Significance
We report a method for dating ancient human samples that uses the recombination clock. To infer the age of ancient genomes, we take advantage of the shared history of Neanderthal gene flow into non-Africans that occurred around 50,000 y ago and measure the amount of “missing evolution” in terms of recombination breakpoints in the ancient genome compared with present-day samples. We show that this method provides age estimates that are highly correlated to radiocarbon dates, thus documenting the promise of this approach. By studying the linear relationship between the dates of Neanderthal admixture and the radiocarbon dates, we obtain, to our knowledge, the first direct estimate of the historical generation interval of 26–30 y.
Keywords: molecular clock, generation interval, ancient DNA, branch shortening
Abstract
The study of human evolution has been revolutionized by inferences from ancient DNA analyses. Key to these studies is the reliable estimation of the age of ancient specimens. High-resolution age estimates can often be obtained using radiocarbon dating, and, while precise and powerful, this method has some biases, making it of interest to directly use genetic data to infer a date for samples that have been sequenced. Here, we report a genetic method that uses the recombination clock. The idea is that an ancient genome has evolved less than the genomes of present-day individuals and thus has experienced fewer recombination events since the common ancestor. To implement this idea, we take advantage of the insight that all non-Africans have a common heritage of Neanderthal gene flow into their ancestors. Thus, we can estimate the date since Neanderthal admixture for present-day and ancient samples simultaneously and use the difference as a direct estimate of the ancient specimen’s age. We apply our method to date five Upper Paleolithic Eurasian genomes with radiocarbon dates between 12,000 and 45,000 y ago and show an excellent correlation of the genetic and 14C dates. By considering the slope of the correlation between the genetic dates, which are in units of generations, and the 14C dates, which are in units of years, we infer that the mean generation interval in humans over this period has been 26–30 y. Extensions of this methodology that use older shared events may be applicable for dating beyond the radiocarbon frontier.
Ancient DNA analyses have transformed research into human evolutionary history, making it possible to directly observe genetic variation patterns that existed in the past, instead of having to infer them retrospectively (1). To interpret findings from an ancient specimen, it is important to have an accurate estimate of its age. The current gold standard is radiocarbon dating, which is applicable for estimating dates for samples up to 50,000 y old (2). This method is based on the principle that, when a living organism dies, the existing 14C starts decaying to 14N with a half-life of ∼5,730 y (3). By measuring the ratio of 14C to 12C in the sample and assuming that the starting ratio of carbon isotopes is the same everywhere in the biosphere, the age of the sample is inferred. A complication is that carbon isotope ratios vary among carbon reservoirs (e.g., marine, freshwater, atmosphere) and over time. Thus, 14C dates must be converted to calendar years using calibration curves based on other sources, including annual tree rings (dendrochronology) or uranium-series dating of coral (2). Such calibrations, however, may not fully capture the variation in atmospheric carbon. In addition, contamination of a sample by modern carbon, introduced during burial or by handling afterwards, can make a sample seem younger than it actually is (2). The problem is particularly acute for samples that antedate 30,000 y ago because they retain very little original 14C.
Here, we describe a genetic approach for dating ancient samples, applicable in cases where DNA sequence data are available, as is becoming increasingly common (1). This method relies on the insight that an ancient genome has experienced fewer generations of evolution compared with the genomes of its living (i.e., extant) relatives. Because recombination occurs at an approximately constant rate per generation, the accumulated number of recombination events provides a molecular clock for the time elapsed or, in the case of an ancient sample, the number of missing generations since it ceased to evolve. This idea is referred to as “branch shortening” and estimates of missing evolution can be translated into absolute time in years by using an estimate of the mean age of reproduction (generation interval) or an independent calibration point such as human–ape divergence time.
Branch shortening has been used in studies of population history, for inferring mutation rates, and for establishing time scales for phylogenic trees in humans and other species (4, 5). It was first applied for dating ancient samples on a genome-wide scale by Meyer et al. (6), who used the mutation clock (instead of the recombination clock as proposed here) to estimate the age of the Denisova finger bone, which is probably older than 50,000 y, and has not been successfully radiocarbon dated (6). Specifically, the authors compared the divergence between the Denisova and extant humans and calibrated the branch shortening relative to human–chimpanzee (HC) divergence time. The use of ape divergence time for calibration, however, relies on estimates of mutation rate that are uncertain (7). In particular, recent pedigree studies have yielded a yearly mutation rate that is approximately twofold lower than the one obtained from phylogenetic methods (7). In addition, comparison with HC divergence relies on branch-shortening estimates that are small relative to the total divergence of millions of years, so that even very low error rates in allele detection can bias estimates. These issues lead to substantial uncertainty in estimated age of the ancient samples, making this approach impractical for dating specimens that are tens of thousands of years old, a time period that encompasses the vast majority of ancient human samples sequenced to date.
