Abstract
The risk of type 2 diabetes (T2D) is increased by abnormalities in sleep quantity and quality, circadian alignment, and melatonin regulation. A common genetic variant in a receptor for the circadian-regulated hormone melatonin (MTNR1B) is associated with increased fasting blood glucose and risk of T2D, but whether sleep or circadian disruption mediates this risk is unknown. We aimed to test if MTNR1B diabetes risk variant rs10830963 associates with measures of sleep or circadian physiology in intensive in-laboratory protocols (n = 58–96) or cross-sectional studies with sleep quantity and quality and timing measures from self-report (n = 4,307–10,332), actigraphy (n = 1,513), or polysomnography (n = 3,021). In the in-laboratory studies, we found a significant association with a substantially longer duration of elevated melatonin levels (41 min) and delayed circadian phase of dim-light melatonin offset (1.37 h), partially mediated through delayed offset of melatonin synthesis. Furthermore, increased T2D risk in MTNR1B risk allele carriers was more pronounced in early risers versus late risers as determined by 7 days of actigraphy. Our results provide the surprising insight that the MTNR1B risk allele influences dynamics of melatonin secretion, generating a novel hypothesis that the MTNR1B risk allele may extend the duration of endogenous melatonin production later into the morning and that early waking may magnify the diabetes risk conferred by the risk allele.
Introduction
Increased risk of type 2 diabetes (T2D) is associated with abnormalities in sleep quantity (1) and quality (1,2), circadian alignment (3,4), and melatonin regulation (5,6). A common variation at MTNR1B (7,8) was identified by genome-wide association studies to associate with diabetes traits. The mechanism whereby these variants lead to elevated T2D risk is unknown.
MTNR1B is one of two high-affinity receptors for the pineal hormone melatonin, which is released exclusively at night and plays a role in glucose homeostasis (6,9). In humans, the release of melatonin occurs concurrent with overnight fasting during sleep. Elevated melatonin levels during an oral glucose load during the day causes impaired glucose tolerance (6,10). Common variants in MTNR1B associate with increased risk of T2D, fasting blood glucose (FBG) levels (7,8), and lower glucose-stimulated insulin secretion in individuals without diabetes (11). Functional studies have established that MTNR1B rs10830963 is the likely causal variant (12). Strong independent association of rare, loss-of-function variants in MTNR1B with increased risk of T2D further implicates MTNR1B as the most likely causal gene in the region (13).
Although MTNR1B plays a role in glucose homeostasis (reviewed in refs. 14 and 15), it is currently unknown how MTNR1B rs10830963 may alter its normal role in glucose metabolism. In this study, we explored the hypothesis that the association of rs10830963 with T2D may be mediated via effects on melatonin endocrinology, sleep timing/physiology, and/or the circadian system. Understanding these intermediate trait associations may lead to further insights into mechanisms by which risk variants influence glycemic traits and point toward new avenues of therapeutic intervention. We tested association of rs10830963 with sleep, circadian, and melatonin traits in two study populations with complementary strengths: 1) intensive in-laboratory protocols (n = 58–96) with participants assessed for precise measures of circadian physiology, and 2) cross-sectional studies (Candidate-gene Association Resource [CARe]) with sleep quantity, quality, and timing measures from questionnaires (n = 4,307–10,332), actigraphy (n = 1,513), and overnight polysomnography (PSG) (n = 3,021).
Research Design and Methods
In-Laboratory Studies
Study Participants
Participants included 193 healthy individuals (a subset of 58–96 for whom measures of circadian physiology were available) from completed research studies in the Intensive Physiologic Monitoring Unit, Center for Clinical Investigation, Brigham and Women’s Hospital from 2001–2011, as previously described, who donated a blood sample for genetic analysis (16). To promote a stable circadian rhythm, all participants maintained an 8-h sleep schedule of their choice at home for 1–3 weeks prior to admission. Compliance was verified with a sleep diary, call-ins, and wrist actigraphy. Participants also completed a morningness-eveningness questionnaire (MEQ) (17). The genetic sample collection and analyses were approved by the Partners HealthCare Human Research Committee. Separate informed consent was obtained for enrollment in the genetic studies.
Circadian Phenotypes
Of the 193 participants studied in the laboratory, a subset of 58–96 was assessed in intensive protocols with precise measures of endogenous circadian physiology. Measures of circadian rhythm timing (phase), magnitude (amplitude), length (period), and melatonin physiology were measured in the in-laboratory samples using hourly plasma melatonin concentrations and core body temperature (CBT) (1-min epochs) collected over a minimum of 24 h (Supplementary Table 1). We used baseline data from individual studies in which subjects had undergone either a constant routine or posture protocol (n = 96) (18–20) or a forced desynchrony protocol (n = 63) (21–23) (Supplementary Fig. 1A–C). Melatonin phase measures collected included dim-light melatonin onset (DLMO) and dim-light melatonin offset (DLMOff) calculated as the time of the melatonin profile fitted curve at which levels crossed 25% of peak upward and downward, respectively; melatonin synthesis offset calculated from a linear model fitted to each melatonin profile (24); and the midpoint of melatonin calculated as the midpoint between DLMO and DLMOff [DLMO + (DLMOff − DLMO)/2]. Phase was also measured by CBT nadir, the time when the fitted circadian curve of CBT was at its minimum (18). To assess the difference between internal and external timing, phase angles were calculated as the time between sleep midpoint and DLMO, DLMOff, or CBT nadir. Circadian amplitude of melatonin and CBT were calculated as 50% of the difference between the minimum and peak of the fitted circadian curve. Measures of melatonin stability were calculated from a linear model fitted to each melatonin profile, generating plasma melatonin clearance rate and half-life. The duration of melatonin secretion was measured as the difference between DLMO and DLMOff. Sleep timing phenotypes included bedtime, wake time, midpoint of sleep, and sleep duration derived from 7 days of time-stamped call-ins during which subjects were required to maintain a self-selected but fixed 8-h sleep schedule prior to laboratory admission. Procedures for the determination of circadian phase, phase angle, amplitude, and period have been previously described (17,19,21,24).
Sample Genotyping
DNA was extracted from whole blood using standard methods (Qiagen). All samples were genotyped for rs10830963 and 58 African American and Hispanic ancestry informative markers to test and correct for population stratification. Genotyping was performed using the Sequenom platform (Broad Institute, Cambridge, MA). Quality control steps excluded samples with <60% call rate and single nucleotide polymorphisms (SNPs) with <90% call rate, departure from Hardy-Weinberg equilibrium (P < 10−7), or minor allele frequency <1%. In-laboratory samples acquired since the original genotyping effort (n = 9) and samples that failed quality control in the previous round (n = 49) were whole-genome amplified and re-genotyped for all SNPs.
Evaluation of Population Stratification
The smartpca feature in EIGENSTRAT software (25) was used to calculate principal components after merging with HapMap 3 CEU, YRI, ASW, and CHB populations, and outliers 4 SD from the mean of the CEU population in the first three principal components were removed. Concordance between self-reported “non-Hispanic white” ancestry and included samples of European ancestry was 90.6%.
