Abstract
AcidoCEST magnetic resonance imaging (MRI) has previously been shown to measure tumor extracellular pH (pHe) with excellent accuracy and precision. This study investigated the ability of acidoCEST MRI to monitor changes in tumor pHe in response to therapy. To perform this study, we used the Granta 519 human mantle cell lymphoma cell line, which is an aggressive B-cell malignancy that demonstrates activation of the phosphatidylinositol-3-kinase/Akt/mammalian target of rapamycin (mTOR) pathway. We performed in vitro and in vivo studies using the Granta 519 cell line to investigate the efficacy and associated changes induced by the mTOR inhibitor, everolimus (RAD001). AcidoCEST MRI studies showed a statistically significant increase in tumor pHe of 0.10 pH unit within 1 day of initiating treatment, which foreshadowed a decrease in tumor growth of the Granta 519 xenograft model. AcidoCEST MRI then measured a decrease in tumor pHe 7 days after initiating treatment, which foreshadowed a return to normal tumor growth rate. Therefore, this study is a strong example that acidoCEST MRI can be used to measure tumor pHe that may serve as a marker for therapeutic efficacy of anticancer therapies.
Keywords: mTOR inhibition, CEST MRI, extracellular pH, therapy response, lymphoma
Introduction
Tumor acidosis is an emerging biomarker for characterizing tumors that is correlated with increased metastatic and invasive potential.1,2 The altered cellular energetics in cancer cells, first described by Otto Warburg in 1919 and therefore called the Warburg effect, result in the overproduction of lactic acid and its excretion into the extracellular tumor microenvironment.3 The overabundance of lactic acid in the microenvironment reduces the extracellular pH (pHe). This link between the cancer cells’ metabolic activity and the tumor pHe indicates that the tumor pHe can serve as a biomarker for measuring the efficacy of metabolism-altering drug therapies.
Several methods have been developed in an attempt to measure tumor pHe. The most rudimentary method uses a microelectrode that is surgically inserted into the tumor tissue.4–6 However, this method is invasive and therefore is seldom used in clinical practice. A positron emission topography method uses a 64Cu-conjugated peptide that inserts into cell membranes within acidic environments. However, the technique is limited by the peptide’s sinusoidal relationship between pH and membrane insertion, which affects the precision of this measurement method.7–10 Optics-based imaging methods can measure pH based on emission spectra or fluorescence lifetimes. However, the dyes used for these imaging studies can associate with extracellular proteins, which causes a shift in their emission wavelengths and fluorescence lifetimes, thereby causing the pH measurement to become inaccurate.11–13 Moreover, optics-based imaging methods are subject to absorbance and light-scattering effects in tissues and therefore have diminished sensitivities in deep tissues where most tumors are found. Magnetic resonance imaging (MRI) and magnetic resonance (MR) spectroscopy have also been used to develop pH measurement methods. Hyperpolarized 13C MR spectroscopic imaging and standard MR spectroscopy of 31P and 19F can accurately measure pHe, but detection of these heteronuclei by MRI requires expensive, specialized resources that are not available in most radiology clinics.14–18 Magnetic resonance spectroscopic imaging of the 1H signal from imidazole derivatives has also been used to measure pHe, but these compounds act as buffers that may alter the endogenous pH value during the MRI scan session.19–21 Finally, T1-relaxation MRI has shown success in measuring pHe using a Gd(III)-based contrast agent. Unfortunately, the concentration of the agent also must be known to accurately determine the pHe value from T1-weighted MRI studies, which has proven difficult to achieve.22–25 Overall, there still exists a need to be able to reliably and robustly measure tumor pHe to rapidly assess the efficacy of metabolism-altering drug therapies.
We have developed a noninvasive MRI-based imaging method to meet this need for assessing tumor pHe. Called acidoCEST MRI, this method takes advantage of chemical exchange saturation transfer (CEST) from an exogenous contrast agent to the bulk water in the tumor environment.26 In comparison to the other molecular imaging methodologies described above, acidoCEST MRI is unique in its robust design and clinical practicality, due in part to its use of abundantly available and Food and Drug Administration–approved contrast agents. Therefore, acidoCEST MRI has a clear pathway toward clinical translation. We have shown that acidoCEST MRI can accurately and precisely measure tumor pHe, can generate parametric maps of tumor pHe at moderately high spatial resolution, and can be used with more than one contrast agent.27 Similar CEST MRI methods with an exogenous agent have been used to measure the pHe in kidney and tumor tissues.28–29 We have also shown that faster growing tumors exhibit higher acidosis and that shrinking lesions of idiopathic lung fibrosis exhibit lower acidosis.30–31 However, acidoCEST MRI and related CEST MRI techniques have not yet investigated the utility of this methodology for detecting the early response to drug therapies.
This study investigated the effects of inhibiting mammalian target of rapamycin complex 1 (mTORC1) with the first-generation mTOR inhibitor, everolimus (RAD001), in a Mantle cell lymphoma (MCL) model with the anticipation of observing modest in vivo activity.
To examine tumor pHe as a biomarker for drug efficacy, we studied the early response of a xenograft model of Granta 519 MCL to a first-generation inhibitor of the mTORC1. Mantle cell lymphoma is a rare B-cell non-Hodgkin lymphoma (NHL) subtype that accounts for 5% to 10% of NHL.31 Although MCL responds well to initial treatment, it invariably relapses and eventually becomes refractory to further therapy, with a median survival of 3 to 5 years after diagnosis.32 Everolimus has also been investigated in MCL in 2 phase 2 studies with an overall response rate ranging from 20% to 32%.33,34 First-generation mTOR inhibitors have had limited clinical success, due to one of several accumulated resistance pathways. Therefore, monitoring the early responses to mTORC1 inhibitors may have great clinical impact by providing positive evidence that the patient should continue the treatment or negative evidence that would lead to a change in the therapeutic regimen for the patient. For these reasons, the investigation of the effect of everolimus on the tumor pHe of the Granta 519 MCL model provides a more compelling and potentially more impactful test of acidoCEST MRI than an investigation with mTOR complex 2 (mTORC2) inhibitors.