Given the challenges associated with the use of the mutation clock, here we explore the possibility of using a molecular clock based on the accumulation of crossover events (the recombination clock), which is measured with high precision in humans (8). In addition, instead of using a distant outgroup, such as chimpanzees, we rely on a more recent shared event that has affected both extant and ancient modern humans and is therefore a more reliable fixed point on which to base the dating. Previous studies have documented that most non-Africans derive 1–4% ancestry from Neanderthals from an admixture event that occurred ∼37,000–86,000 y before present (yBP) (9, 10), with some analyses proposing a second event (around the same time) into the ancestors of East Asians (11, 12). Because the vast majority of ancient samples sequenced to date were discovered in Eurasia (with estimated ages of ∼2,000–45,000 yBP), postdate the Neanderthal admixture, and show evidence of Neanderthal ancestry, we used the Neanderthal gene flow as the shared event.
The idea of our method is to estimate the date of Neanderthal gene flow separately for the extant and ancient genomes. Because the ancient sample is closer in time to the shared Neanderthal admixture event, we expect that the inferred dates of Neanderthal admixture will be more recent in ancient genomes (by an amount that is directly determined by the sample’s age) compared with the dates in the extant genomes. The difference in the dates thus provides an estimate of the amount of missing evolution: that is, the age of the ancient sample. An illustration of the idea is shown in SI Appendix, Fig. S1. An assumption in our approach is that the Neanderthal admixture into the ancestors of modern humans occurred approximately at the same time and that the same interbreeding events contributed to the ancestry of all of the non-African samples being compared. Deviations from this model could lead to incorrect age estimates. Our method is not applicable for dating genomes that do not have substantial Neanderthal ancestry, such as sub-Saharan African genomes.
To date the Neanderthal admixture event, we used the insight that gene flow between genetically distinct populations, such as Neanderthals and modern humans, introduces blocks of archaic ancestry into the modern human background that break down at an approximately constant rate per generation as crossovers occur (13–15). Thus, by jointly modeling the decay of Neanderthal ancestry and recombination rates across the genome, we can estimate the date of Neanderthal gene flow, measured in units of generations. Similar ideas have been used to estimate the time of admixture events between contemporary human populations (14–16), as well as between Eurasians and Neanderthals (9, 17). An important feature of our method is that it is expected to give more precise results for samples that are older because these samples are closer in time to the Neanderthal introgression event, thus it is easier to accurately estimate the time of the admixture event for them. Thus, unlike 14C dating, the genetic approach becomes more reliable with age and, in that regard, complements 14C dating.
Results
Model and Simulations.
Although a number of approaches exist for dating admixture when multiple genomes are available from the target (9, 14, 15), none are applicable to single diploid genomes (as required here for ancient specimens). Thus, we took advantage of our recent method introduced in Fu et al. (17), which measures the extent of covariance across pairs of alleles of putative Neanderthal ancestry: that is, sites where Neanderthals carry at least one derived allele (relative to chimpanzees) and all individuals in a panel of sub-Saharan Africans [which have little or no evidence of Neanderthal ancestry (18) carry the ancestral allele (17)]. We chose this ascertainment (referred to as “ascertainment 0”) because it minimizes the signal of background correlation, while amplifying the signal of Neanderthal ancestry (9). This statistic (referred to as the “single-sample statistic”) is expected to decay approximately exponentially with genetic distance, and the rate of decay is informative of the time of mixture (17). Assuming that the gene flow occurred instantaneously and by fitting a single exponential to the decay pattern, we estimate the average date of the Neanderthal gene flow in the target genome.
To assess the reliability of our approach, we performed coalescent simulations generating data for Neanderthals, present-day west Africans, and Europeans, with Europeans deriving 3% ancestry from Neanderthal gene flow that occurred between 100 and 2,500 generations ago (the range of time-depths relevant to our analysis) (SI Appendix, Note S2). Our simulations find that the estimated ages of Neanderthal gene flow are accurate when the admixture occurred between 100 and 1,500 generations ago. However, for samples older than 2,000 generations, our method underestimates the true ages. A downward bias was also observed for older admixture dates (∼2,500 generations) in simulations of complex demographic scenarios, including severe bottlenecks and recent expansion (17). To avoid complications due to the bias, we restricted our application of the single-sample statistic to ancient genomes where the expected date of Neanderthal gene flow is less than 1,500 generations. For older dates of Neanderthal admixture as expected in extant samples, where we have access to multiple genomes, we applied the “population-sample statistic” from ref. 9. This method measures the extent of admixture linkage disequilibrium (LD) by computing the covariance for each pair of ascertained single nucleotide polymorphisms (SNPs) and thus requires data from more than one diploid genome (making it inapplicable when only a single ancient genome is available). For extant genomes, we verified that the application of this statistic removes the bias observed in ref. 17 (SI Appendix, Note S2b).
To test the utility of our method for estimating the age of ancient genomes (and not just dating Neanderthal gene flow), we simulated data for both extant and ancient Europeans (sampled between 500 and 1,750 generations ago) and set the date of the shared Neanderthal gene flow to 2,000 generations ago (9). Our simulations show that the estimated ages of ancient genomes are accurate and that, as expected, the dates are more precise for older samples (SI Appendix, Note S2c).