Association Testing
Genetic association analyses were performed in PLINK (26), using an additive genetic model and adjusting for age, sex, and five significant principal components that capture ancestry information. The significance threshold was set at P = 0.05. No correction was performed for multiple phenotypes tested.
CARe Study
Study Participants
Briefly, participants in the CARe study included >40,000 multiethnic individuals from nine National Institute of Heart, Lung, and Blood Institute (NHLBI) cohorts with genotype and phenotype data, described by Musunuru et al. (27). We used data from up to 10,322 individuals of European ancestry from the Atherosclerosis Risk in Communities (ARIC) study (28), the Coronary Artery Risk Development in Young Adults (CARDIA) study (29), the Cardiovascular Health Study (CHS) (30), the Framingham Heart Study (FHS) (31), the Multi-Ethnic Study of Atherosclerosis (MESA) (32), and the ancillary Sleep Heart Health Study (SHHS) (33), selected based on the availability of data on genotyping, glycemic traits, sleep questionnaires, and PSG (n = 3,021). We also used data from the ancillary MESA Sleep Study conducted at visit 5 (n = 1,513), based on the availability of data on genotyping, wrist actigraphy (34), and glycemic traits.
Sleep Phenotypes
Self-reported sleep measures were assessed via questionnaires covering sleep behavior over the month leading up to the study in each parent cohort (32,33,35). Individual cohort questions (Supplementary Table 2) were harmonized across CARe cohorts into the following self-reported sleep phenotypes: weekday and weekend bedtime; wake time; midpoint of sleep; weekday, weekend, and weekly sleep duration; average sleep latency; and the binary questions of difficulty falling asleep, wake after sleep onset, early morning awakenings, frequent napping, and excessive daytime sleepiness.
PSG sleep measures were available in the SHHS cohorts (n = 3,021). PSG was conducted during an unattended overnight home session as previously described (36). Participants were fitted with sensors by a certified technician, and data were captured overnight. Sleep stages were scored using guidelines described by Kales and Rechtschaffen (37). Total sleep time and total time in bed were available from the FHS component of SHHS (n = 556), and percentage of sleep time in each stage was available for all three SHHS cohorts (FHS, CHS, and ARIC, n = 3,021). The percentage of sleep time in each stage was computed by dividing time in the sleep stage by the recorded sleep time.
The MESA Sleep Study protocol included 7-day actigraphy (Actiwatch Spectrum, Philips Respironics, Murrysville, PA) together with sleep diaries and questionnaires (n = 1,513). Actigraphy data during 30-s intervals were scored as sleep or wake by Actiware-Sleep v.5.59 analysis software. Subject bedtime, sleep midpoint, and wake time from weekday, weekend, and weekly averaged data were calculated from actigraphy using the sleep log as upper and lower bounds. Sleep duration was defined as the average duration of sleep between sleep onset (sleep start time) and morning wakening (sleep end time) while in bed after “lights off.”
T2D Phenotypes
Information on demographics, age, sex, and racial/ethnic group was obtained by questionnaire. Height, weight, and FBG levels were measured at visit 5. The use of diabetes medications was determined by questionnaire and from medication containers (32). T2D was defined as an FBG ≥7.0 mmol/L (126 mg/dL) or use of insulin/oral hypoglycemia medications.
Sample Genotyping
The ITMAT/Broad/CARE (IBC) array v2 genotype data included rs10830963 (27,38). SNPs were clustered into genotypes using Illumina BeadStudio software. Quality control filters for SNPs and samples were applied separately within each cohort using PLINK (26). SNPs were excluded for Hardy-Weinberg equilibrium P < 10−7 and call rates <95% and samples for individual call rates <90%, sex mismatch, and duplicate discordance. To control for relatedness, estimates of pairwise identity-by-descent were calculated, and individuals with values >0.125 were pruned from the sample.
Evaluation of Population Stratification
Self-reported ethnicity was verified by multidimensional scaling analysis of identity-by-state distances as implemented in PLINK, including HapMap panels as reference standards. SNPs in linkage disequilibrium (r2 >0.3) were pruned and EIGENSTRAT was used to compute 10 principal components on the subset of individuals passing quality control for use as covariates in the regression analyses (25).
Association Testing
Power calculations were performed using Quanto in independent subjects using the gene-only setting (39). Linear and logistic regression analyses were performed in PLINK adjusting for age, sex, BMI, and principal components of ancestry (26). A fixed-effects, inverse-variance meta-analysis was performed in METAL (40). For primary analysis, significance threshold was set at P = 0.05 (as only one hypothesis was tested). No correction was performed for the multiple phenotypes tested. For interaction analyses, the significance was set at P < 0.05, as only one hypothesis was tested. Interaction analysis adjusting for age, sex, and BMI was performed in PLINK (26). Interaction plots were generated in R using the effects package.
Results
Association With Later DLMOff and Longer Melatonin Duration in the In-Laboratory Studies
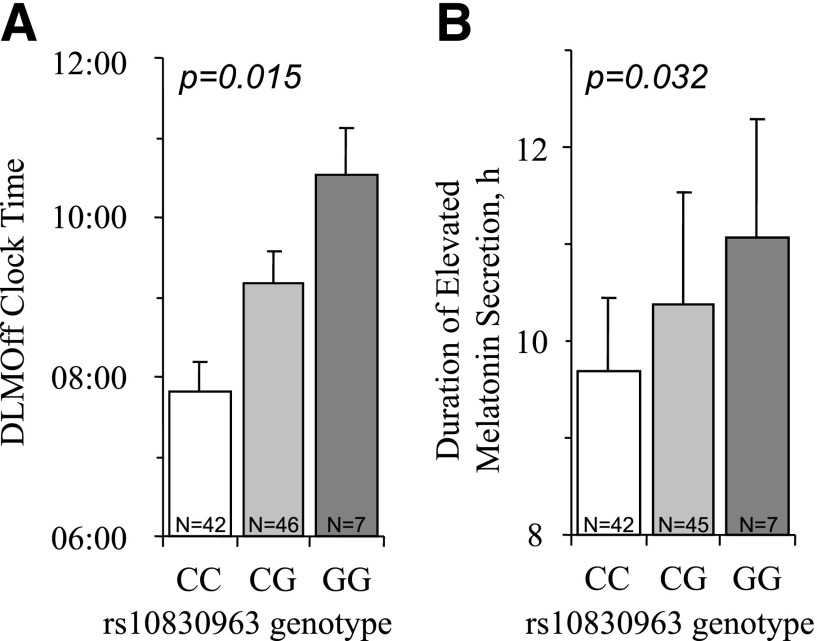
Descriptive characteristics of the laboratory study population are shown in Table 1. We tested rs10830963 for association with sleep and circadian traits (Table 2). In the in-laboratory studies, we found significant associations between MTNR1B diabetes risk variant rs10830963G and timing of the melatonin rhythm: an allelic dose-dependent delayed DLMOff by 1 h and 22 min (β = 1.36 h, 95% CI 0.28–2.44, N = 95, P = 0.015, r2 = 19%) and a longer duration of elevated melatonin levels by 41 min during constant routine protocols (defined as the difference between DLMO and DLMOff) (β = 41 min, 95% CI 4.2–78, N = 94, P = 0.032, r2 = 2.5%). This association is driven by the delay in DLMOff, as we did not see an association with DLMO (P = 0.236) (Table 2 and Fig. 1).