To perform this study, we first investigated whether everolimus affects mTOR in the Granta 519 cell line. We then investigated the downstream effects of mTOR inhibition, including the expression of proteins that are responsible for key metabolic steps, the production of adenosine triphoshpate (ATP), and the generation of lactic acid. We then determined the effect of everolimus on the tumor growth of the Granta 519 xenograft model. Finally, we performed acidoCEST MRI to investigate changes in tumor pHe in response to daily everolimus treatment of the Granta 519 tumor model. The acidoCEST MRI studies were then compared with the evaluations of the effect of everolimus on tumor metabolism to assess whether acidoCEST MRI can be used as an early response indicator for drug therapy.
Materials and Methods
Tissue Culture Procedures
Granta 519 cells were obtained from the American Type Culture Collection (Manassas, Virginia). This cell line has been previously well characterized as a model of MCL. Cells were tested for mycoplasma contamination using the MycoAlert mycoplasma detection assay kit from Lonza (Basel, Switzerland) and found to be negative. The cell line was verified to be authentic by the University of Arizona Genomics Core. Cells were routinely grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (HyClone Thermo Scientific, Waltham, Massachusetts) in a humidified incubator at 37°C and 5% CO2. Per test parameters, the DMEM either had 4.5 or 1.0 g/L of glucose. Cells were treated with 10 nmol/L everolimus dissolved in biological grade 100% dimethyl sulfoxide (DMSO).
Western Blot Analyses
The cells were washed with phosphate-buffered saline (PBS), suspended in lysis buffer (50 mmol/L Tris-HCl, pH 7.4, 1% Triton X-100, 150 mmol/L NaCl, 2 mmol/L EDTA, 5 mmol/L Na3VO4, 200 μmol/L NaF, 21 μmol/L leupeptin, 230 nmol/L aprotinin, and 1 mmol/L phenylmethane sulfonyl fluoride [PMSF]), and further lysed by sonication. Protein concentration of the supernate was determined using the 660 nM Protein Assay kit (Thermo Scientific, Rockford, Illinois). Lysates were separated by 10% Tris-glycine gel electrophoresis (Invitrogen, Carlsbad, California), transferred to 0.2 μm Polyvinylidene fluoride (PVDF) membranes (Bio Rad Hercules, CA), and then blocked for 30 minutes in 5% nonfat dry milk/tris-buffered saline and tween 20 (TBST). Membranes were immunoblotted with primary antibodies at concentrations recommended by the manufacturer. Antibodies against P-mTOR (Ser2448), mTOR, P-p70 S6 K (Thr289), p70 S6 K, full-length caspase 3, and cleaved caspase 3 were obtained from Cell Signaling Technology (Beverly, Massachusetts). Antibodies against monocarboxylate transporter 1 (MCT-1) and MCT-4 were purchased from Santa Cruz Biotechnology (Santa Cruz, California), and antibodies against α-tubulin were purchased from Calbiochem (La Jolla, California). Following primary antibody incubation, the membranes were then washed in TBST and then probed with a horseradish peroxidase (HRP)-conjugated antirabbit antibody or antimouse antibody purchased from Jackson ImmunoResearch (West Grove, Pennsylvania). Membranes were then visualized with HyGLO chemiluminescent HRP antibody detection reagent (Denville Scientific Inc, Metuchen, New Jersey). Blots were stripped with OneMinute Plus Western Blot Stripping Buffer (GM Biosciences, Rockville, Maryland), according to the manufacturer’s instructions.
Flow Cytometry
Cell death was analyzed with the annexin V–fluorescein isothiocyanate (FITC) apoptosis detection kit (Sigma-Aldrich, St Louis, Missouri), according to the manufacturer’s protocol. Cells were seeded with treatment media in 6-well plates and incubated for 24 hours. The cells were then collected and washed twice in PBS. The cells were analyzed by flow cytometry after incubation with annexin V–FITC and propidium iodide (PI; FACSCanto II; BD Biosciences, San Jose, California).
Cell Viability
Cell viability was determined by quantitation of ATP. Granta 519 cells were seeded in 96-well plates with various treatment media. After a 24-hour incubation, the CellTiter-Glo kit (Pro-mega Madison, WI) was used according to the manufacturer’s instructions. Luminescence was measured by a Beckman Coulter Multimode DTX 880 microplate reader (Bechman Coulter, Brea, CA). Values are presented as a percentage of control.
Gene Expression Analysis
Isolation of RNA from cultured cells was performed using the RNeasy Plus Mini Kit (Qiagen, Germantown MD), according to the manufacturer’s protocol. Final RNA elution was performed using 50 μL of RNase-free water. RNA concentrations were determined using a NanoDrop 2000c (Thermo Scientific, Waltham, Massachusetts) microvolume spectrophotometer. Complementary DNA (cDNA) for reverse transcription–polymerase chain reaction (RT-PCR) was synthesized using the qScript cDNA synthesis kit (Quanta Biosciences, Gaithersburg, Maryland), according to the manufacturer’s protocol using 500 ng of RNA as template per reaction. The thermal cycler conditions for cDNA synthesis were 1 cycle at 22°C for 5 minutes, 1 cycle at 42°C for 30 minutes, and 1 cycle at 85°C for 5 minutes. The RT-PCR reactions were performed using 2× TaqMan Universal PCR Master Mix (Applied Biosystems, Branchburg, New Jersey) and 20× TaqMan Gene Expression Assays (Applied Biosystems), which contain both the target primers and probe, and 2 μL of cDNA template were used per reaction according to the manufacturer’s protocol. Each FAM-labeled gene of interest was analyzed in triplicate and multiplexed with the primer-limited, VIC-labeled control gene RPLPO as an endogenous control. The RT-PCR amplification reactions were performed using an ABI Prism 7000 Sequence Detection System (Applied Biosystems). The cycling conditions for RT-PCR were 1 cycle at 50°C for 2 minutes, 1 cycle at 95°C for 10 minutes, 40 cycles at 95°C for 15 seconds, followed by 60°C for 1 minute. Relative gene expression was determined using the 2−ΔΔCT method.35
Lactate and Protein Concentration Assays
Lactate production was analyzed in media from cells treated with vehicle control or everolimus using the L-Lactate Assay Kit I (Eton Bioscience, San Diego, California) per manufacturer’s instructions. Conditioned media were collected by centrifuging cells at 2000g and removing supernatant. Media were immediately frozen at −80°C and thawed immediately prior to analysis. A standard curve was generated using lactate and bovine serum albumin standards provided in the respective kits. The lactate concentration of samples was determined by plotting results on the standard curve, normalized to protein concentration and accounting for the lactate concentration of media.