Thus far, we have assumed that admixture occurred instantaneously as a single pulse of gene flow. However, in real populations, admixture could occur as multiple pulses or continuously over an extended period. To explore how this scenario affects our results, we performed simulations based on a similar setup as before, with the modification that the admixture occurred continuously for a period of either 10 or 500 generations, starting at 2,000 generations ago. Fitting a single exponential to the ancestry covariance patterns, we found that the estimated dates of admixture were intermediate between the start and end of the period of gene flow. The magnitude of the effect was similar for both ancient and extant samples and thus there is no reason to think that this complication would bias the date estimates (SI Appendix, Note S2d).
Accounting for Uncertainty in Parameters in Real Data.
Our simulations relied on the accurate modeling of the recombination rate across the genome. In applications to real data, we used the “shared” African American genetic map (“S map”) from ref. 8, which was inferred by combining information from the deCODE pedigree map in Europeans [based on ∼500,000 crossovers identified in ∼15,000 Icelandic meioses (19)] and the African American genetic map [based on ∼2.1 million crossovers detected using ancestry switch points observed between African and European ancestry in 30,000 unrelated African Americans (8)]. The S map, which focuses on the part of the landscape of recombination in African Americans that is shared with Europeans, is one of the most accurate genetic maps for Europeans currently available (8).
Despite the high resolution, even the best available genetic maps are not perfectly accurate at the short distances [tens of kilobases (kb)] that are relevant for some of our analysis. Notably, Sankararaman et al. (9) showed that the fine scale errors in the genetic map can underestimate the date of Neanderthal gene flow (9). To account for the errors in the genetic map, we applied the Bayesian “genetic map correction” developed by ref. 9 that estimates the map uncertainty by comparing the genetic distances in the S map with the crossover distribution observed in an independently generated map [in this case the deCODE pedigree dataset, based on 71,929 meioses (20)]. This correction is a function of the date of Neanderthal gene flow ( and a scalar parameter ( that is related to the precision of the genetic map (larger values of indicate a more precise map) (SI Appendix, Note S1). Using this approach, we estimated that the for the S map is 3,109 ± 308 per Morgan. The effect of this level of map uncertainty is likely to be minimal for ancient samples, in which the ancestry covariance extends to large distances (greater than hundreds of kb). In contrast, for extant samples where the blocks are an order of magnitude smaller, the resulting bias can be substantial, as shown in ref. 9. Thus, we applied the map correction separately for ancient and extant samples to obtain corrected dates of Neanderthal gene flow in generations (tn in generations).
To convert the dates of gene flow from generations to years (tn in years) while accounting for uncertainty in the generation interval, we assumed a uniform prior probability distribution on the generation interval between 25 and 33 y (21–24). The mean generation interval in ancient humans is not known and is likely to have some cultural variability but (21, 24) showed that, at least in modern humans over a wide variety of cultures and degrees of economic development, the mean age of reproduction falls within this range. The difference in the dates of gene flow in ancient and extant genomes translates to an estimate of branch shortening or the age of the ancient genome (tc).
Case Studies.
To illustrate the utility of our method, we applied our approach to ancient genomes that have radiocarbon dates of at least 10,000 y. This threshold was chosen so that the expected date of Neanderthal admixture in the ancient genome is less than 1,500 generations (thus not affected by the bias seen in simulations) and that the difference between the dates of admixture in extant and ancient samples is significant (beyond statistical error). We broadly matched the ancestry of the ancient and extant samples, comparing ancient Eurasian samples with northern European samples from the 1000 Genomes project (designated “CEU”) (25).
Using the population-sample statistic for SNPs matching ascertainment 0 (SI Appendix, Note S1) and the genetic map correction described above, we estimated that the Neanderthal admixture in CEU occurred between 1,569 and 1,700 generations or 40,510–54,454 yBP (95% credible interval). This date is within the previously published estimate of 37,000–86,000 yBP (most likely range of 47,000–65,000 yBP) based on a different ascertainment scheme and genetic map correction (9). The broader confidence interval in ref. 9 is extremely conservative, reflecting an attempt to account for biases observed in simulations of complex demographic scenarios. Our simulations indicate that the use of the population-sample statistic and ascertainment 0 should not provide biased dates under demographic scenarios that are applicable to Europeans, and thus we believe that the additional bias correction is too aggressive (SI Appendix, Note S2). If our assumptions are valid, dates in the range of 40,510–54,454 y ago are important because they suggest that the main Neanderthal interbreeding with modern humans may have occurred in the context of the Upper Paleolithic expansion of modern humans, rather than at earlier times (26).
We applied our method to estimate the age of five ancient samples. Because many of these samples (Clovis, Mal’ta1, Kostenki14, Oase1) were sequenced to medium depth coverage, we could not reliably call heterozygous sites, and thus we restricted analysis to pseudohomozygous genotypes [where we sampled the single majority allele observed in the reads mapped to each site (SI Appendix, Note S1)]. Below and in SI Appendix, Table S1, we discuss the dating results for each sample using the S map and ascertainment 0. In SI Appendix, Note S3, we show that our results are robust to other genotype-calling approaches, SNP ascertainments, and comparison to high coverage west Eurasian samples from Simons Genome Diversity Panel (instead of 1000 Genomes CEU).