Table 1.
Cohort characteristics
| In-laboratory studies | |
|
N |
198 |
| Females, n (%)† |
70 (35) |
| Age, years† |
25.41 (9.58) |
| Owl/lark questionnaire, numeric score (n = 193)† |
52.18 (12.15) |
| Bedtime, clock time (n = 151)* |
23:49 (1.49) |
| Sleep midpoint, clock time (n = 151)* |
03:59 (1.41) |
| Wake time, clock time (n = 151)* |
08:10 (1.46) |
| Calculated sleep duration, h (n = 151)* |
8.04 (0.13) |
| Phase of DLMO, clock time (n = 96) |
10:31 (1.86) |
| Phase angle between melatonin onset and sleep midpoint, h (n = 93) |
5.58 (1.09) |
| Midpoint of melatonin secretion, clock time (n = 95) |
03:36 (1.82) |
| Phase of DLMOff, clock time (n = 95) |
08:38 (1.98) |
| Phase angle between melatonin offset and sleep midpoint, h (n = 93) |
4.53 (1.20) |
| Phase of melatonin synthesis offset, clock time (n = 82) |
6:31 (1.95) |
| Duration of melatonin secretion, h (n = 94) |
10.11 (1.08) |
| Plasma melatonin clearance rate, min−1 (n = 80) |
0.03 (0.02) |
| Plasma melatonin clearance half-life, min (n = 80) |
34.17 (20.57) |
| Circadian melatonin amplitude, pg/mL (n = 95) |
38.30 (23.02) |
| Circadian period of melatonin, h (n = 58) |
24.17 (0.19) |
| Phase of circadian CBT nadir, clock time (n = 90) |
04:59 (2.09) |
| Phase angle between CBT nadir and sleep midpoint, h (n = 88) |
0.77 (1.37) |
| Circadian CBT amplitude, °F (n = 89)† |
0.52 (0.15) |
| Circadian period of CBT, h (n = 64)† |
24.15 (0.20) |
| CARe study | |
|
N |
10,332 |
| Females, n (%) |
5,683 (55) |
| Age, years |
64.65 (12.49) |
| BMI, kg/m2 |
27.34 (5.04) |
| Self-report average weekly sleep duration, h (n = 8,380) |
7.24 (2.77) |
| Self-report average weekday sleep duration, h (n = 6,508) |
7.1 (1.15) |
| Self-report average weekend sleep duration, h (n = 4,517) |
7.48 (1.22) |
| Calculated average weekly sleep duration, h (n = 4,476)** |
7.48 (1.05) |
| Calculated average weekday sleep duration, h (n = 4,505)** |
7.39 (1.10) |
| Calculated average weekend sleep duration, h (n = 4,488)** |
7.72 (1.19) |
| Bedtime, weekday, clock time (n = 4,542) |
22:56 (1.05) |
| Bedtime, weekend, clock time (n = 4,528) |
23:11 (1.07) |
| Sleep midpoint, weekday, clock time (n = 4,511) |
02:32 (0.84) |
| Sleep midpoint, weekend, clock time (n = 4,502) |
02:55 (0.84) |
| Wake time, weekday, clock time (n = 4,527) |
06:19 (1.16) |
| Wake time, weekend, clock time (n = 4,530) |
06:54 (1.24) |
| Sleep latency, min (n = 4,495) |
16.61 (17.14) |
| Total sleep time, PSG measured, h (n = 556) |
6.39 (0.97) |
| Total time in bed, PSG measured, h (n = 556) |
7.50 (0.84) |
| REM sleep percent, PSG measured (n = 3,026) |
19.51 (6.64) |
| Stage 1 sleep percent, PSG measured (n = 3,026) |
5.26 (3.87) |
| Stage 2 sleep percent, PSG measured (n = 3,026) |
57.02 (13.13) |
| Stage 3/4 sleep percent, PSG measured (n = 3,026) |
18.22 (12.24) |
| Self-reported cases, n (%) |
|
| Frequent daytime sleepiness |
2,778 (26.97) |
| Frequent difficulty falling asleep |
3,455 (33.49) |
| Frequent wake after sleep onset |
5,760 (55.79) |
| Frequent early awakening |
3,747 (36.56) |
| Frequent naps |
2,415 (42.41) |
| MESA | |
|
N |
1,513 |
| Females, n (%) |
853 (56) |
| Age, years |
69.18 (9.21) |
| BMI, kg/m2 |
28.73 (5.63) |
| Objectively measured bedtime, clock time |
23:31 (1.40) |
| Objectively measured sleep midpoint, clock time |
03:07 (1.19) |
| Objectively measured wake time, clock time | 06:42 (1.38) |
Data are shown as mean (SD) or n (%).
*Measures were collected via call-ins during a 1-week schedule of 8 h of sleep prior to in-laboratory studies. All in-laboratory measures, except for those indicated with †, were from subjects on a study protocol with restricted 8-h time in bed.
**Sleep duration was calculated from self-reported bedtime and wake time. REM, rapid eye movement.
Table 2.
MTNR1B rs10830963 association with sleep, circadian, and melatonin traits in the in-laboratory studies
| N | β | SE | P | |
|---|---|---|---|---|
| Owl/lark questionnaire, numeric score |
193 |
−0.345 |
2.389 |
0.885 |
| Bedtime, clock time |
151 |
30.36 |
19.38 |
0.119 |
| Sleep midpoint, clock time |
151 |
31.8 |
19.38 |
0.103 |
| Wake time, clock time |
151 |
33.12 |
19.5 |
0.091 |
| Calculated sleep duration, h |
151 |
0.046 |
0.033 |
0.165 |
| Phase of DLMO, clock time |
96 |
0.648 |
0.542 |
0.236 |
| Phase angle between melatonin onset and sleep midpoint, h |
93 |
0.165 |
0.338 |
0.627 |
| Midpoint of melatonin secretion, clock time |
95 |
1.014 |
0.514 |
0.052 |
| Phase of DLMOff, clock time |
95 |
1.361 |
0.55 |
0.015 |
| Phase angle between melatonin offset and sleep midpoint, h |
93 |
0.45 |
0.382 |
0.242 |
| Phase of melatonin synthesis offset, clock time |
82 |
1.047 |
0.624 |
0.097 |
| Duration of melatonin secretion, h |
94 |
0.684 |
0.314 |
0.032 |
| Plasma melatonin clearance rate, min−1 |
80 |
−0.006 |
0.007 |
0.382 |
| Plasma melatonin clearance half-life, min |
80 |
4.753 |
7.048 |
0.502 |
| Circadian melatonin amplitude, pg/mL |
95 |
0.058 |
7.258 |
0.994 |
| Circadian period of melatonin, h |
57 |
0.01 |
0.067 |
0.885 |
| Phase of circadian CBT nadir, clock time |
90 |
0.627 |
0.575 |
0.279 |
| Phase angle between CBT nadir and sleep midpoint, h |
88 |
−0.242 |
0.413 |
0.559 |
| Circadian CBT amplitude, °F |
89 |
−0.027 |
0.042 |
0.521 |
| Circadian period of CBT, h | 63 | 0.003 | 0.069 | 0.964 |
Results are from linear regression analysis in whites adjusted for age, sex, and five principal components of ancestry. Significant results are shown in boldface type, no correction was applied for multiple phenotypes. Allele frequency of rs10830963 in the in-laboratory studies was 0.32.