Xenograft Tumor Growth Studies
Animals were handled according to the guidelines of the Institutional Animal Care and Use Committee (IACUC) of the University of Arizona. A total of 10 × 106 Granta 519 cells were mixed 1:1 with Matrigel (BD Bioscience, Franklin Lakes, New Jersey) and injected subcutaneously into 4- to 6-week-old severe combined immunodeficiency (SCID) mice. Xenograft tumors grew to approximately 150 mm3 prior to stratification into groups of 12 animals having equal mean tumor volumes. Everolimus was dissolved into DMSO, and animals were administered everolimus at 5 mg/kg/d intraperitoneally (IP) with a maximum of 50 μL of volume. Measurements were performed every 3 to 4 days using a digital caliper with tumor volume calculated as π/6 × (short axis in mm)2 × (long axis in mm). Animals were euthanized when tumors reached ≥2000 mm3. The analysis of the timing for reaching this tumor size was assessed using GraphPad (La Jolla, CA).
AcidoCEST MRI and Drug Administration
The same animal model used for xenograft tumor growth studies was used for acidoCEST MRI studies. An imaging cohort of 16 mice was used, split into 2 groups of 8 control and 8 treated animals. Xenograft tumors grew to approximately 300 to 400 mm3 prior to drug treatment. Everolimus dissolved in DMSO was injected IP in mice at a dose of 5 mg/kg/d for 7 days during the imaging study. The acidoCEST MRI scans were performed 1 day prior to the first drug dose, 1 day after the first drug dose, and 7 days after the first dose.
The acidoCEST MRI protocol was detailed previously.26 To initiate an acidoCEST MRI scan session, a mouse was anesthetized with 1.5% to 2.5% isoflurane delivered in 1 L/min oxygen gas ventilation and then secured in a custom cradle. A 27-g catheter was inserted into a tail vein, which was used to inject a 200 μL bolus of clinical-grade iopromide at 788 mmol/L (300 mg iodine/mL Ultravist; Bayer Healthcare, Inc, Leverkusen, Germany). The mouse in the cradle was inserted into a 7T Biospec MRI scanner with a 72-mm-diameter quadrature transceiver coil (Bruker Biospin, Inc, Billerica, Massachusetts). The breathing rate was monitored and maintained at approximately 40 to 60 breaths per minute by adjusting the level of isoflurane anesthetic. Body temperature was also monitored and maintained at 37.0°C ± 0.2°C using an automated feedback loop between the temperature probe and an air heater (SA Instruments, Inc, Stony Brook, New York). The catheter was connected to a perfusion pump to deliver iopromide at 150 μL/h during imaging. A FLASH MRI acquisition sequence was performed to determine the location of the tumor (500 ms relaxation time [TR], 10 ms TE, 625 × 625 μm in-plane resolution, 2.0-mm slice thickness, 64 × 64 pixels, 4.0 × 4.0 cm FOV, one average, 32-second total acquisition time). A spin-echo MRI acquisition sequence was performed to obtain images with good contrast for determining the tumor volume (2000 ms TR, 35 ms echo time [TE], 312.5 × 312.5 μm in-plane resolution, 1.0-mm slice thickness, 128 × 128 pixels, 4.0 × 4.0 cm field of view [FOV], one average, 4:20-minute total acquisition time). Our acidoCEST MRI protocol consisted of 52 fast imaging with steady state precession (FISP) MR images acquired with selective saturation applied for 5 seconds at 2.8 μT saturation power at different saturation frequencies ranging from −10 to 10 ppm, which required 4:50 of acquisition time. The parameters used were a 90° flip angle, 3.218 milliseconds TR, 1.609 milliseconds TE, 60° excitation angle, with a 2-mm slice thickness, 250 × 250 mm in-plane resolution, 3.2 × 3.2 cm field of view, linear coding order, unbalanced free induction decay (FID) mode, and a 418.54 milliseconds scan time. This process was repeated 6 times, for a total acidoCEST MRI acquisition time of 29 minutes. At the conclusion of the imaging scan, the mouse was removed from the scanner and cradle and allowed to recover.