Clovis.
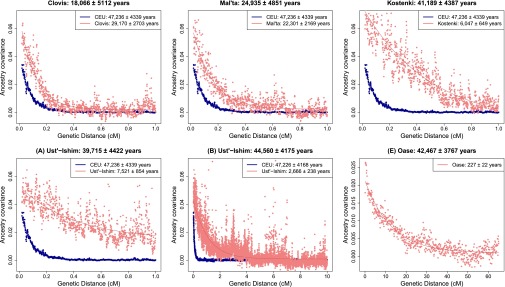
The Clovis genome from North America sequenced to an average coverage of 14.0× has a radiocarbon date of 12,556–12,707 (95% confidence) calibrated years BP (calBP) (27). Using the single-sample statistic, we estimated that the Neanderthal gene flow in Clovis occurred 29,170 ± 2,703 (one SE) y before he lived. Considering the difference between the dates of Neanderthal admixture in Clovis and CEU provides an estimate for its age of 18,066 ± 5,112 y. After accounting for uncertainty, this estimate is consistent with its radiocarbon date (Fig. 1).
Fig. 1.
Estimated age of ancient genomes. Estimated dates of Neanderthal gene flow in extant Europeans shown in blue and ancient Eurasians shown in pink (for details, see SI Appendix, Note S1). Estimated ages of the ancient genome (mean ± SE) are shown in the titles. For Ust’-Ishim, we show two plots: (Lower Left, marked A) single exponential fit up to the genetic distance of 1 cM and (Lower Center, marked B) single exponential fit up to the genetic distance of 10 cM. For Oase1 (Lower Right), we show single exponential fit up to the genetic distance of 65 cM and bin size of 0.1 cM. We do not show CEU because the analysis was based on a different bin size and maximum distance.
Mal’ta1.
The Mal’ta1 individual dated as 23,891–24,423 calBP old was sampled in south-central Siberia and was sequenced to an average coverage of 1.0× (28). We applied the single-sample statistic and estimated that the Neanderthal gene flow occurred 22,301 ± 2,169 y before he lived. In turn, this difference translates into an estimated age of 24,935 ± 4,851 y, which is consistent with its radiocarbon date (Fig. 1).
Kostenki14.
The Kostenki14 (K14) genome from European Russia sequenced to an average coverage of 2.8× has a radiocarbon date of 36,262–38,684 calBP (29). Applying our inference procedure, we estimated that the Neanderthal gene flow in K14 occurred 6,047 ± 649 y before he lived. This date is not consistent with the recently published estimate of ∼15,000 y before he lived (29). However, the details of method used in ref. 29 are unpublished so we cannot evaluate what the source of the discrepancy might be. Considering the difference with CEU provides an estimated age of 41,189 ± 4,387 y (Fig. 1), which is statistically consistent with its radiocarbon date.
Ust’-Ishim.
The Ust’-Ishim (UI) genome from western Siberia was sequenced to 42-fold coverage and has been dated twice by 14C to be ∼43,210–46,880 calBP (17). Because the coverage for this sample is high enough, we were able to make reliable heterozygous calls and thus we used diploid genotypes for the inference. We note that the dates based on diploid and pseudohomozygous calls are concordant (SI Appendix, Note S3). Applying the single-sample statistic, we estimated that the date of the Neanderthal admixture was 7,521 ± 854 y before the individual lived, consistent with the date reported in ref. 17 (which used a different genetic map and did not correct for genetic map errors). Considering the difference with CEU provides an age estimate of 39,715 ± 4,422 y, similar to its radiocarbon date (Fig. 1).
Unlike the previously discussed ancient genomes, UI contains many Neanderthal segments longer than 1 cM, which are poorly fit by the exponential distribution (because the intercept at 1 cM is substantially greater than 0) (Fig. 1). A plausible explanation for this pattern is that UI may not have received all its Neanderthal ancestry from a single pulse of gene flow—or even the same event that affected the extant European populations, as assumed by our model (SI Appendix, Fig. S1). To explore this possibility, we reran the method up to longer genetic distances until the intercept of exponential was close to 0. Applying our analysis up to 10 cM, we estimated that the Neanderthal gene flow occurred 47,226 ± 4,168 yBP in CEU and 2,666 ± 238 y before UI lived. The difference provides an estimated age of UI as 44,560 ± 4,175 y (Fig. 1), which is also consistent with its radiocarbon date.