Figure 1.
Circadian phase of DLMOff and duration of elevated melatonin levels vary by MTNR1B genotype in the in-laboratory cohort. Adjusted mean and standard error shown by rs10830963 genotype (T2D risk allele G). P value derived from multiple linear regression tests between genotype and phenotype adjusted for age, sex, and principal components of ancestry. A: Circadian phase of DLMOff (n = 95, adjusted mean [SE] in clock time, CC 07:49 [23 min], CG 09:10 [24 min], and GG 10:32 [35 min]). B: Duration of melatonin production (n = 94, adjusted means [SE] in clock time, CC 9.70 h [0.74], CG 10.38 h [1.15], and GG 11.07 h [1.21]).
We then asked if melatonin synthesis offset accounts for the relationship between rs10830963 and delayed DLMOff. A suggestive association of the risk allele with delayed melatonin synthesis offset (β = 1.05 h, 95% CI −0.17 to −2.28, N = 82, P = 0.097) was observed, and after conditioning on melatonin synthesis offset, the effect of rs10830963 on DLMOff was halved (βconditional = 0.65 h, 95% CI −0.066 to 1.366, P = 0.079, PANOVA < 0.001), suggesting partial mediation. Additional adjustment for season of study had no effect (data not shown).
We tested if chronotype or sleep timing contributes to the association between rs10830963 and DLMOff. We found significant mediation by sleep timing (bedtime 82.3%, P = 0.05; midpoint 84.4%, P = 0.04; wake time 85.9%, P = 0.05; chronotype (MEQ) 47.7%, P = 0.13), although a small portion of the effect is independent of sleep timing. The relationship between rs10830963 and melatonin duration is not mediated by sleep duration (P = 0.24).
Association With Glycemic Traits and Modification of T2D Risk by Sleep Timing in the CARe Study
Descriptive characteristics of the CARe cohort are shown in Table 1. MTNR1B variant rs10830963 was significantly associated with T2D and FBG in the CARe study (T2D: odds ratio [OR] 1.08, 95% CI 1.01–1.16, N = 2,516 participants with diabetes/17,293 participants without diabetes, P = 0.01; FBG: β = 1.52 mmol/L, 95% CI 1.30–1.74, N = 17,252 participants without diabetes, P = 1.41 × 10−41).
No significant association was observed between MTNR1B rs10830963 and self-reported, 7-day actigraphy, or PSG measures of sleep timing, quality, or duration (Table 3). Notably, no comparable measures of melatonin secretion were available in the CARe cohorts, therefore our laboratory findings could not be evaluated in this study population.
Table 3.
MTNR1B rs10830963 association with sleep traits in CARe study
| rs10830963G |
|||||
|---|---|---|---|---|---|
| N | Effect/OR (95%CI) | SE | P | Minimum effect detectable† | |
| Bedtime, weekday, min |
4,359 |
−0.72 |
1.50 |
0.63 |
3.93 |
| Bedtime, weekend, min |
4,346 |
−0.84 |
1.56 |
0.60 |
4.2 |
| Sleep midpoint, weekday, min |
4,329 |
−0.78 |
1.20 |
0.51 |
3.3 |
| Sleep midpoint, weekend, min |
4,321 |
−0.66 |
1.20 |
0.58 |
3.3 |
| Wake time, weekday, min |
4,344 |
−1.08 |
1.62 |
0.51 |
4.5 |
| Wake time, weekend, min |
4,347 |
−0.78 |
1.80 |
0.66 |
4.8 |
| Self-report average weekly sleep duration, h |
6,406 |
0.01 |
0.03 |
0.62 |
0.135 |
| Self-report average weekday sleep duration, h |
6,321 |
0.02 |
0.02 |
0.45 |
0.065 |
| Self-report average weekend sleep duration, h |
4,333 |
0.01 |
0.03 |
0.69 |
0.08 |
| Calculated average weekly sleep duration, h |
4,295 |
0.00 |
0.03 |
0.99 |
0.07 |
| Calculated average weekday sleep duration, h |
4,323 |
−0.01 |
0.03 |
0.85 |
0.07 |
| Calculated average weekend sleep duration, h |
4,307 |
0.01 |
0.03 |
0.83 |
0.08 |
| REM sleep percent, PSG measured |
3,021 |
0.22 |
0.19 |
0.23 |
0.53 |
| Stage 1 sleep percent, PSG measured |
3,021 |
0.18 |
0.11 |
0.08 |
0.31 |
| Stage 2 sleep percent, PSG measured |
3,021 |
0.15 |
0.35 |
0.67 |
1.045 |
| Stage 3/4 sleep percent, PSG measured |
3,021 |
-0.55 |
0.32 |
0.08 |
0.97 |
| Sleep latency, min |
6,316 |
0.99 |
0.01 |
0.38 |
1.15 |
| Objectively measured bedtime, min |
1,513 |
−2.20 |
3.54 |
0.54 |
9.7 |
| Objectively measured sleep midpoint, min |
1,513 |
−1.92 |
0.75 |
0.53 |
8.1 |
| Objectively measured wake time, min |
1,513 |
−1.64 |
3.50 |
0.64 |
9.6 |
| Frequent difficulty falling asleep |
9,846 |
1.01 (0.94–1.07) |
0.88 |
1.052 |
|
| Frequent early awakening |
9,808 |
0.98 (0.92–1.05) |
0.66 |
1.051 |
|
| Frequent daytime sleepiness |
9,977 |
0.98 (0.91–1.05) |
0.65 |
1.05 |
|
| Frequent naps |
6,457 |
1.06 (0.98–1.15) |
0.15 |
1.0875 |
|
| Frequent wake after sleep onset | 9,855 | 0.97 (0.91–1.04) | 0.43 | 1.064 | |
Results are from linear or logistic regression analyses adjusting for age, sex, BMI, and ancestry. Suggestive results are shown in boldface type. Allele frequency was 0.27.
