Abstract
In the nematode model organisms Caenorhabditis elegans and Pristionchus pacificus, a new class of natural products based on modular assembly of primary-metabolism-derived building blocks control organismal development and behavior. We report identification and biological activities of the first pentamodular metabolite, pasa#9, and the 8-oxoadenine-containing npar#3 from P. pacificus. These structures suggest co-option of nucleoside and tryptophan metabolic pathways for the biosynthesis of endogenous metabolite libraries that transcend the dichotomy between “primary” and “secondary” metabolism.
Graphical Abstract
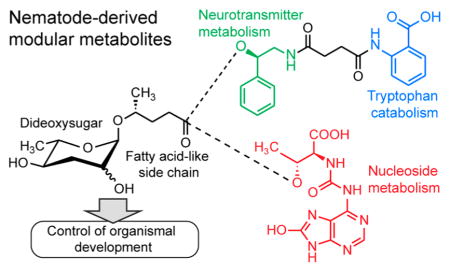
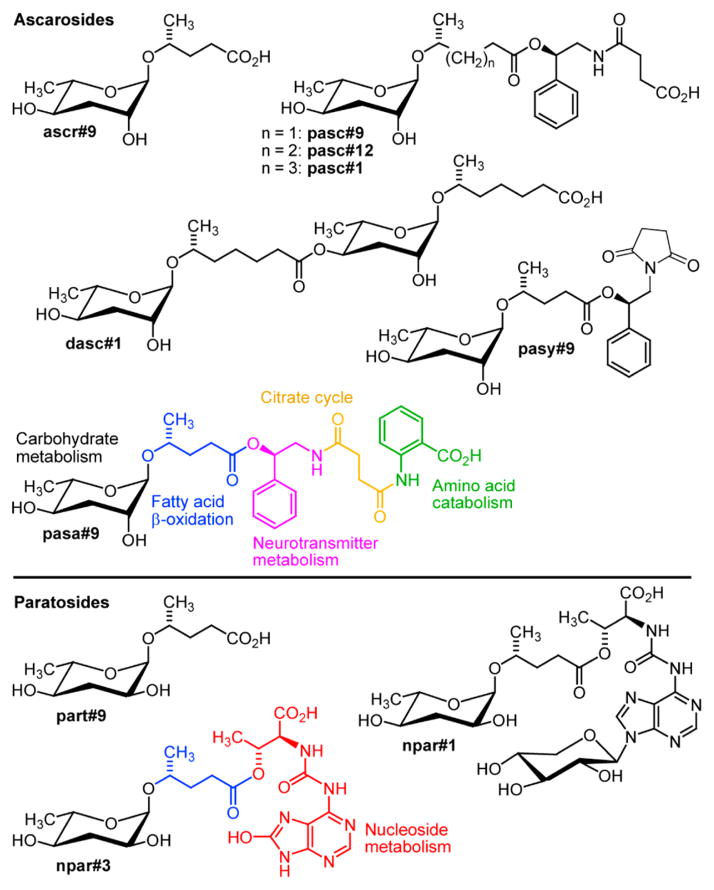
Small molecules play a central role for many aspects of inter-and intraorganismal signaling; however, the chemical structures of small-molecule signals in animal model systems have remained largely uncharacterized. Recent studies have shown that in the model organisms Caenorhabditis elegans and Pristionchus pacificus, a novel family of signaling molecules based on the dideoxy sugars ascarylose and paratose, play a central role for almost any aspect of life history, including lifespan, development, reproduction, and a wide range of behaviors.1–5 Their structures appear to be derived from modular assembly of building blocks from major primary metabolic pathways, including lipid, amino acid, and nucleoside metabolism, using ascarylose- and paratose-derived glycosides as central scaffolds (Figure 1).
Figure 1.
Examples for ascarylose- and paratose-based signaling molecules recently identified from C. elegans and P. pacificus and structures of novel metabolites pasa#9, pasy#9, and npar#3.