The inference of different dates of gene flow in UI depending on the genetic distance threshold used (unlike in CEU) is consistent with our hypothesis that there may have been at least two pulses of Neanderthal admixture into the ancestors of UI, with the curve fitted up to 1 cM mostly sensitive to the older events and the curve fitted up 10 cM more sensitive to recent events. To test formally whether UI has a history of multiple Neanderthal genetic inputs, we applied the likelihood ratio test (LRT) described in ref. 30 that analyzes whether a single exponential or a sum of exponentials provides a better fit to the observed ancestry covariance patterns. This approach found overwhelming support for the two-pulse model of Neanderthal admixture (P < 10−20). Indeed, visualization of the putative Neanderthal ancestry blocks present in the UI genome exhibits a broadly bimodal pattern, with some regions containing greater than 5- to 10-Mb-long blocks (17), which would not be expected unless some of the gene flow occurred recently. By explicitly fitting a model of two Neanderthal gene flow events, we estimated that the admixture events occurred 6,600 ± 618 and 1,258 ± 113 y before UI lived (SI Appendix, Fig. S3). Because it was not clear which of these events might be shared with extant Europeans, we estimated the age of the UI genome based on each of the two admixture events separately, obtaining 40,626 ± 4,214 and 45,968 ± 4,170 y (SI Appendix, Fig. S3). Both of these estimates are consistent with the radiocarbon date of the sample.
To test whether we can replicate these patterns in simulation, we generated data for a 1,750-generation-old ancient sample that had a similar history as UI (two pulses of Neanderthal gene flow that occurred at 2,000 and 1,800 generations ago, where the older pulse was shared with the extant samples). Fitting a single exponential to the ancestry decay patterns in the ancient genome provided a date of Neanderthal admixture that was intermediate between the date of the first and second pulse of mixture. Similar to UI, this sample contained many Neanderthal segments that were longer than 1 cM. Thus, we ran the analysis to longer distances and then applied LRT to confirm the history of multiple pulse of admixture (P < 10−20). By fitting a sum of exponentials, we reliably inferred the dates of the two admixture events (SI Appendix, Note S2e).
Oase1.
The age of the Oase1 genome from Romania has been estimated to be ∼37,000–42,000 calBP by radiocarbon dating (31). Because the specimen contained tiny amounts of highly contaminated human DNA, it was not feasible to whole genome sequence this individual. Instead, this sample was captured on panels of known SNPs, including the Archaic panel (panel 4), where at least one Neanderthal allele differs from the majority allele in a panel of 24 West African Yoruba samples (31). This ascertainment, however, contains SNPs where Yoruba is derived and archaic samples contain the ancestral allele. Such sites will likely amplify some background LD, biasing the dates of Neanderthal admixture. Thus, we removed these SNPs from our analysis. Using this ascertainment for CEU, we estimated that Neanderthal gene flow occurred 42,694 ± 3,767 yBP in CEU, which is consistent with the previous estimate. Based on the recommendation in ref. 31, we ran our single-sample statistic for Oase1 up to 65 cM (where the intercept of exponential is almost 0) (SI Appendix, Note S1). We estimated that the Neanderthal gene flow in Oase1 occurred 227 ± 22 y before he lived, similar to estimates in ref. 31. Considering the difference with CEU provided an estimated age of 42,467 ± 3,767 y, consistent with the radiocarbon date of this specimen. Oase1, like UI, has a bimodal distribution of Neanderthal ancestry segments (31). Applying LRT provided strong support for the two-pulse model of Neanderthal admixture (P < 10−12). By explicitly fitting a model of two Neanderthal gene flow events, we estimated that the admixture occurred 2,012 ± 385 y and 164 ± 14 y before he lived, translating to age estimates of 40,682 ± 3,787 and 42,530 ± 3,767 y, respectively (SI Appendix, Figs. S2 and S3); both of these dates are consistent with the radiocarbon dates of this specimen. To provide further confidence that our restriction to SNPs on the Archaic panel provides reliable estimates, we reestimated the age of K14 and UI for the same set of SNPs as Oase1. We estimated the age of K14 and UI (maximum distance of 10 cM as described earlier) as 39,855 ± 3,917 and 39,122 ± 3,785 y, respectively, similar to our genome-wide estimates. These results document how sparse genome-wide data are sufficient to provide reliable age estimates.
Robustness of Age Estimates.
A central assumption of our method is that recombination has not changed over time and across populations. Recombination rates, however, are known to have evolved over the course of human evolution as reflected in the observation that the alleles of PRDM9, which are the major determinants of recombination hotspots in humans, are changing rapidly (32). Present-day human hotspots seem to have been active for ∼10% of time since divergence from chimpanzees (∼650,000–1.3 million y) (33), suggesting that our assumption is likely to be valid over the time scale of interest here. Nonetheless, some variation in hotspot usage is known to exist across human populations that separated ∼50,000–100,000 y ago (8). Ideally, then, our analysis should be based on a map that reflects the average recombination rate over the time since Neanderthal introgression in the ancestry of each sample being analyzed. Because such data are unavailable and unlikely to become available for ancient samples, we verified that our inferences are robust to the choice of existing maps by repeating the analysis with an African-American map (8) that includes hotspots in Africans as well as shared hotspots between Africans and Europeans, and the Oxford CEU LD map (34) that reflects historical recombination rates in Europeans valid over tens of thousands of years. Although there is significant variation across maps [as indicated by differences in map uncertainty (] (SI Appendix, Note S1), the age estimates based on the three different maps are qualitatively similar (within two standard errors) (SI Appendix, Table S2).