†Minimum detectable effect at 80% power, α = 0.05. REM, rapid eye movement.
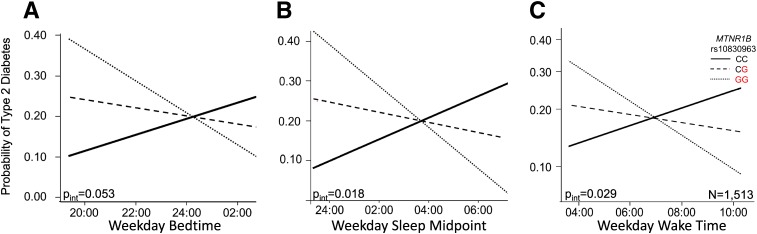
However, given that sleep timing under a controlled sleep duration schedule largely mediated the association with DLMOff, we tested if rs10830963 association with T2D is modulated by sleep timing in the CARe study. If true, this would be consistent with the hypothesis that risk allele carriers with earlier wake times would be more likely to have elevated melatonin levels than noncarriers at times of T2D diagnostic testing and morning meal consumption, and this difference between genotypes would be less apparent in participants with later wake times. Objectively measured sleep timing (7-day actigraphy) significantly modified the effect of rs10830963 on T2D risk, such that earlier sleep timing in combination with the G allele carries an increased risk compared with later sleep timing (N = 1,513, bedtime Pinteraction = 0.053, sleep midpoint Pint = 0.0176, wake time Pinteraction = 0.024) (Table 4 and Fig. 2). In analyses of participants of European descent stratified by median bedtime, midpoint, and wake time, a significant association between rs10830963 genotype and T2D was seen in early sleep timing (bedtime <23:12, N = 310, OR [95% CI] 1.48 [1.01–2.18], P = 0.044; midpoint <02:58, N = 310, 1.80 [1.00–3.22], P = 0.0492; wake time <06:41, N = 303, 1.88 [1.04–3.40], P = 0.035) but not in late sleep timing (bedtime ≥23:12, N = 310, 1.29 [0.77–2.16], P = 0.337; midpoint ≥02:58, N = 310, 1.34 [0.79–2.29], P = 0.277; wake time ≥06:41, N = 320, 1.25 [0.74–2.13], P = 0.406), independent of sleep duration. Thus, the effect of rs10830963 on risk of T2D may be modified by sleep timing, with risk allele carriers with earlier sleep timing at an increased risk.
Table 4.
Interaction results of rs10830963 with sleep timing on T2D risk
| Interaction phenotype | Effect allele | Ethnicity | MAF | N | Early timing |
Late timing |
Pinteraction | |||
|---|---|---|---|---|---|---|---|---|---|---|
| CHR | SNP | OR (95% CI) | OR (95% CI) | SE | ||||||
| Bedtime |
11 |
rs10830963 |
G |
Whites |
0.26 |
619 |
1.48 (1.01–2.18) |
1.29 (0.77–2.16) |
0.1446 |
0.1907 |
| Asians |
0.42 |
167 |
2.38 (0.88–6.46) |
0.70 (0.24–2.07) |
0.2356 |
0.1170 |
||||
| Blacks |
0.08 |
370 |
1.11 (0.56–2.19) |
1.34 (0.60–3.00) |
0.1867 |
0.9468 |
||||
| Hispanics |
0.19 |
357 |
1.07 (0.57–2.04) |
1.02 (0.55–1.89) |
0.1490 |
0.2639 |
||||
| Meta-analysis |
0.22 |
1,513 |
1.38 (1.03–1.83) |
1.14 (0.81–1.60) |
0.0846 |
0.0533 |
||||
| Sleep midpoint |
11 |
rs10830963 |
G |
Whites |
0.26 |
619 |
1.80 (1.00–3.22) |
1.34 (0.79–2.29) |
0.1783 |
0.0549 |
| Asians |
0.42 |
167 |
2.45 (0.89–6.70) |
0.97 (0.37–2.49) |
0.3007 |
0.0626 |
||||
| Blacks |
0.08 |
370 |
1.07 (0.57–2.02) |
1.02 (0.55–1.89) |
0.2118 |
0.5954 |
||||
| Hispanics |
0.19 |
357 |
1.30 (0.59–2.90) |
1.05 (0.53–2.07) |
0.1668 |
0.4665 |
||||
| Meta-analysis |
0.22 |
1,513 |
1.49 (1.05–2.13) |
1.13 (0.81–1.57) |
0.0996 |
0.0180 |
||||
| Wake time |
11 |
rs10830963 |
G |
Whites |
0.26 |
619 |
1.88 (1.04–3.40) |
1.25 (0.74–2.13) |
0.1720 |
0.0320 |
| Asians |
0.42 |
167 |
2.09 (0.85–5.06) |
0.84 (0.37–2.63) |
0.2672 |
0.1616 |
||||
| Blacks |
0.08 |
370 |
1.33 (0.61–2.94) |
0.94 (0.47–1.87) |
0.1808 |
0.3368 |
||||
| Hispanics |
0.19 |
357 |
0.84 (0.44–1.60) |
1.26 (0.67–2.37) |
0.1490 |
0.8488 |
||||
| Meta-analysis | 0.22 | 1,513 | 1.32 (0.95–1.84) | 1.13 (0.82–1.57) | 0.0900 | 0.0290 |
Results are from logistic regression with an interaction term. Model is adjusted for age, sex, and BMI. Wake time was measured by actigraphy across a minimum of 3 days. CHR, chromosome; MAF, minor G allele frequency. Odds of T2D are shown stratified by the median bedtime, midpoint, and wake time into “early” and “late.” Significant results (P < 0.05) are shown in boldface type.
Figure 2.
Sleep timing modifies the effect of MTNR1B variant rs10830963 on risk of T2D in MESA (n = 1,513). The effect of sleep timing on T2D risk is shown by rs10830963 genotype. The rs10830963G T2D risk allele is shown in red. Lines represent the genotype-specific linear regression of rs10830963 × 7-day actigraphy measured at bedtime (A), midpoint (B), and wake time (C) in 1,513 subjects of multiethnic ancestry, adjusted for age, sex, and BMI.
Discussion
We hypothesized that a common T2D risk variant in MTNR1B would be associated with melatonin, sleep, or circadian traits. We found the MTNR1B diabetes risk variant (rs10830963G) was associated with a later melatonin offset and a longer duration of elevated melatonin levels in highly controlled laboratory studies. Furthermore, we demonstrated that the increased T2D risk in rs10830963G carriers is more pronounced in early sleep timing and almost absent in late sleep timing, in which an extended morning melatonin profile would be obscured by the later rise time. Thus, taken together, our data suggest that MTNR1B rs10830963G extends the duration of melatonin production later into the morning, and waking up earlier in the morning magnifies the diabetes risk with MTNR1B genotype.
