjah P, Ahmad SR, Karwoski C, et al. Estimates of acetaminophen (paracetomal)-associated overdoses in the United States. Pharmacoepidemiol Drug Saf. 2006;15:398–405. doi: 10.1002/pds.1191. [DOI] [PubMed] [Google Scholar]
- 25.Bari K, Fontana RJ. Acetaminophen overdose: What practitioners need to know. Clin Liver Dis. 2014;4:17–21. doi: 10.1002/cld.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaeschke H, McGill MR, Ramachandran A. Oxidant stress, mitochondria, and cell death mechanisms in drug-induced liver injury: Lessons learned from acetaminophen hepatotoxicity. Drug Metab Rev. 2012;44:88–106. doi: 10.3109/03602532.2011.602688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winek CL. Winek’s Toxicological Annual. Pittsburgh, PA: Allegheny County Department Laboratories; 1994. Drug and chemical blood-level data. [Google Scholar]
- 28.Dollery C. Therapeutic Drugs. Vols 1 and 2. London, U.K.: Churchill Livingstone; 1993. [Google Scholar]
- 29.Harrison PM, Keays R, Bray GP, et al. Improved outcome of paracetamol-induced fulminant hepatic failure by late administration of acetylcysteine. Lancet. 1990;335:1572–1573. doi: 10.1016/0140-6736(90)91388-q. [DOI] [PubMed] [Google Scholar]
- 30.Bailey B, McGuigan MA. Management of anaphylactoid reactions to intravenous N-acetylcysteine. Ann Emerg Med. 1998;31:710–715. doi: 10.1016/s0196-0644(98)70229-x. [DOI] [PubMed] [Google Scholar]
- 31.Appelboam AV, Dargan PI, Knighton J. Fatal anaphylactoid reaction to N-acetylcysteine: Caution in patients with asthma. Emerg Med J. 2002;19:594–595. doi: 10.1136/emj.19.6.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pakravan N, Waring WS, Sharma S, et al. Risk factors and mechanisms of anaphylactoid reactions to acetylcysteine in acetaminophen overdose. Clin Toxicol (Phila) 2008;46:697–702. doi: 10.1080/15563650802245497. [DOI] [PubMed] [Google Scholar]
- 33.Medine CN, Lucendo-Villarin B, Storck C, et al. Developing high-fidelity hepatotoxicity models from pluripotent stem cells. Stem Cells Translational Medicine. 2013;2:505–509. doi: 10.5966/sctm.2012-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cameron K, Tan R, Schmidt-Heck W, et al. Recombinant laminins drive the differentiation and self-organization of hESC-derived hepatocytes. Stem Cell Rep. 2015;5:1250–1262. doi: 10.1016/j.stemcr.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu X, Dhakal IB, Beggs M, et al. Functional genetic variants in the 3′-untranslated region of sulfotransferase isoform 1A1 (SULT1A1) and their effect on enzymatic activity. Toxicol Sci. 2010;118:391–403. doi: 10.1093/toxsci/kfq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thong VD, Akkarathamrongsin S, Poovorawan K, et al. Hepatitis C virus genotype 6: Virology, epidemiology, genetic variation and clinical implication. World J Gastroenterol. 2014;20:2927–2940. doi: 10.3748/wjg.v20.i11.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heidet L, Gubler MC. The renal lesions of Alport syndrome. J Am Soc Nephrol. 2009;20:1210–1215. doi: 10.1681/ASN.2008090984. [DOI] [PubMed] [Google Scholar]
- 38.Daige CL, Wiggins JF, Priddy L, et al. Systemic delivery of a miR34a mimic as a potential therapeutic for liver cancer. Mol Cancer Ther. 2014;13:2352–2360. doi: 10.1158/1535-7163.MCT-14-0209. [DOI] [PubMed] [Google Scholar]
- 39.Kalscheuer S, Zhang X, Zeng Y, et al. Differential expression of microRNAs in early-stage neoplastic transformation in the lungs of F344 rats chronically treated with the tobacco carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis. 2008;29:2394–2399. doi: 10.1093/carcin/bgn209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao R, Sun J, Zhang L, et al. MicroRNAs play a role in the development of human hematopoietic stem cells. J Cell Biochem. 2008;104:805–817. doi: 10.1002/jcb.21668. [DOI] [PubMed] [Google Scholar]
- 41.To KK, Zhan Z, Litman T, et al. Regulation of ABCG2 expression at the 3′ untranslated region of its mRNA through modulation of transcript stability and protein translation by a putative microRNA in the S1 colon cancer cell line. Mol Cell Biol. 2008;28:5147–5161. doi: 10.1128/MCB.00331-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan YZ, Morris ME, Yu AM. MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol Pharmacol. 2009;75:1374–1379. doi: 10.1124/mol.108.054163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baek D, Villén J, Shin C, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Witkos TM, Koscianska E, Krzyzosiak WJ. Practical aspects of microRNA target prediction. Curr Mol Med. 2011;11:93–109. doi: 10.2174/156652411794859250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.