These nematode-derived modular metabolites (NDMMs) exert their diverse biological functions via conserved signaling cascades, including insulin signaling, TGF-β signaling, and steroid biosynthesis converging on the nuclear hormone receptor DAF-12, a liver-X and vitamin D receptor homologue.6,7 While the first NDMMs were identified from C. elegans,8–11 subsequent studies revealed even greater structural diversity in the satellite model organism P. pacificus, a nematode known for its necromenic association with scarab beetles.12 Small molecules in P. pacificus not only exhibit greater structural complexity but also revealed additional functional diversity. As in C. elegans, specific NDMMs, including the xylopyrano-adenosine-derived paratoside npar#1, trigger formation of dauer larvae, a state of metabolic diapause adapted to survival under conditions of environmental stress and starvation.5 Other NDMMs, for example the dimeric ascaroside dasc#1, affect adult body shape and physiology, triggering development of an alternate mouth form optimized for predacious feeding on other nematodes instead of bacteria.13,14
Although several dozen NDMMs have been identified, it is unclear whether the so-far detected structures are representative of the full chemical diversity of nematode-signaling molecules. For example, all known NDMMs are derived from attachment of primary metabolism-derived modules directly to the ascaroside or paratoside scaffold, as opposed to attachment to other parts of the structures.1 Moreover, recent studies demonstrated that small-molecule-based nematode signals often consist of several different NDMMs, with additive or sometimes synergistic activities that can vary significantly between different genotypes of C. elegans or P. pacificus.15,16 As a result, NDMMs are often missed by identification efforts based on activity-guided fractionation.8 Therefore, we conducted a targeted metabolomic screen for additional ascaroside and paratoside derivatives in the P. pacificus metabolome, using 2D NMR spectroscopy and high-resolution HPLC–MS/MS, which revealed the first pentamodular nematode metabolite, pasa#9, suggesting integration of building blocks from five different primary metabolic pathways as well as the 8-oxoadenine-containing paratoside npar#3 (Figure 1).
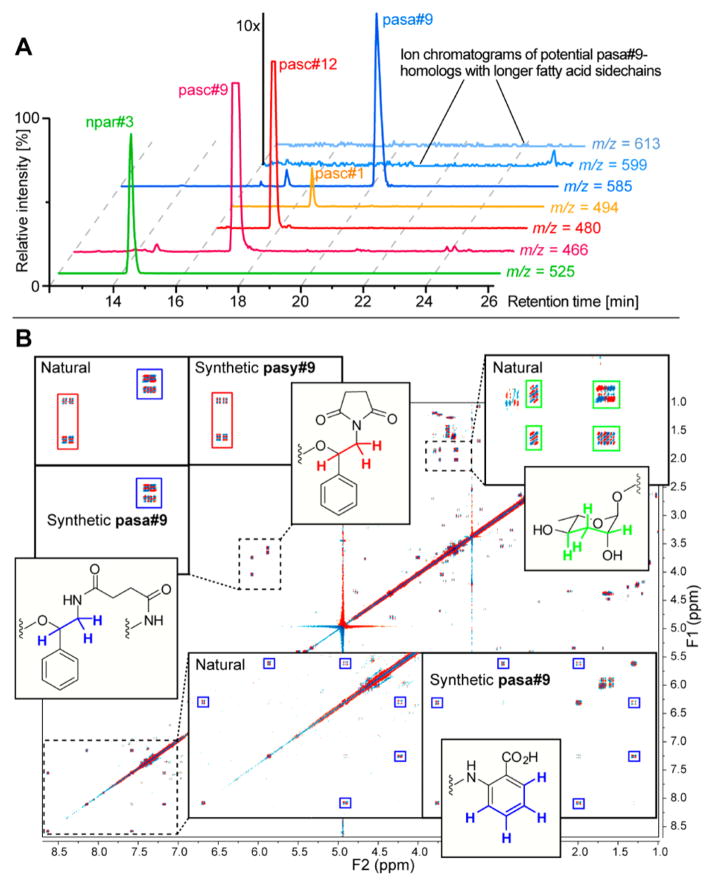
To survey the P. pacificus metabolome for ascaroside-based NDMMs, we employed HPLC–MS/MS in negative ionization mode, monitoring the formation of the fragment m/z 73.03 (C3H5O2−; calcd m/z 73.0294), which has been shown to be highly characteristic for ascarosides and paratosides.4 In addition to previously reported compounds,5 we detected two prominent peaks with molecular ions at m/z 525.1963 (C21H29N6O10−; calcd m/z 525.1951) and m/z 585.2453 (C30H37N2O10−; calcd m/z 585.2454), which appeared to represent novel metabolites (Figure 2A). We named these compounds npar#3 and pasa#9, respectively (see Supporting Information section 1.1 and www.smid-db.org for NDMM nomenclature). For further characterization of npar#3 and pasa#9, the P. pacificus metabolome was minimally fractionated via reversed-phase chromatography and HPLC followed by 2D NMR spectroscopic analysis of npar#3- and pasa#9-containing fractions. This approach relies on the demonstrated utility of high-resolution 2D NMR spectra for characterizing novel structures from complex mixtures and thereby avoids extensive chromatographic fractionation, which is particularly advantageous for the characterization of highly potent signaling molecules that are often produced only at nanomolar (or lower) concentrations.17,18
Figure 2.