The impact of MTNR1B rs10830963G on DLMOff is significantly mediated by sleep timing, suggesting that MTNR1B variation may influence DLMOff through changes in sleep timing or that MTNR1B variation may influence sleep timing through changes in the timing of the melatonin profile. No significant associations with other sleep and circadian traits were observed, consistent with previous studies of narrower scope showing no MTNR1B risk allele effect on self-reported sleep disturbances (41,42). Although our observations must be regarded as preliminary due to the limited sample size with detailed circadian measures and melatonin profile, they collectively add new insights linking MTNR1B to T2D. Melatonin receptor 1B (known as Mel1B or MT2) is one of two trans-membrane receptors for melatonin, a hormone that acts as a signal for the biological night. MTNR1B rs10830963G allele carriers have been reported to show increased Mel1B receptor expression in the pancreatic β-cell (11). Melatonin signaling during the night, when diurnal humans are fasting, inhibits basal and glucose-stimulated insulin secretion (5,43,44). Delayed DLMOff and a longer duration of melatonin in risk allele carriers may result in an increased risk for food intake to coincide with elevated melatonin levels in the morning, leading to decreased glucose tolerance and possibly elevated diabetes risk. Consistently, risk allele carriers with earlier sleep timing have an increased T2D risk, possibly due to concomitant food intake and elevated melatonin levels in the morning. In addition to the adverse effects of an increase in melatonin levels into daytime, a reduction in nighttime melatonin signaling also appears to be deleterious. Reduced nighttime melatonin signaling, either by MTNR1B receptor rare loss-of-function variants (13) or reduced nighttime melatonin levels (5), is associated with an increased risk of T2D. Future studies are warranted to test causality and to assess how the impact of rs10830963G on melatonin offset and duration alters the proper timing and magnitude of basal and postprandial insulin secretion and glucose control.
The strength of our study comes from the depth and breadth of phenotypes available in our cohorts. The laboratory study was limited in size due to the nature of the intensive physiological studies required to obtain precise phenotypes. This sample is one of the largest of its kind and contains precisely measured endogenous circadian measures that require multiday studies under highly controlled laboratory conditions to assess endogenous circadian control of plasma melatonin and CBT. These results need to be interpreted in light of the biases of this study, however, as only young healthy subjects were studied and held to self-selected sleep schedules prior to the in-laboratory portion of the study (8-h sleep duration) that may not reflect biological preference. Although the sample size is limited, the phenotypes were measured in great depth, minimizing misclassification and maximizing specificity to biological processes of interest. Sleep timing in MESA is measured objectively across multiple days, minimizing phenotype measurement error. The sample size, however, is limited. In all future studies, it will also be important to measure melatonin levels at the time of glucose assessment in the morning by genotype.
The CARe study is a large well-powered study encompassing one of the largest epidemiological studies of sleep habits (SHHS) with self-reported and objective overnight PSG-measured sleep phenotypes. Notably, our study did not identify significant associations with the available measures of sleep quality or quantity in the CARe study, consistent with previous studies (41,42). It is important to recognize, however, that the indices of sleep quality, duration, and timing available in these cohort studies likely are measured with modest-to-moderate error, and misclassification would attenuate any true associations. Previous studies demonstrated a 50% reduction in power associated with measurement error equivalent to one SD of the trait (45). This may even be true for PSG-measured phenotypes in the CARe study, where first-night effects influence sleep measures taken during a single unsupervised overnight PSG episode. This study establishes that diabetes risk variants in MTNR1B are unlikely to play a role in central sleep behaviors, and thus, future research should emphasize the evaluation of their role in peripheral tissues of relevance to T2D.
Given the clear and adverse effects of sleep disruption and circadian disruption on glucose control and diabetes risk, our new evidence linking sleep and the circadian-related MTNR1B gene variant with altered melatonin physiology and indicating how this might impact glucose control and diabetes risk is an important advance. Moving forward, the association of MTNR1B rs10830963 with melatonin rhythm phenotypes should be followed up with further in-depth mechanistic studies on a tissue level and phenotyping in individuals preselected based on genotypes of interest. In general, circadian metabolic assessments as well as targeted interventions (e.g., with light intervention or pharmacological doses of melatonin) may be useful strategies for probing the functional consequences of the variant on circadian rhythms, sleep physiology, and metabolism. Ultimately, this research could lead us toward new therapeutic interventions that reduce the impact of extended elevated melatonin levels into the morning perhaps via alterations to melatonin dynamics, melatonin-mediated insulin secretion, or timing of food intake.
Article Information
Acknowledgments. The authors thank the study participants for each cohort and the genotyping and central CARe statistical analysis group at the Broad Institute for generating IBC array genotypes and quality control filtering. The authors also thank Elizabeth Lydon for help with participant recruiting and Dan Cohen for data collection.
Funding. This study was conducted with support from the National Institutes of Health (NIH) National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (DK089378 to R.S. and F.A.J.L.S.), support from Harvard Catalyst of the Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, NIH Award 8UL1TR00017005 to F.A.J.L.S. and R.S.), support from the NHLBI (T32HL07901) and the National Institute on Aging (NIA) (F32 AG316902) to K.S., and financial contributions from Harvard University and its affiliated academic health care centers. J.M.L., S.R., and R.S. were further supported by the following grants: NHLBI R21 HL121728 (R.S.), NIDDK F32 DK102323 (J.M.L.), and NHLBI R01 HL113338 and NHLBI R01 HL098433 (S.R.). F.A.J.L.S. was further supported by the NHLBI (R01HL094806 and R01HL118601) and the NIDDK (R01DK099512). A.-M.C. was also supported by the NHLBI (K01 HL115458). The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic health care centers, or the NIH. The CARe study and SHHS are supported by the NHLBI cooperative agreements U01HL53941 (Boston University), U01HL53916 (University of California, Davis), U01HL53934 (University of Minnesota), U01HL53937 and U01HL63429 (Johns Hopkins University), and U01HL63463 (Case Western Reserve University). The following funding contributed to phenotype data collection in in-laboratory studies: NIA (R01AG06072, R01AG06072, and P01AG009975), NHLBI (R01HL077453, R01HL080978, R01HL093279, R01HL094654, and R01HL077399), Air Force Office of Scientific Research (FA9550), NIH (R21AT002571, R01NS054277, and R01MH45130), National Space Biomedical Research Institute (HFP01601), Brain & Behavior Research Foundation Young Investigator Award, and General Clinical Research Center (M01RR02635). J.F.D. was supported by the NIA (R01AG044416), the NHLBI (R01HL093279 and R01HL094654), and the Brigham and Women's Hospital Brigham Research Institute Fund to Sustain Research Excellence. Investigators were also funded by financial contributions from Brigham and Women's Hospital and from Harvard University and its affiliated academic health care centers. This study was also funded by the NHLBI (01EHLE114088, K24EHL105664, R01HL09327, R01HL09465, and RC2EHL101340), the National Space Biomedical Research Institute (HFP02802), and the NIH (M01RR02635 and R01EGME10501).