AcidoCEST MRI Image Analyses
The acidoCEST MR images were processed with MATLAB R2012B (Mathworks, Inc, Natick, Massachusetts). All 6 CEST-FISP images were averaged. To improve signal to noise ratio, groups of 3 × 3 adjacent pixels were binned. A CEST spectrum was generated, smoothed with cubic splines, and interpolated. The CEST spectrum was then fitted with a sum of 3 Lorentzian line shapes to account for the direct saturation of water and to measure the amplitudes of the CEST effects at 4.2 and 5.6 ppm. The pHe of each binned pixel with 2 CEST effects greater than × noise was determined using an empirical calibration based on identical imaging of chemical solutions. This empirical calibration can determine pH between 6.2 and 7.0 units with a precision of 0.07 pH units and has been shown to have an in vivo accuracy of 0.034 pH units. These pixels were represented as colored pixels in the pHe map. Pixels with only a single CEST effect at 4.2 ppm greater than × noise were set to be pH 7.0. These pixels were represented as white pixels in the pHe map. Using a × noise threshold ensured that the CEST contrast was due to the agent with a 95% probability.26
Results
Everolimus Abrogates mTOR Activity in Grant 519 Cells
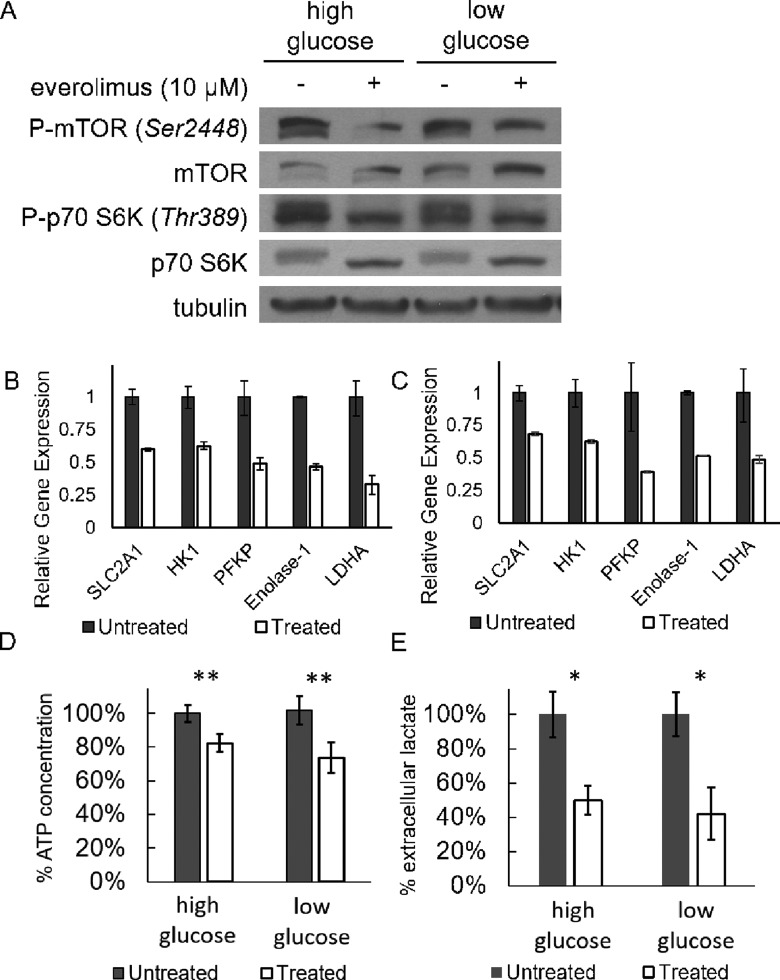
Constitutive activation of the phosphorylated-mTOR (P-mTOR) in the Granta 519 cell line has been previously observed and found to be due to a loss of active phosphatase and tensin homolog.7,8 To investigate whether everolimus treatment suppresses endogenous mTOR activity in this cell line, the Granta 519 cells were treated with 10 μmol/L everolimus for 24 hours in both low- (1 g/L) and high- (4.5 g/L) glucose conditions (Figure 1A). Low- and high-glucose concentrations were used to investigate whether target expression and subsequent modulation were influenced by culture conditions. Western blot analysis showed that the P-mTOR concentration decreased after everolimus treatment, with the high-glucose condition demonstrating a more marked decrease than the low-glucose condition. Similarly, the concentration of phosphorylated p70 S6 K decreased upon treatment with everolimus, whereas the total p70 S6 K concentration showed an increase in concentration. This result further substantiated that everolimus inhibited the activation of mTOR, so that mTOR was unable to phosphorylate p70 S6 K.
Figure 1.
The effect of 10 μmol/L everolimus treatment on tumor cell metabolism after 24 hours of treatment. A, Everolimus caused a decrease in phosphorylation of mammalian target of rapamycin (mTOR) and p70 S6 kinase in Granta 519 cells as evaluated with a Western blot. B, The differences in gene expression levels between the everolimus-treated and control groups in a low-glucose (1 g/L) media. C, The relative differences in gene expression levels in a high-glucose (4.5 g/L) media. Vehicle control data were normalized to 1. D, Intracellular ATP concentration (normalized to untreated groups) decreased with everolimus treatment in both high- and low-glucose conditions. E, Lactate normalized to total protein (normalized to control groups) decreased by approximately 50% in both high- and low-glucose conditions when treated with everolimus. *P < 0.05; **P < 0.0001.
Everolimus Decreases Expression of Genes That Regulate Glycolytic Proteins and Enzymes
In a previous study in pancreatic cancer cells, everolimus was shown to downregulate hexokinase (HK) 2 gene expression.36 We observed a significant decrease in gene expression of glucose transporter 1 (SLC2A1), HK 1, phosphofructose kinase 1, enolase 1, and lactate dehydrogenase A in cells cultured both in low-glucose and high-glucose conditions (Figure 1B and C). There was however no change in the expression of the lactate transport proteins MCT-1 or MCT-4 (data not shown). The downregulation of these key proteins and enzymes that participate in glycolytic metabolism supports the role of everolimus in the suppression of metabolism and the downstream product of metabolism, lactic acid.
Everolimus Decreases Intracellular ATP Production and Extracellular Lactate Accumulation
The effects of everolimus on the metabolism of the tumor cell were evaluated by assessing ATP and extracellular lactate during the in vitro studies (Figures 1D and E). Both the intracellular ATP concentration and the extracellular lactate concentration significantly decreased during treatment under conditions of high and low glucose. There was no additional decrease in lactate production when cells were treated with higher doses (data not shown). These results suggested that the inhibition of mTOR with everolimus slowed glycolysis and may have also slowed aerobic respiration. This general slowing of metabolism may be due to slower ribosomal biogenesis, which is known to be a consequence of mTOR inhibition.37
Everolimus Has Minimal Effect on Apoptosis In Vitro After 24 hours of Treatment
Despite evidence that everolimus affects a specific metabolic pathway and the general metabolism of the tumor cells, everolimus did not induce apoptosis in vitro (Figure 2). Western blot analyses showed that treatment with everolimus caused no change in cleaved caspase 3 levels within cells. No significant apoptosis or necrosis was observed by flow cytometry, as evidenced by low levels of annexin-V and PI. These results are consistent with observations from others that inhibition of the PI3K/AKT pathway in the Granta 519 cell line requires up to 48 hours of treatment before markers of apoptosis are observed.38
Figure 2.