(A) HPLC–MS analysis of P. pacificus exometabolome showing ion chromatograms for npar#3 and pasa#9 as well as previously described pasc#9, which is accompanied by two homologues, pasc#12 and pasc#1, with longer fatty acid side chains. In contrast, no homologues of pasa#9 were detected. (B) Analysis of dqfCOSY spectra of P. pacificus metabolome fractions (“natural”) containing pasa#9 and pasy#9 and comparison with spectra of synthetic compounds.
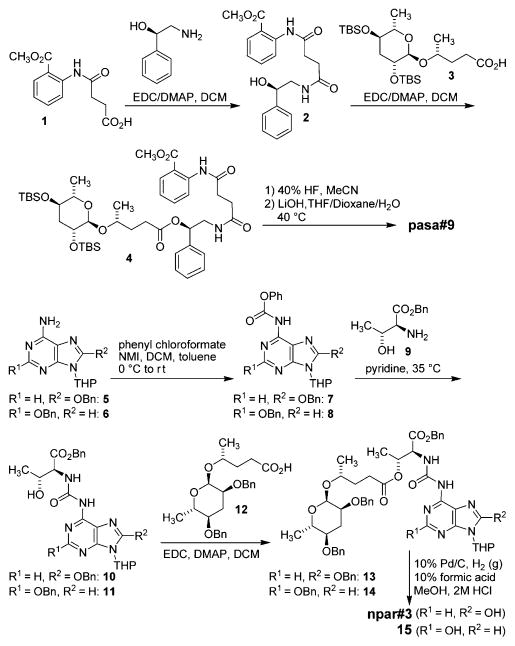
dqfCOSY spectra of metabolome fractions containing pasa#9 revealed a 4-hydroxypentanoic acid based ascaroside as well as phenylethanolamine, anthranilic acid, and succinate moieties (Figure 2B). 1H chemical shift and spin–spin coupling data suggested that the succinyl moiety linked the anthranilate and phenylethanolamine units as a diamide and that the 4-hydroxypentanoic acid based ascaroside is attached to the phenylethanolamine as an ester. Stereochemical assignments were made on the basis of comparison of dqfCOSY crosspeaks of the ascaroside moiety of pasa#9 to those of previously reported ascarosides. To confirm the proposed structure and to obtain pure samples for bioassays, we adapted previously published syntheses of ascr#9 and pasc#9 (Scheme 1). Starting from commercially available N-succinyl methyl anthranilate 1, we successively added modules in a biomimetic fashion, first connecting the phenyl ethanolamine moiety, which was subsequently linked to bis-TBS protected ascaroside 3. Following TBS deprotection and hydrolysis of the methyl ester, comparison of dqfCOSY spectra (Figure 2B) and HPLC retention times of the synthetic compound with those of natural pasa#9 confirmed our assignments (Supporting Information, Table S1).
Scheme 1.
Synthesis of pasa#9 and npar#3
In addition to signals representing pasa#9, 2D NMR spectra of pasa#9-containing metabolome fractions consistently showed signals representing another ascaroside derivative (Figure 2B). This compound, named pasy#9, was identified as the imide variant of pasc#9 (Figure 1 and 2B) and thus lacks a carboxy group, explaining that it was not detected in the m/z 73.03 MS/MS screen which uses negative ionization. pasc#9 and pasa#9 do not spontaneously cyclize to form pasy#9; however, pasy#9 could be derived from cyclization of pasc#9-coenzyme A ester, a plausible biosynthetic precursor of pasa#9.