Another concern is that previous studies have shown that Neanderthal ancestry proportion varies across chromosomes, with unexpectedly large regions devoid of any Neanderthal ancestry and correlation in Neanderthal ancestry proportion to B-statistic (a measure of linked selection) (12, 35), implying a role for natural selection in removing Neanderthal-derived alleles from the modern human gene pool (36). The B-statistic or B-score measures the reduction in diversity levels at a site due to linked selection, with smaller values implying higher selective constraint in the region (37). To assess the effect of natural selection, we estimated the age of each sample by removing all ascertained SNPs in regions that are documented targets of natural selection, including conserved elements across primates and coding regions in humans (38). We observed that the age estimates were similar to the results reported earlier (SI Appendix, Table S3). We also studied the effect on the estimated date of Neanderthal admixture as a function of the B-score. Because many samples have limited coverage, we divided the genome into two bins: regions with low B (0–4) and regions with high B (5–9). We observed that the dates of Neanderthal admixture for all samples, except K14, were qualitatively similar in both bins. For the lower coverage samples K14 and Mal’ta1, the results are less reliable because the fit was very noisy, given limited data. In addition, we observed no systematic difference in dates in the two B-score bins (P = 0.5, based on permutation of labels in the two bins) (SI Appendix, Table S4). We conclude that, within the limits of our resolution, the effect of selection on our dates is not significant.
Historical Generation Interval in Humans.
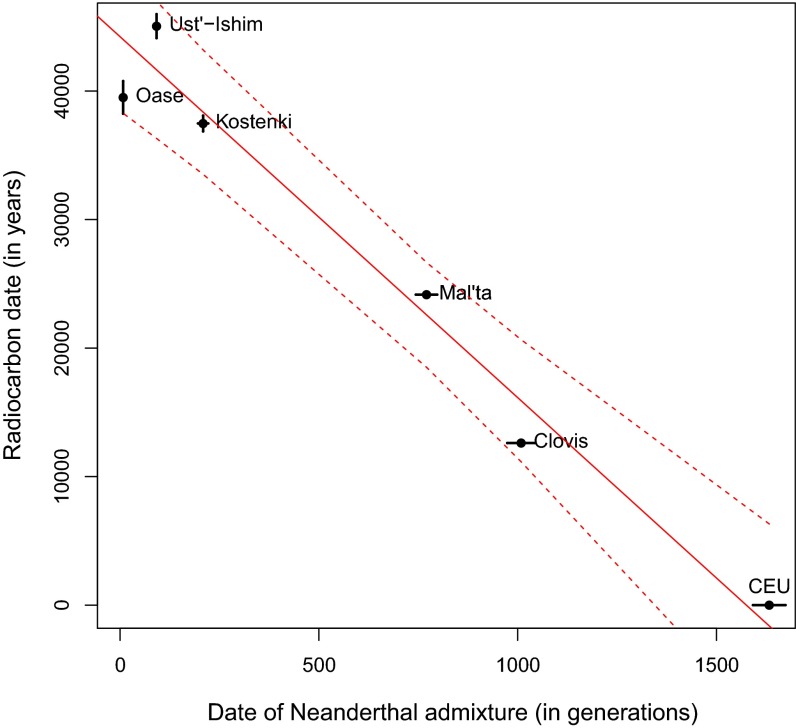
A feature of our method is that it estimates dates in generations (because it is based on the recombination clock) whereas 14C dates are determined in years. By fitting a linear model to the relationship between these dates, we can jointly estimate the generation interval (reflected by the slope) and the time of the shared pulse of Neanderthal admixture in modern humans (reflected by the intercept). To estimate these parameters while accounting for the uncertainty in dates, we implemented a Bayesian approach using importance sampling (39) (SI Appendix, Note S4). Under the simplifying assumption that males and females have the same generation interval and assuming it has remained constant since the Neanderthal introgression, we estimated that the historical generation interval in humans is 28.1 ± 0.7 y and that the shared pulse of Neanderthal admixture occurred 44,301 ± 591 y ago (Fig. 2), consistent with the date in present-day West Eurasians. This result is robust to choice of priors of the slope and intercept and assumptions about the complex history of Neanderthal admixture in UI and Oase1 (SI Appendix, Note S4).
Fig. 2.
Estimation of historical generation interval. Relationship between dates of Neanderthal admixture (in generations) and radiocarbon dates (in years). We show mean ± SE of dates for each sample. To estimate the generation interval and time of shared Neanderthal admixture event, we fit a line and use importance sampling to infer the mean and uncertainty of the slope and intercept (SI Appendix, Note S4).
Discussion
We have developed a genetic approach for dating ancient human specimens that is applicable for dating ancient non-African samples that share a history of Neanderthal admixture with extant non-Africans. By studying the linear relationship between the dates of Neanderthal admixture and the radiocarbon dates, we infer that the historical generation interval in humans is 26–30 y, consistent with direct estimates of the current sex-averaged generation intervals from genealogical surveys and pedigree studies (21, 22, 24), suggesting that the generation interval has not changed substantially over the past 45,000 y. To our knowledge, this is the first direct estimate of the human generation interval deep in the past.
Comparison with Radiocarbon Dating.