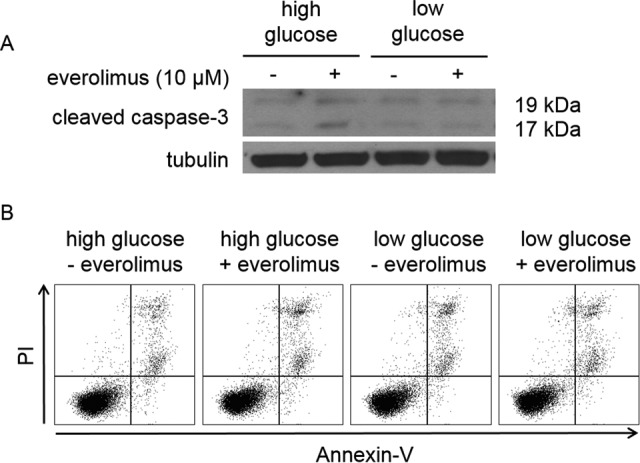
Effect of 10 μmol/L everolimus treatment on apoptosis following 24 hours of treatment. A, Everolimus did not induce a large cleaved caspase 3 response in Granta 519 cells. B, Everolimus caused minimal apoptosis or necrosis while modulating metabolism.
Everolimus Causes In Vivo Growth Inhibition
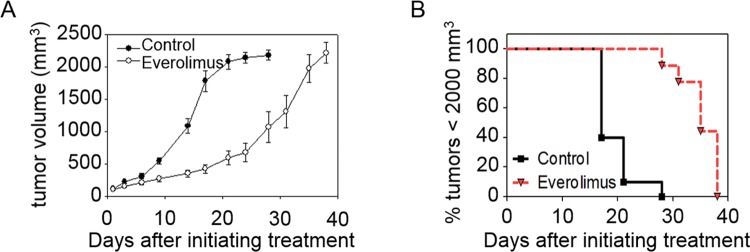
The in vivo Granta 519 flank xenograft model was treated with daily administration of everolimus. Initial daily administration of the drug did not significantly affect tumor growth for the first 7 days of treatment (Figure 3A). However, the daily treatment caused a latent tumor growth that resulted in a statistically significant difference in the average tumor volume by day 9 (P < 0.001). This difference in tumor growth was used to estimate the tumor cell doubling rates. The natural log of tumor volumes up to 1000 mm3 in each group were plotted and fit with a straight line. The slope of the DMSO group (Mc) was 0.1658 with R 2 = 0.9781, which was roughly twice the slope of the everolimus-treated group (Mt) = 0.077 with R 2 = 0.9812. Therefore, the control group grew in volume twice as fast as the treated group. Additionally, the tumor growth delay (T/C) was calculated by a ratio of the volumes at day 21. The division of cancer treatment considers a T/C < 44% as significant, and our T/C rate was calculated to be 29%.39 Tumor volumes reached 2000 mm3 in an average of 28 days without treatment, whereas everolimus treatment increased the time to reach 2000 mm3 tumor volume to an average of 40 days. Treatment with everolimus resulted in a statistically significant increase in time to reach a tumor size of 2000 mm3 (P < 0.0001; Figure 3B).
Figure 3.
Effect of 5 mg/kg/d everolimus treatment on tumor growth. A, Everolimus-treated mice showed a statistically slower tumor growth rate than DMSO control-treated counterparts after 9 days of treatment. B, The time to reach a tumor volume ≤2000 mm3 increased with everolimus treatment.
Serial Assessment of pHe Using AcidoCEST MRI in Mice Treated With Everolimus
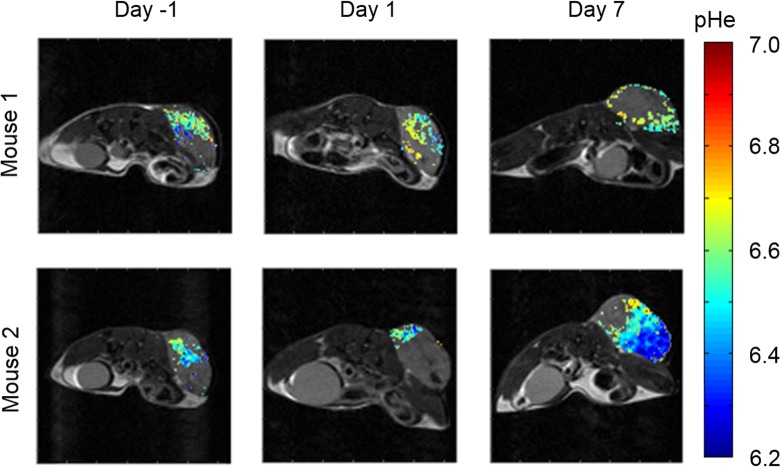
To assess the effects of everolimus on the metabolism of the Granta 519 xenografts, we used acidoCEST MRI to longitudinally evaluate tumor pHe of this lymphoma model (Figure 4). The mice experienced no distress during this noninvasive imaging procedure and showed no behavioral symptoms after each imaging session. AcidoCEST MRI generated pixel-wise maps of tumor pHe, which were then used to determine the average pHe of the measured tumor region. Before treatment, the average pHe was 6.82 ± 0.06 (Figure 5A), which matched previous in vivo pHe assessments of this tumor model.40 One day after initiating the administration of everolimus, the average tumor pHe significantly increased to pHe 6.92 ± 0.07 (P < 0.02 from day −1 to 1). By 7 days of treatment, the tumor had returned to an acidic state, with an average pHe of 6.70 ± 0.11 (P < 0.001 from day 1 to 7). There was no statistical significant difference in average tumor pHe from day −1 to 7. For comparison, our previous studies showed that the Granta 519 xenograft tumor model consistently decreased tumor pHe during a 2-week time period.41 Therefore, these imaging studies supported the assertion that everolimus initially caused a decrease in general metabolism, which decreased extracellular production of lactic acid, and this decrease in general metabolism was temporary.
Figure 4.
AcidoCEST magnetic resonance imaging (MRI) produced parametric maps of the extracellular pH (pHe) in a Granta 519 lymphoma flank xenograft model treated daily with everolimus at 5 mg/kg/d starting at day 0.
Figure 5.