Duality of Interest. C.A. reports receiving a research award/prize from Sanofi and lecturing fees from Brown Medical School/Rhode Island Hospital, Ausmed Education, and Rio Tinto. She has also received contract research funding from Pacific Brands and VicRoads through an agreement with Monash University. She has served as consultant to the Rail, Tram and Bus Union, the National Transport Commission, the Transport Accident Commission, and the Victoria Police. She is a participant in the Cooperative Research Centre for Alertness, Safety and Productivity. C.A.C. has received consulting fees from or served as a paid member of scientific advisory boards for the Boston Celtics, the Boston Red Sox, Citgo, the Cleveland Browns, Merck, Novartis, Purdue Pharma LP, Quest Diagnostics, Inc., Teva Pharmaceuticals Industries Ltd., Valero Inc., and Vanda Pharmaceuticals, Inc. He also owns an equity interest in LifeTrac, Inc., Somnus Therapeutics, Inc., and Vanda Pharmaceuticals, Inc. and has received royalties from McGraw Hill, Penguin Press/Houghton Mifflin Harcourt, and Philips Respironics, Inc. He has also received grants and research support from Cephalon, the National Football League Charities, Philips Respironics, ResMed Foundation, the San Francisco Bar Pilots, and Sysco. He has received lecture fees from the American Academy of Sleep Medicine, American Academy of Dental Sleep Medicine, Harvard School of Public Health, Integritas Communications Group, Montefiore Medical Center, Stanford Center for Sleep Sciences and Medicine, and the University of Buffalo. The Harvard Medical School Division of Sleep Medicine (HMS/DSM), which C.A.C. directs, has received gifts from many outside organizations and individuals including the Concord Music Company, Delos Living, Flux Software, Jordan’s Furniture, King Koil, Leggett & Platt, Merck Neurosciences, Metro Naps, Novartis Consumer Health, Optum, Patient Point, Philips Home Healthcare Solutions, ResMed, Simmons Bedding, Sleep Apnea Treatment Centers of America, Sleep Med, Turner Broadcasting, UnitedHealthcare Clinical Services, and Vanda Pharmaceuticals. The HMS/DSM Sleep and Health Education Program has received educational grant funding from Cephalon, Takeda Pharmaceuticals, Sanofi, and Sepracor. C.A.C. is the incumbent of an endowed professorship provided to Harvard University by Cephalon and holds a number of process patents in the field of sleep/circadian rhythms (e.g., photic resetting of the human circadian pacemaker). Since 1985, he has also served as an expert witness on various legal cases related to sleep and/or circadian rhythms including Bombardier, Inc., Citgo, Greyhound, Michael Jackson’s mother and children, Purdue Pharma, United Parcel Service (UPS), and Valero Inc. O.M.B. reports investigator-initiated research grant support from Cephalon (now Teva) that partially supported this work. Unrelated to this work, O.M.B. discloses investigator-initiated research grant support from Sepracor (now Sunovion), personal fees from Takeda Pharmaceuticals North America, expert witness testimony for Dinsmore LLC, serving on the scientific advisory board for Matsutani America, and receiving travel support and/or honoraria from the Wake Forest University Medical Center, American Academy of Craniofacial Pain, NHLBI, NIDDK, National Postdoctoral Association, Oklahoma State University, Oregon Health & Science University, SUNY Downstate Medical Center, American Diabetes Association, New York University, and Academy of Nutrition and Dietetics outside of the submitted work.
Author Contributions. J.M.L., A.C.B., and R.S. performed genetic analyses. J.M.L., F.A.J.L.S., and R.S. wrote the manuscript and all the coauthors helped interpret data and reviewed and edited the manuscript before approving its submission. The study was designed by A.-M.C., S.A.S., J.F.D., O.M.B., F.A.J.L.S., and R.S. The following coauthors contributed to phenotype data collection in study cohorts: in-laboratory studies (D.A., C.A., S.W.C., C.A.C., J.J.G., E.B.K., S.W.L., M.M., S.M.W.R., M.R., N.S., K.S., E.V.R., J.F.D., and O.M.B.) and CARe cohorts University of Pennsylvania (S.F.A.G.), SHHS (B.C.E., S.P., and S.R.), CHS (S.A.G.), FHS (D.J.G.), CARDIA (D.S.L.), ARIC (N.M.P.), and MESA (P.C.Z.). A.-M.C. and M.A.S.H. performed analyses to generate in-laboratory phenotypes. R.S. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented at the 29th Annual Meeting of the Associated Professional Sleep Societies, Seattle, WA, 6–10 June 2015, and the American Society of Human Genetics Annual Meeting, Baltimore, MD, 6–10 October 2015.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db15-0999/-/DC1.
M.M. is currently affiliated with Charité–University Medicine Berlin, Germany Institute of Physiology, Group Sleep Research and Clinical Chronobiology, Berlin, Germany. J.J.G. is currently affiliated with the Neuroscience and Behavioral Disorders Programme, Duke–National University of Singapore Graduate Medical School, Singapore, Singapore. S.M.W.R. is currently affiliated with the School of Psychological Sciences and Monash Institute of Cognitive and Clinical Neurosciences, Monash University, Victoria, Australia. N.S. is currently affiliated with the Division of Sleep Medicine, Surrey Sleep Research Centre, University of Surrey, Surrey, U.K. K.S. is currently affiliated with the Wits Sleep Laboratory, Brain Function Research Group, School of Physiology, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, Johannesburg, South Africa. E.V.R. is currently affiliated with the Department of Psychiatry and Human Behavior, Alpert Medical School and Sleep for Science Research Laboratory, Brown University, Providence, RI.
See accompanying article, p. 1490.