Analysis of 1H NMR and dqfCOSY spectra of fractions containing npar#3 suggested a paratose sugar attached to a 4-hydroxypentanoic acid, a threonine-derived moiety, as well as a 2- or 8-hydroxyadenine derivative (Supporting Information, Figure S3). Like many other nematode-signaling molecules, npar#3 is produced only in minute quantities, which were insufficient to obtain carbon NMR spectroscopic data via HSQC and HMBC. Moreover, the exact structure of the putative oxoadenine is difficult to determine by NMR spectroscopy alone because of the scarcity of protons in this moiety. Therefore, we synthesized both the 2- and 8-hydroxyadenine variants (Scheme 1 and Supporting Information sections 4.3 and 4.4). Protected 2- and 8-hydroxyadenine (5 and 6) were converted into the corresponding phenyl carbamates 7 and 819 and then linked to threonine benzyl ester 9. The remaining free hydroxy group in the threonine moieties of 10 and 11 was then coupled to benzyl-protected paratoside 12. Following deprotection, comparison of spectroscopic data and HPLC retention times of the two synthetic isomers with those of natural npar#3 unambiguously demonstrated that natural npar#3 corresponds to the 8-oxoadenine-based isomer (see the Supporting Information, Figures S2–S4).5
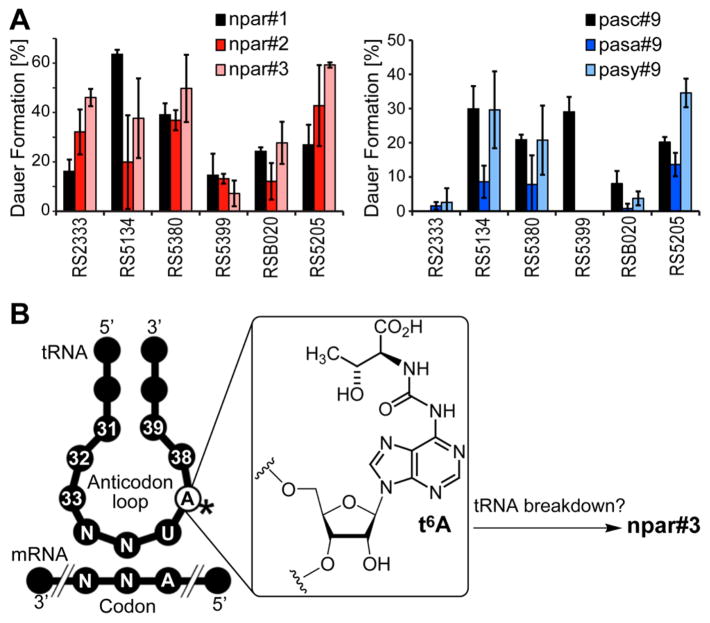
Next, we tested the biological activity of the newly identified NDMMs in the dauer larva formation assay. In P. pacificus and other nematodes, dauer larvae represent a stage of developmental arrest adapted to survival under harsh environmental conditions,20 and specific NDMMs, e.g., npar#1 and pasc#9 in the case of P. pacificus, have been shown to induce the transitioning of developing larvae into the dauer stage.5,16 This dauer formation assay is highly reproducible and thus most appropriate for the comparison of the activity of the newly identified compounds with that of known NDMMs. Because recent studies have revealed strong natural variation of the potency of signaling molecules between different wild-type strains of P. pacificus,16 we performed dauer induction assays in six different wild-type strains, spanning three of the four P. pacificus clades. We found that the newly identified compounds induce dauer larva formation in some, but not all strains (Figure 3A). For example, npar#3 strongly induces dauer in the clade C strain RS5380, whereas it is largely inactive in strain RS5399, another clade C strain. Overall, dauer-inducing activity was markedly dependent on molecular structure as well as genetic background, suggesting that the receptors involved in perception of these NDMMs have significantly diverged between different P. pacificus strains. Previous studies in C. elegans have shown that dauer-inducing ascarosides bind to G protein-coupled receptors (GPCRs) that are expressed in several chemosensory neurons in the head of the worm,21,22 and ascaroside-sensing GPCRs have been shown to evolve rapidly even under laboratory conditions,23 providing a rationale for the starkly divergent responses observed for the set of P. pacificus wild-type strains tested here.
Figure 3.