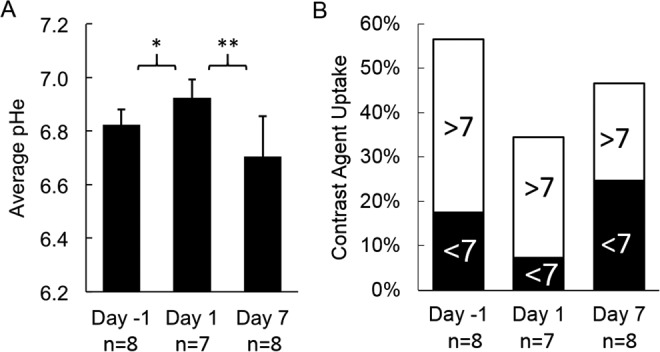
Everolimus treatment results in time-dependent modulation of the extracellular pH (pHe) in lymphoma microenvironment. A, Statistically significant changes in pHe values were observed with acidoCEST magnetic resonance imaging (MRI) after 1 day of treatment. The pHe returned to a more acidic state after 7 days of treatment. B, The fraction of the tumor volume with pHe < 7 decreased 1 day after initiating therapy, which then increased by day 7 after initiating therapy. *P < 0.02; **P < 0.001.
The pixel-wise maps generated by acidoCEST MRI only showed a pHe value if a tumor region had sufficiently high uptake of the imaging agent to generate statistically significant CEST signals (Figure 4). Therefore, acidoCEST MRI also provided a qualitative assessment of “percentage contrast agent uptake” in the entire tumor region studied with acidoCEST MRI (Figure 5B). The average percentage uptake was assessed for tumor regions that showed values above and below pHe 7.0. This assessment showed a substantial decrease in percentage uptake for the acidic portion of the tumor after 1 day of treatment and a substantial increase in percentage uptake for the acidic tumor portion after 7 days of treatment. Therefore, the percentage uptake measurement paralleled the measurements of tumor pHe following treatments with everolimus.
Discussion
We investigated the effects of everolimus using acidoCEST MRI. Everolimus has been investigated in MCL in previous literature, which provided a basis for this study. This therapeutic agent was chosen based on its potential to directly interfere with metabolism and result in changes in acidosis. We observed an early increase in pHe in the lymphoma microenvironment following treatment with everolimus. This statistically significant increase in pHe due to everolimus treatment is different than the behavior of an untreated control group from our previous study, which showed a consistent decrease in tumor pHe in the Granta 519 xenograft model. This reflects the decrease in the production of lactate due to cellular metabolism inhibition, as validated by our initial in vitro investigations of the levels of lactic acid, ATP, and the expression of key proteins that drive metabolism that are affected by everolimus treatment.
Our results showed that the Granta 519 xenograft model decreased tumor pHe by 7 days of treatment. This increased acidosis may reflect metabolic adaptation due to negative feedback mechanisms that contribute to resistance to mTORC1-directed therapies.42,43 Although everolimus significantly delayed tumor growth in the Granta 519 model, the treated tumors eventually grew to maximally allowable tumor volumes. Our results suggest that the inability to sustain a suppression of tumor acidosis may foreshadow drug resistance. This observation shows that acidoCEST MRI can be used for noninvasive longitudinal monitoring of early response to treatment by monitoring changes in cellular metabolism.
Conventionally, lymphoma response is assessed by changes in tumor volume on computed tomography scans. However, many targeted agents cause tumor stasis rather than tumor regression, which can limit the value of using only tumor volume measurements to monitor response. Lymphoma response is also commonly assessed by glucose avidity with 2-deoxy-2-[18F]-fluoro-D-glucose positron emission tomography (FDG PET). However, FDG PET may not reflect the metabolism of tumors that rely on metabolic pathways other than glycolysis, such as glutaminolysis. Also, FDG PET has a 20% to 30% false-positive rate due to background inflammation, and attempts to change therapy based on interim FDG PET imaging have been mixed.43 A previous imaging study used 13C MR spectroscopy to monitor everolimus treatment in preclinical models of glioblastoma and lymphoma.44,45 Yet 13C MRS fails to provide spatially encoded information in the same capacity that is provided by MRI.
We have previously shown that acidoCEST MRI can measure an increase in tumor pHe in a xenograft model after administering an alkalinizing treatment, 200 mmol/L sodium bicarbonate provided in drinking water. We have also used acidoCEST MRI to measure a decrease in tumor pHe in a xenograft model after administering metaiodobenzylguanidine (MIBG), a metabolic poison that potentiates lactic acid production. Our current study adds to the evidence that acidoCEST MRI can detect changes in tumor pHe by measuring an increase in pHe in the Granta 519 xenograft model. These 2 previous studies measured large changes in tumor pHe (a change of 0.65 pH units after administering bicarbonate and 0.35 pH units after administering MIBG). For comparison, our current study measured a small increase in tumor pHe of 0.10 pH unit within 1 day of initiating everolimus treatment. Despite the small magnitude, this change was statistically significant, which demonstrates the value of our highly precise acidoCEST MRI method. Based on the successful outcome of this study, acidoCEST MRI is a promising method for evaluating the early response to other therapies that affect tumor metabolism during preclinical studies and may also provide impact for assessments of patients undergoing therapy when acidoCEST MRI is translated to the radiology clinic. Second-generation mTOR inhibitors that target mTORC2 and mTORC1 are in development and have demonstrated promising activity in preclinical studies of MCL.46,47
One limitation of our study is the incomplete uptake of the contrast agent into the entire extracellular microenvironment of the tumor. Many solid tumors show high uptake of many types of imaging contrast agents because tumor angiogenesis typically generates highly permeable blood vessels. Yet some regions of the tumor may have relatively poor vascular perfusion, such as the tumor core of a xenograft tumor, which limits delivery of the contrast agent to these tumor regions. This problem is exacerbated by the low detection sensitivity of CEST MRI relative to other MRI methods. Our results showed that our contrast agent accumulated to sufficient concentrations for pHe measurements in an average of 55%, 35%, and 45% of the tumor region on days −1, 1, and 7 (Figure 5B). Therefore, our measurements of average tumor pHe are biased toward tumor regions with high vascular perfusion and permeability. Yet the combination of a rise in pHe with lower uptake of contrast agent may provide a multiparametric assessment of chemotherapeutic effects that can further improve the monitoring of drug effects in patients with lymphoma. A second limitation of the study is the number of acidoCEST MRI scans each mouse is capable of withstanding during each project. Most animals can be imaged for 3 acidoCEST MRI imaging sessions until tail vein catheterizations reduce the tissue integrity of the tail, limiting the number of sessions. Secondly, per our IACUC animal protocol, animals are not allowed to be imaged more than once in a 48-hour time period. Timing of scans during/after treatment is an optimization parameter that must be investigated per each project, warranting future studies.