We show that our results are consistent with radiocarbon dates for all studied specimens (with correlation of 0.98, P value = 0.002). Although radiocarbon dates are in general more precise than genetic age estimates, our method is complementary to 14C dating in that it uses independent information based on the molecular clock. In addition, although in this study, we have focused on Neanderthal admixture as our calibration point for dating, there is nothing unique about this event from the perspective of dating, and, in fact, other shared LD-generating events such as other introgression events (e.g., the Denisova admixture into the ancestors of Southeast Asians and Oceanians) (6) or founder events (e.g., out-of-Africa migration) (1) could be used as alternative calibration points through extensions of the methodology reported here. Importantly, if one were to use an older calibration point than the date of Neanderthal gene flow, genetic data could allow estimation of dates for skeletal remains that are beyond the limits of radiocarbon dating but for which sequence data exist, such as the Altai/Mezmaiskaya Neanderthals (18), or the three Denisova samples (6, 40) that are too small or too old to have enough preserved carbon for radiocarbon dating. A limitation of our method is that it is not applicable for dating samples that do not share a history of Neanderthal gene flow with non-Africans, such as the recently published ancient Ethiopian genome (41). In addition, unlike 14C dating, the genetic method is unstable for very young samples that are less than 10,000 y old. This problem reflects the fact that, for a single genome with an old admixture date, it is hard to reliably identify very short segments of Neanderthal ancestry. However, the use of a more recent calibration point should make it possible to obtain accurate estimates of the age of young ancient genomes.
Outlook.
In this paper we have estimated the age of ancient samples by comparing the dates of Neanderthal admixture to extant samples, which is challenging and the main reason for the large uncertainty of our age estimates. As more ancient samples become available, it should be possible to estimate the age of ancient genomes by building a calibration entirely from other genomes for which both radiocarbon dates and genetic dates are available (similar to Fig. 2), and interpolating the age of the studied genome based on its inferred date of Neanderthal admixture. Preliminary results for predicting the age of ancient samples in this way gives promising results (SI Appendix, Fig. S5).
Materials and Methods
We applied our method to estimate the age of five ancient samples: Clovis (27), Mal’ta1 (28), Kostenki14 (29), Ust’-Ishim (17), and Oase1 (31). To estimate the age () of each ancient genome, we quantified the difference in dates of Neanderthal admixture in an ancient genome () (estimated using the single-sample statistic) (17) and extant CEU genomes () (estimated using the population-sample statistic) (9). We estimated SEs based on the Bayesian framework described in ref. 9. For details, see SI Appendix, Note S1.
Supplementary Material
Acknowledgments
We thank Claude Bherer and Mark Lipson for helpful discussions about characterizing the uncertainty in the genetic maps. We thank Bridget Alex, Bence Viola, Katerina Douka, Thomas Higham, Svante Pääbo, and David Pilbeam for comments on the manuscript. We thank Thomas Higham for helpful discussions about the biases and reliability of radiocarbon dates. P.M. was supported by the National Institutes of Health (NIH) under Ruth L. Kirschstein National Research Service Award F32 GM115006-01. S.S. was supported in part by NIH Grants 5K99GM111744-02 and 4R00GM111744-03. Q.F. was supported by National Natural Science Foundation of China Grant L1524016 and Chinese Academy of Sciences Discipline Development Strategy Project Grant 2015-DX-C-03. M.P. was supported by US National Institutes of Health Grant R01 GM83098. D.R. and N.P. were supported by US National Science Foundation HOMINID Grant BCS-1032255 and US National Institutes of Health Grant GM100233. D.R. is a Howard Hughes Medical Institute Investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1514696113/-/DCSupplemental.