(A) Biological activity of pasa#9, pasy#9, and npar#3 in the dauer formation assay. (B) Putative biogenetic origin of npar#3 from 6-threonylcarbamoyladenosine (t6A), a highly conserved nucleoside that forms part of the anticodon loop of a family of tRNAs.24
The structures of pasa#9 and npar#3 suggest that these compounds are derived from straightforward assembly of conserved primary metabolic building blocks, whereby the anthranilate and hydroxyadenine moieties in pasa#9 and npar#3 likely originate from tryptophan25 and nucleoside26,27 metabolism, respectively (Figure 3B).24 Although enzymes that link different building blocks via ester and amide bonds have not been identified, assembly of pasa#9 and npar#3 is remarkably specific, strongly suggesting that these compounds are the products of enzyme-controlled biosynthesis. For example, neither pasa#9 nor npar#3 is accompanied by homologues with longer or shorter fatty acid like side chains, although the previously identified pasc#9, which differs from pasa#9 only in that it lacks the anthranilate moiety, is consistently accompanied by large quantities of pasc#12, a homologue including 5-hydroxyhexanoic acid instead of 4-hydroxypentanoic acid (Figure 1 and 2A).5 With regard to incorporation of 8-hydroxyadenine in npar#3, it should be noted that 8-hydroxyadenine is produced primarily as a result of oxidative damage to nucleic acids,26,27 although similar oxidized derivatives of the tRNA nucleoside t6A, from which the 6-threonylcarbamoyladenine moiety in npar#3 may be derived (Figure 3B), have not been described.
In conclusion, we report the identification, synthesis, and biological activities of the first pentamodular NDMM, pasa#9, its derivative pasy#9, and the nucleobase-derived paratoside npar#3. These compounds function as dauer-inducing signaling molecules in P. pacificus, and their structures suggest that NDMMs may be more structurally diverse than previously suspected, with pasa#9 demonstrating that additional modules are not only incorporated via attachment to the ascaroside scaffold, but may also get incorporated via linkages to other modules. Since the chemical structures of pasa#9 and other multimodular NDMMs appear to integrate input from several primary metabolic pathways, more detailed analysis of the biological functions of NDMMs and their biosyntheses may reveal how nematode metabolic state is transduced into a “language” of signaling molecules that transcend the dichotomy between “primary” and “secondary” metabolism. Given the suspected primary metabolic origin of the NDMMs, the discovery of additional structural diversity that does not require the nematode-specific sugar ascarylose may provide additional motivation for a comprehensive reanalysis of vertebrate metabolism and associated small-molecule signaling pathways.
Supplementary Material
Acknowledgments
Financial support for this project was provided by the Max Planck Society (R.J.S.) and the National Institutes of Health (GM088290 and AT008764 to F.C.S.). N.B. is grateful to the Cornell/Rockefeller/Sloan-Kettering Tri-Institutional Training Program in Chemical Biology fellowship.
Footnotes
Notes
The authors declare no competing financial interest.
Experimental procedures, supplemental figures, supplemental tables, and compound characterization. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Schroeder FC. Chem Biol. 2015;22:7. doi: 10.1016/j.chembiol.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ludewig AH, Izrayelit Y, Park D, Malik RU, Zimmermann A, Mahanti P, Fox BW, Bethke A, Doering F, Riddle DL, Schroeder FC. Proc Natl Acad Sci USA. 2013;110:5522. doi: 10.1073/pnas.1214467110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ludewig AH, Schroeder FC. Worm Book. 2013:1. doi: 10.1895/wormbook.1.155.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Reuss SH, Bose N, Srinivasan J, Yim JJ, Judkins JC, Sternberg PW, Schroeder FC. J Am Chem Soc. 2012;134:1817. doi: 10.1021/ja210202y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bose N, Ogawa A, von Reuss SH, Yim JJ, Ragsdale EJ, Sommer RJ, Schroeder FC. Angew Chem, Int Ed Engl. 2012;51:12438. doi: 10.1002/anie.201206797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu PJ. Worm Book. 2007. p. 1. [Google Scholar]