Acknowledgments
The authors thank the University of Arizona Cancer Center Experimental Mouse Shared Resource, the University of Arizona Genomics Core, and the Flow Cytometry Shared Resource for assistance with conduct of these studies.
Footnotes
Authors’ Note: P.J.A. and A.F.B. designed the study. N.T.H. and A.G. performed in vitro studies. P.J.A. and C.M.H. performed the in vivo imaging studies. P.J.A., D.O.P., M.D.P., and A.F.B. prepared and refined the manuscript. All coauthors reviewed and approved the manuscript.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by the 1PO-CA130805, 1RO1CA167183, 5P30CA023074, and T32CA009213-36. P.J.A. was supported through the Anne Rita Monahan Foundation and T32CA009213-36 Cancer Biology Training Grant.
References
- 1. Gatenby RA, Gawlinski ET, Gmitro AF, Kaylor B, Gillies RJ. Acid-mediated tumor invasion: a multidisciplinary study. Cancer Res. 2006;66(10):5216–5223. [DOI] [PubMed] [Google Scholar]
- 2. Estrella V, Chen T, Lloyd M, et al. Acidity generated by the tumor microenvironment drives local invasion. Cancer Res. 2013;73(5):1524–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2005;134(5):703–707. [DOI] [PubMed] [Google Scholar]
- 4. Ashby BS, Cantab MB. pH studies in human malignant tumours. Lancet. 1996;2(7458):312–315. [DOI] [PubMed] [Google Scholar]
- 5. Prescott DM, Charles HC, Poulson JM, et al. The relationship between intracellular and extracellular pH in spontaneous canine tumors. Clin Cancer Res. 2000;6(6):2501–2505. [PubMed] [Google Scholar]
- 6. Wike-Hooley JL, van den Berg AP, van der Zee J, Reinhold HS. Human tumour pH and its variation. Eur J Cancer Clin Oncol. 1985;21(7):785–791. [DOI] [PubMed] [Google Scholar]
- 7. Reshetnyak YK, Segala M, Andreev OA, Engelman DM. A monomeric membrane peptide that lives in three worlds: in solution, attached to, and inserted across lipid bilayers. Biophys J. 2007;93(7):2363–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vavere AL, Biddlecombe GB, Spees WM, et al. A novel technology for the imaging of acidic prostate tumors by positron emission tomography. Cancer Res. 2009;69(10):4510–4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weerakkody D, Moshnikova A, Thakur MS, et al. Family of pH (low) insertion peptides for tumor targeting. Proc Natl Acad Sci U S A. 2013;110(15):5834–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Andreev OA, Dupuy AD, Segala M, et al. Mechanism and uses of a membrane peptide that targets tumors and other acidic tissues in vivo. Proc Natl Acad Sci U S A. 2007;104(19):7893–7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li C, Xia J, Wei X, Yan HH, Si Z, Ju S. PH-Activated near-infrared fluorescence nanoprobe imaging tumors by sensing the acidic microenvironment. Adv Funct Mater. 2010;20(14):2222–2230. [Google Scholar]
- 12. Dellian M, Helmlinger G, Yuan F, Jain RK. Fluorescence ratio imaging of interstitial pH in solid tumours: effect of glucose on spatial and temporal gradients. Br J Cancer. 1996;74(8):1206–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martin GR, Jain RK. Noninvasive measurement of interstitial pH profiles in normal and neoplastic tissue using fluorescence ratio imaging microscopy. Cancer Res. 1994;54(21):5670–5674. [PubMed] [Google Scholar]
- 14. Gillies RJ, Liu Z, Bhujwalla Z. 31P-MRS measurements of extracellular pH of tumors using 3-aminopropylphosphonate. Am J Physiol. 1994;267(1 pt 1):C195–C203. [DOI] [PubMed] [Google Scholar]
- 15. Aoki Y, Akagi K, Tanaka Y, Kawai J, Takahashi M. Measurement of intratumor pH by pH indicator used in 19Fmagnetic resonance spectroscopy: measurement of extracellular pH decrease caused by hyperthermia combined with hydralazine. Invest Radiol. 1996;31(11):680–689. [DOI] [PubMed] [Google Scholar]
- 16. Bhujwalla ZM, McCoy CL, Glickson JD, Gillies RJ, Stubbs M. Estimations of intra- and extracellular volume and pH by 31P magnetic resonance spectroscopy: effect of therapy on RIF-1 tumours. Br J Cancer. 1998;78(5):606–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lutz NW, Fur YW, Chiche J, Pouysse J, Cozzone PJ. Quantitative in vivo characterization of intracellular and extracellular pH profiles in heterogeneous tumors: a novel method enabling multiparametric pH analysis. Cancer Res. 2013;73(15):4616–4628. [DOI] [PubMed] [Google Scholar]
- 18. Zhou R, Bansal N, Leeper DB, Glickson JD. Intracellular acidification of human melanoma xenografts by the respiratory inhibitor m-iodobenzylguanidine plus hyperglycemia: a 31P magnetic resonance spectroscopy study. Cancer Res. 2000;60(13):3532–3536. [PubMed] [Google Scholar]
- 19. García-Martín ML, H, érigualt G, Rémy C, et al. Mapping extracellular pH in rat brain gliomas in vivo by 1 H magnetic resonance spectroscopic imaging: comparison with maps of metabolites. Cancer Res. 2001;61(17):6524–6531. [PubMed] [Google Scholar]
- 20. Gillies RJ, Morse DL. In vivo magnetic resonance spectroscopy in cancer. Annu Rev Biomed Eng. 2005;7:287–326. [DOI] [PubMed] [Google Scholar]
- 21. Vermathen P, Capizzano AA, Maudsley A. Administration and (1)H MRS detection of histidine in human brain: application to in vivo pH measurement. Magn Reson Med. 2000;43(5):665–675. [DOI] [PubMed] [Google Scholar]
- 22. Garcia-Martin ML, Martinez GV, Raghunand N, Sherry AD, Zhang S, Gillies RJ. High resolution pHe imaging of rat glioma using pH-dependent relaxivity. Magn Reson Med. 2006;55(2):309–315. [DOI] [PubMed] [Google Scholar]