References
- 1.Pickrell JK, Reich D. Toward a new history and geography of human genes informed by ancient DNA. Trends Genet. 2014;30(9):377–389. doi: 10.1016/j.tig.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bronk Ramsey C. Radiocarbon dating: Revolutions in understanding. Archaeometry. 2008;50(2):249–275. [Google Scholar]
- 3.Godwin H. Half-life of radiocarbon. Nature. 1962;195(4845):984. [Google Scholar]
- 4.Fu Q, et al. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr Biol. 2013;23(7):553–559. doi: 10.1016/j.cub.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stadler T, Yang Z. Dating phylogenies with sequentially sampled tips. Syst Biol. 2013;62(5):674–688. doi: 10.1093/sysbio/syt030. [DOI] [PubMed] [Google Scholar]
- 6.Meyer M, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338(6104):222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ségurel L, Wyman MJ, Przeworski M. Determinants of mutation rate variation in the human germline. Annu Rev Genomics Hum Genet. 2014;15:47–70. doi: 10.1146/annurev-genom-031714-125740. [DOI] [PubMed] [Google Scholar]
- 8.Hinch AG, et al. The landscape of recombination in African Americans. Nature. 2011;476(7359):170–175. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sankararaman S, Patterson N, Li H, Pääbo S, Reich D. The date of interbreeding between Neandertals and modern humans. PLoS Genet. 2012;8(10):e1002947. doi: 10.1371/journal.pgen.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328(5979):710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim BY, Lohmueller KE. Selection and reduced population size cannot explain higher amounts of Neandertal ancestry in East Asian than in European human populations. Am J Hum Genet. 2015;96(3):454–461. doi: 10.1016/j.ajhg.2014.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vernot B, Akey JM. Resurrecting surviving Neandertal lineages from modern human genomes. Science. 2014;343(6174):1017–1021. doi: 10.1126/science.1245938. [DOI] [PubMed] [Google Scholar]
- 13.Chakraborty R, Weiss KM. Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc Natl Acad Sci USA. 1988;85(23):9119–9123. doi: 10.1073/pnas.85.23.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hellenthal G, et al. A genetic atlas of human admixture history. Science. 2014;343(6172):747–751. doi: 10.1126/science.1243518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moorjani P, et al. The history of African gene flow into Southern Europeans, Levantines, and Jews. PLoS Genet. 2011;7(4):e1001373. doi: 10.1371/journal.pgen.1001373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loh P-R, et al. Inferring admixture histories of human populations using linkage disequilibrium. Genetics. 2013;193(4):1233–1254. doi: 10.1534/genetics.112.147330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu Q, et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature. 2014;514(7523):445–449. doi: 10.1038/nature13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prüfer K, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505(7481):43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong A, et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467(7319):1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- 20.Kong A, et al. Common and low-frequency variants associated with genome-wide recombination rate. Nat Genet. 2014;46(1):11–16. doi: 10.1038/ng.2833. [DOI] [PubMed] [Google Scholar]
- 21.Fenner JN. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am J Phys Anthropol. 2005;128(2):415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- 22.Helgason A, Hrafnkelsson B, Gulcher JR, Ward R, Stefánsson K. A populationwide coalescent analysis of Icelandic matrilineal and patrilineal genealogies: Evidence for a faster evolutionary rate of mtDNA lineages than Y chromosomes. Am J Hum Genet. 2003;72(6):1370–1388. doi: 10.1086/375453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amster G, Sella G. Life history effects on the molecular clock of autosomes and sex chromosomes. Proc Natl Acad Sci USA. 2015;113(6):1588–1593. doi: 10.1073/pnas.1515798113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun JX, et al. A direct characterization of human mutation based on microsatellites. Nat Genet. 2012;44(10):1161–1165. doi: 10.1038/ng.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. The 1000 Genomes Project Consortium (2010) A map of human genome variation from population-scale sequencing. Nature 467(7319):1061–1073. [DOI] [PMC free article] [PubMed]
- 26.Higham T, et al. The timing and spatiotemporal patterning of Neanderthal disappearance. Nature. 2014;512(7514):306–309. doi: 10.1038/nature13621. [DOI] [PubMed] [Google Scholar]
- 27.Rasmussen M, et al. The genome of a Late Pleistocene human from a Clovis burial site in western Montana. Nature. 2014;506(7487):225–229. doi: 10.1038/nature13025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raghavan M, et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature. 2014;505(7481):87–91. doi: 10.1038/nature12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seguin-Orlando A, et al. Paleogenomics. Genomic structure in Europeans dating back at least 36,200 years. Science. 2014;346(6213):1113–1118. doi: 10.1126/science.aaa0114. [DOI] [PubMed] [Google Scholar]
- 30.Moorjani P, et al. Genetic evidence for recent population mixture in India. Am J Hum Genet. 2013;93(3):422–438. doi: 10.1016/j.ajhg.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu Q, et al. An early modern human from Romania with a recent Neanderthal ancestor. Nature. 2015;524(7564):216–219. doi: 10.1038/nature14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327(5967):836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lesecque Y, Glémin S, Lartillot N, Mouchiroud D, Duret L. The red queen model of recombination hotspots evolution in the light of archaic and modern human genomes. PLoS Genet. 2014;10(11):e1004790. doi: 10.1371/journal.pgen.1004790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310(5746):321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 35.Sankararaman S, et al. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507(7492):354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Juric I, Aeschbacher S, Coop G. The strength of selection against Neanderthal introgression. 2015 doi: 10.1371/journal.pgen.1006340. bioRxiv:030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McVicker G, Gordon D, Davis C, Green P. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 2009;5(5):e1000471. doi: 10.1371/journal.pgen.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai JJ, Macpherson JM, Sella G, Petrov DA. Pervasive hitchhiking at coding and regulatory sites in humans. PLoS Genet. 2009;5(1):e1000336. doi: 10.1371/journal.pgen.1000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bishop C. 2007. Pattern Recognition and Machine Learning, Information Science and Statistics (Springer, New York), corrected 2nd Ed.
- 40.Sawyer S, et al. Nuclear and mitochondrial DNA sequences from two Denisovan individuals. Proc Natl Acad Sci USA. 2015;112(51):15696–15700. doi: 10.1073/pnas.1519905112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Llorente MG, et al. Ancient Ethiopian genome reveals extensive Eurasian admixture in Eastern Africa. Science. 2015;350(6262):820–822, and erratum (2016) 351(6275):aaf3945. doi: 10.1126/science.aad2879. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.