References
- 1.Cappuccio FP, D’Elia L, Strazzullo P, Miller MA. Quantity and quality of sleep and incidence of type 2 diabetes: a systematic review and meta-analysis. Diabetes Care 2010;33:414–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spiegel K, Tasali E, Leproult R, Van Cauter E. Effects of poor and short sleep on glucose metabolism and obesity risk. Nat Rev Endocrinol 2009;5:253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science 2010;330:1349–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheer FAJL, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A 2009;106:4453–4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McMullan CJ, Schernhammer ES, Rimm EB, Hu FB, Forman JP. Melatonin secretion and the incidence of type 2 diabetes. JAMA 2013;309:1388–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rubio-Sastre P, Scheer FA, Gómez-Abellán P, Madrid JA, Garaulet M. Acute melatonin administration in humans impairs glucose tolerance in both the morning and evening. Sleep 2014;37:1715–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouatia-Naji N, Bonnefond A, Cavalcanti-Proença C, et al. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet 2009;41:89–94 [DOI] [PubMed] [Google Scholar]
- 8.Prokopenko I, Langenberg C, Florez JC, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet 2009;41:77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karamitri A, Renault N, Clement N, Guillaume JL, Jockers R. Minireview: Toward the establishment of a link between melatonin and glucose homeostasis: association of melatonin MT2 receptor variants with type 2 diabetes. Mol Endocrinol 2013;27:1217–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cagnacci A, Arangino S, Renzi A, et al. Influence of melatonin administration on glucose tolerance and insulin sensitivity of postmenopausal women. Clin Endocrinol (Oxf) 2001;54:339–346 [DOI] [PubMed] [Google Scholar]
- 11.Lyssenko V, Nagorny CL, Erdos MR, et al. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet 2009;41:82–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaulton KJ, Ferreira T, Lee Y, et al.; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium . Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet 2015;47:1415–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonnefond A, Clément N, Fawcett K, et al.; Meta-Analysis of Glucose and Insulin-Related Traits Consortium (MAGIC) . Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat Genet 2012;44:297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nedeltcheva AV, Scheer FA. Metabolic effects of sleep disruption, links to obesity and diabetes. Curr Opin Endocrinol Diabetes Obes 2014;21:293–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Depner CM, Stothard ER, Wright KP Jr. Metabolic consequences of sleep and circadian disorders. Curr Diab Rep 2014;14:507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang AM, Buch AM, Bradstreet DS, Klements DJ, Duffy JF. Human diurnal preference and circadian rhythmicity are not associated with the CLOCK 3111C/T gene polymorphism. J Biol Rhythms 2011;26:276–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horne JA, Ostberg O. A self-assessment questionnaire to determine morningness-eveningness in human circadian rhythms. Int J Chronobiol 1976;4:97–110 [PubMed] [Google Scholar]
- 18.Brown EN, Czeisler CA. The statistical analysis of circadian phase and amplitude in constant-routine core-temperature data. J Biol Rhythms 1992;7:177–202 [DOI] [PubMed] [Google Scholar]
- 19.Chang AM, Scheer FA, Czeisler CA. The human circadian system adapts to prior photic history. J Physiol 2011;589:1095–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duffy JF, Dijk DJ. Getting through to circadian oscillators: why use constant routines? J Biol Rhythms 2002;17:4–13 [DOI] [PubMed] [Google Scholar]
- 21.Czeisler CA, Duffy JF, Shanahan TL, et al. Stability, precision, and near-24-hour period of the human circadian pacemaker. Science 1999;284:2177–2181 [DOI] [PubMed] [Google Scholar]
- 22.Duffy JF, Dijk DJ, Klerman EB, Czeisler CA. Later endogenous circadian temperature nadir relative to an earlier wake time in older people. Am J Physiol 1998;275:R1478–R1487 [DOI] [PubMed] [Google Scholar]
- 23.Kleitman N. Sleep and Wakefulness. University of Chicago Press, 1963 [Google Scholar]
- 24.St. Hilaire MA, Gronfier C, Zeitzer JM, Klerman EB. A physiologically based mathematical model of melatonin including ocular light suppression and interactions with the circadian pacemaker. J Pineal Res 2007;43:294–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006;38:904–909 [DOI] [PubMed] [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musunuru K, Lettre G, Young T, et al.; NHLBI Candidate Gene Association Resource . Candidate-gene Association Resource (CARe): design, methods, and proof of concept. Circ Cardiovasc Genet 2010;3:267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ARIC Investigators. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am J Epidemiol 1989;129:687–702 [PubMed] [Google Scholar]
- 29.Friedman GD, Cutter GR, Donahue RP, et al. CARDIA: study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol 1988;41:1105–1116 [DOI] [PubMed] [Google Scholar]
- 30.Fried LP, Borhani NO, Enright P, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol 1991;1:263–276 [DOI] [PubMed] [Google Scholar]
- 31.Dawber TR, Meadors GF, Moore FE Jr. Epidemiological approaches to heart disease: the Framingham Study. Am J Public Health Nations Health 1951;41:279–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bild DE, Bluemke DA, Burke GL, et al. Multi-Ethnic Study of Atherosclerosis: objectives and design. Am J Epidemiol 2002;156:871–881 [DOI] [PubMed] [Google Scholar]
- 33.Quan SF, Howard BV, Iber C, et al. The Sleep Heart Health Study: design, rationale, and methods. Sleep 1997;20:1077–1085 [PubMed] [Google Scholar]
- 34.Castro-Diehl C, Diez Roux AV, Redline S, Seeman T, Shrager SE, Shea S. Association of sleep duration and quality with alterations in the hypothalamic-pituitary adrenocortical axis: the Multi-Ethnic Study of Atherosclerosis (MESA). J Clin Endocrinol Metab 2015;100:3149–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buysse DJ, Reynolds CF 3rd, Monk TH, Berman SR, Kupfer DJ. The Pittsburgh Sleep Quality Index: a new instrument for psychiatric practice and research. Psychiatry Res 1989;28:193–213 [DOI] [PubMed] [Google Scholar]
- 36.Redline S, Kapur VK, Sanders MH, et al. Effects of varying approaches for identifying respiratory disturbances on sleep apnea assessment. Am J Respir Crit Care Med 2000;161:369–374 [DOI] [PubMed] [Google Scholar]
- 37.Kales A, Rechtschaffen A. A Manual of Standardized Terminology, Techniques and Scoring System for Sleep Stages of Human Subjects. Los Angeles, Brain Information Service at the University of California, 1968 [Google Scholar]
- 38.Keating BJ, Tischfield S, Murray SS, et al. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS One 2008;3:e3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gauderman W, Morrison JM. Quanto 1.1: a computer program for power and sample size calculations for genetic-epidemiology studies [Internet], 2006. Avaliable from http://hydra.usc.edu/gxe. Accessed 3 January 2012
- 40.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu C, Wu Y, Li H, et al. MTNR1B rs10830963 is associated with fasting plasma glucose, HbA1C and impaired beta-cell function in Chinese Hans from Shanghai. BMC Med Genet 2010;11:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsson L, Pettersen E, Ahlbom A, Carlsson S, Midthjell K, Grill V. No effect by the common gene variant rs10830963 of the melatonin receptor 1B on the association between sleep disturbances and type 2 diabetes: results from the Nord-Trøndelag Health Study. Diabetologia 2011;54:1375–1378 [DOI] [PubMed] [Google Scholar]
- 43.Picinato MC, Haber EP, Cipolla-Neto J, Curi R, de Oliveira Carvalho CR, Carpinelli AR. Melatonin inhibits insulin secretion and decreases PKA levels without interfering with glucose metabolism in rat pancreatic islets. J Pineal Res 2002;33:156–160 [DOI] [PubMed] [Google Scholar]
- 44.Stumpf I, Mühlbauer E, Peschke E. Involvement of the cGMP pathway in mediating the insulin-inhibitory effect of melatonin in pancreatic beta-cells. J Pineal Res 2008;45:318–327 [DOI] [PubMed] [Google Scholar]
- 45.Liao J, Li X, Wong TY, et al. . Impact of measurement error on testing genetic association with quantitative traits. PLoS One 2014;9:e87044. [DOI] [PMC free article] [PubMed] [Google Scholar]