- 23. Raghunand N, Howison C, Sherry AD, Zhang S, Gillies RJ. Renal and systemic pH imaging by contrast-enhanced MRI. Magn Reson Med. 2003;49(2)249–257. [DOI] [PubMed] [Google Scholar]
- 24. Raghunand N, Zhang S, Sherry AD, Gillies RJ. In vivo magnetic resonance imaging of tissue pH using a novel pH-sensitive contrast agent, GdDOTA-4AmP. Acad Radiol. 2002;9(2):S481–S483. [DOI] [PubMed] [Google Scholar]
- 25. Martinez GV, Zhang X, García-Martín ML, et al. Imaging the extracellular pH of tumors by MRI after injection of a single cocktail of T1 and T2 contrast agents. NMR Biomed. 2011;24(10):1380–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen LQ, Howison C, Jeffery J, Robey IF, Kuo PH, Pagel MD. Evaluations of extracellular pH within in vivo tumors using acidoCEST MRI. Magn Reson Med. 2014;72(5):1408–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moon B, Jones K, Chen LQ, et al. A comparison of iopromide and iopamidol, two acidoCEST MRI contrast media that measure tumor extracellular pH. Contrast Media Mol Imaging. 2015;10(6):446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Longo D, Busato A Lanzardo S, Antico F, Aime S. Imaging the pH evolution of an acute kidney injury model by means of iopamidol, a MRI-CEST pH-responsive contrast agent. Magn Reson Med. 2013;70(3):859–864. [DOI] [PubMed] [Google Scholar]
- 29. Sheth VR, Li Y, Chen LQ, Howison CM, Flask CA, Pagel MD. Measuring in vivo tumor pHe with CEST-FISP MRI. Magn Reson Med. 2012;67(3):760–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen LQ, Randtke EA, Jones KM, Moon BF, Howison CM, Pagel MD. Evaluations of tumor acidosis within in vivo tumor models using parametric maps generated with AcidoCEST MRI. Mol Imaging Biol. 2015;17(4):488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jones KM, Randtke EA, Howison CM, et al. Measuring extracellular ph in a lung fibrosis model with acidoCEST MRI. Mol Imaging Biol. 2014;17(2):177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mussetti A, Kumar A, Dahi PB, Perales MA, Sauter CS. Lifting the mantle: unveiling new treatment approaches in relapsed or refractory Mantle cell lymphoma. Blood Rev. 2015;29(3):143–152. [DOI] [PubMed] [Google Scholar]
- 33. Witzig TE, Reeder CB, LaPlant BR, et al. A phase II trial of the oral mTOR inhibitor everolimus in relapsed aggressive lymphoma. Leukemia. 2011;25(2):341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Renner C, Zinzani PL, Gressin R, et al. A multicenter phase II trial (SAKK 36/06) of single-agent everolimus (RAD001) in patients with relapsed or refractory Mantle cell lymphoma. Haematologica. 2012;97(7):1085–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- 36. Liu L, Gong L, Zhang Y, Li N. Glycolysis in Panc-1 human pancreatic cancer cells is inhibited by everolimus. Exp Ther Med. 2013;5(1):338–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jastrzebski K, Katarzyna K, Tchoubrieva E, Hannan RD, Pearson RB. Coordinate regulation of ribosome biogenesis and function by the ribosomal protein S6 kinase, a key mediator of mTOR function. Growth Factors. 2007;25(4):209–226. [DOI] [PubMed] [Google Scholar]
- 38. Rudelius M, Pittaluga S, Nishizuka S, et al. Constitutive activation of Akt contributes to the pathogenesis and survival of Mantle cell lymphoma. Blood. 2006;108(5):1668–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Corbett TH, Leopold WR, Dykes DJ, Roberts BJ, Griswold DP, Jr, Schabel FM., Jr Toxicity and anticancer activity of a new triazine antifolate (NSC 127755). Cancer Res. 1982;42(5):1707–1715. [PubMed] [Google Scholar]
- 40. Chen LQ, Howison CM, Spier C, et al. Assessment of carbonic anhydrase IX expression and extracellular pH in B-cell lymphoma cell line models. Leuk Lymphoma. 2014;56(5):1432–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. El-Galaly TC, Hutchings M, Mylam KJ, et al. Impact of 18F-fluorodeoxyglucose positron emission tomography/computed tomography staging in newly diagnosed classical Hodgkin lymphoma: fewer cases with stage I disease and more with skeletal involvement. Leuk Lymphoma. 2014;55(10):2349–2355. [DOI] [PubMed] [Google Scholar]
- 42. Eyre TA, Collins GP, Goldstone AH, Cwynarski K. Time now to TORC the TORC? New developments in mTOR pathway inhibition in lymphoid malignancies. Br J Haematol. 2014;166(3):336–351. [DOI] [PubMed] [Google Scholar]
- 43. Rosich L, Montraveta A, Xargay-Torrent S, et al. Dual PI3K/mTOR inhibition is required to effectively impair microenvironment survival signals in Mantle cell lymphoma. Oncotarget. 2014;5(16):6788–6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee SC, Marzec M, Liu X, et al. Decreased lactate concentration and glycolytic enzyme expression reflect inhibition of mTOR signal transduction pathway in B-cell lymphoma. NMR Biomed. 2013;26(1):106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Venkatesh HS, Chaumeil MM, Ward CS, Haas-Kogan DA, James CD, Ronen SM. Reduced phosphocholine and hyperpolarized lactate provide magnetic resonance biomarkers of PI3K/Akt/mTOR inhibition in glioblastoma. Neuro Oncol. 2012;14(3):315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rosich L, Colomer D, Roue G. Autophagy controls everolimus (RAD001) activity in Mantle cell lymphoma. Autophagy. 2013;9(1):115–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rosich L, Xargay-Torrent S, Lopez-Guerra M, Campo E, Colomer D, Roué G. Counteracting autophagy overcomes resistance to everolimus in Mantle cell lymphoma. Clin Cancer Res. 2013;1(8):5278–5289. [DOI] [PubMed] [Google Scholar]