- 7.Lee SS, Schroeder FC. PLoS Biol. 2012;10:e1001307. doi: 10.1371/journal.pbio.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pungaliya C, Srinivasan J, Fox BW, Malik RU, Ludewig AH, Sternberg PW, Schroeder FC. Proc Natl Acad Sci USA. 2009;106:7708. doi: 10.1073/pnas.0811918106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butcher RA, Ragains JR, Clardy J. Org Lett. 2009;11:3100. doi: 10.1021/ol901011c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srinivasan J, von Reuss SH, Bose N, Zaslaver A, Mahanti P, Ho MC, O’Doherty OG, Edison AS, Sternberg PW, Schroeder FC. PLoS Biol. 2012;10:e1001237. doi: 10.1371/journal.pbio.1001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Artyukhin AB, Yim JJ, Srinivasan J, Izrayelit Y, Bose N, von Reuss SH, Jo Y, Jordan JM, Baugh LR, Cheong M, Sternberg PW, Avery L, Schroeder FC. J Biol Chem. 2013;288:18778. doi: 10.1074/jbc.C113.477000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dieterich C, Clifton SW, Schuster LN, Chinwalla A, Delehaunty K, Dinkelacker I, Fulton L, Fulton R, Godfrey J, Minx P, Mitreva M, Roeseler W, Tian H, Witte H, Yang SP, Wilson RK, Sommer RJ. Nat Genet. 2008;40:1193. doi: 10.1038/ng.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bento G, Ogawa A, Sommer RJ. Nature. 2010;466:494. doi: 10.1038/nature09164. [DOI] [PubMed] [Google Scholar]
- 14.Ragsdale EJ, Müller MR, Rödelsperger C, Sommer RJ. Cell. 2013;155:922. doi: 10.1016/j.cell.2013.09.054. [DOI] [PubMed] [Google Scholar]
- 15.Choe A, von Reuss SH, Kogan D, Gasser RB, Platzer EG, Schroeder FC, Sternberg PW. Curr Biol. 2012;22:772. doi: 10.1016/j.cub.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bose N, Meyer JM, Yim JJ, Mayer MG, Markov GV, Ogawa A, Schroeder FC, Sommer RJ. Curr Biol. 2014;24:1536. doi: 10.1016/j.cub.2014.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taggi AE, Meinwald J, Schroeder FC. J Am Chem Soc. 2004;126:10364. doi: 10.1021/ja047416n. [DOI] [PubMed] [Google Scholar]
- 18.Forseth RR, Schroeder FC. Curr Opin Chem Biol. 2011;15:38. doi: 10.1016/j.cbpa.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho JH, Coats SJ, Schinazi RF. Org Lett. 2012;14:2488. doi: 10.1021/ol300777p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogawa A, Sommer RJ. Science. 2009;326:944. doi: 10.1126/science.1183272. [DOI] [PubMed] [Google Scholar]
- 21.Park D, O’Doherty I, Somvanshi RK, Bethke A, Schroeder FC, Kumar U, Riddle DL. Proc Natl Acad Sci USA. 2012;109:9917. doi: 10.1073/pnas.1202216109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim K, Sato K, Shibuya M, Zeiger D, Butcher R, Ragains J, Clardy J, Touhara K, Sengupta P. Science. 2009;326:994. doi: 10.1126/science.1176331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGrath PT, Xu Y, Ailion M, Garrison JL, Butcher RA, Bargmann CI. Nature. 2011;477:321. doi: 10.1038/nature10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deutsch C, El Yacoubi B, de Crecy-Lagard V, Iwata-Reuyl D. J Biol Chem. 2012;287:13666. doi: 10.1074/jbc.M112.344028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Goot AT, Zhu W, Vázquez-Manrique RP, Seinstra RI, Dettmer K, Michels H, Farina F, Krijnen J, Melki R, Buijsman RC, Ruiz Silva M, Thijssen KL, Kema IP, Neri C, Oefner PJ, Nollen EAA. Proc Natl Acad Sci USA. 2012;109:14912. doi: 10.1073/pnas.1203083109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arczewska KD, Tomazella GG, Lindvall JM, Kassahun H, Maglioni S, Torgovnick A, Henriksson J, Matilainen O, Marquis BJ, Nelson BC, Jaruga P, Babaie E, Holmberg CI, Burglin TR, Ventura N, Thiede B, Nilsen H. Nucleic Acids Res. 2013;41:5368. doi: 10.1093/nar/gkt225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanvah S, Joseph J, Schuster GB, Barnett RN, Cleveland CL, Landman U. Acc Chem Res. 2009;43:280. doi: 10.1021/ar900175a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.