Significance
During 150 million years of mammalian evolution, the membrane-bound mannose 6-phosphate receptor evolved high-affinity binding loops for insulin-like growth factor 2. It remains unknown whether this highly evolved ligand interaction is optimal, and whether it can be further evolved. We addressed these questions using a mutation and selection strategy that incorporated surface display and protein structure. Multiple mutations of all the binding loops were identified and improved affinity by 100-fold when combined, yet retained IGF2 specificity. Structurally, IGF2 surface interactions with binding loops were reshaped, indicating that binding site evolution could not be predicted. High IGF2 affinity binding domains could selectively inhibit IGF2-dependent cell signaling, and may be applied in therapeutic IGF2 targeting in cancer.
Keywords: growth factor receptor, protein evolution, insulin-like growth factor 2, binding kinetics, biological therapy
Abstract
Among the 15 extracellular domains of the mannose 6-phosphate/insulin-like growth factor-2 receptor (M6P/IGF2R), domain 11 has evolved a binding site for IGF2 to negatively regulate ligand bioavailability and mammalian growth. Despite the highly evolved structural loops of the IGF2:domain 11 binding site, affinity-enhancing AB loop mutations suggest that binding is modifiable. Here we examine the extent to which IGF2:domain 11 affinity, and its specificity over IGF1, can be enhanced, and we examine the structural basis of the mechanistic and functional consequences. Domain 11 binding loop mutants were selected by yeast surface display combined with high-resolution structure-based predictions, and validated by surface plasmon resonance. We discovered previously unidentified mutations in the ligand-interacting surface binding loops (AB, CD, FG, and HI). Five combined mutations increased rigidity of the AB loop, as confirmed by NMR. When added to three independently identified CD and FG loop mutations that reduced the koff value by twofold, these mutations resulted in an overall selective 100-fold improvement in affinity. The structural basis of the evolved affinity was improved shape complementarity established by interloop (AB-CD) and intraloop (FG-FG) side chain interactions. The high affinity of the combinatorial domain 11 Fc fusion proteins functioned as ligand-soluble antagonists or traps that depleted pathological IGF2 isoforms from serum and abrogated IGF2-dependent signaling in vivo. An evolved and reengineered high-specificity M6P/IGF2R domain 11 binding site for IGF2 may improve therapeutic targeting of the frequent IGF2 gain of function observed in human cancer.
The functional evolution of proteins is largely considered to occur by chance, frequently because of unpredictable and specific events that confer a structure-based change in function sufficient for subsequent selection or “gain of fitness” (1). One such evolutionary biochemical example is the initial acquisition and subsequent gain of affinity between the insulin-like growth factor 2 (IGF2) ligand and a single domain of a nonsignaling mannose 6-phosphate (M6P)/IGF2 receptor (IGF2R) (domain 11). The structural and functional basis of this evolutionary path, which has occurred over 150 million years of mammalian evolution, has been reported previously (2). The questions that we address in the present work are whether the IGF2:domain 11 interaction has reached an optimal state in the context of IGF2 activation of signaling receptors and in the ligand clearance function of M6P/IGF2R, and how far can we extend the binding interaction in terms of structural, biophysical, and functional properties.
Functionally, and unlike products of other mammalian imprinted genes, domain 11 is unusual because it specifically evolved to bind to an evolutionary conserved IGF2 ligand with high affinity (3–5). After binding, clearance of extracellular IGF2 occurs by receptor internalization followed by IGF2 degradation in the lysosomal compartment (3). This mechanism supports the parental conflict theory of imprinting (6), because loss of M6P/IGF2R function results in IGF2 ligand oversupply and embryonic and placental overgrowth (7–10). Moreover, IGF2:domain 11 binding underpins M6P/IGF2R function as a tumor suppressor, and is the basis for the application of domain 11 as a soluble IGF2 ligand antagonist (11–14). The relative affinity of IGF2 for the signaling receptors IGF1R and isoform A of insulin receptor (IR-A) is slightly lower (2–20 nM) (15) than the affinity for full-length M6P/IGF2R (1–2 nM) (16).
IGF-binding proteins (IGFBPs) 1–6 can achieve higher affinity (between 0.01 and 20 nM), but bind IGF1 and IGF2 and are proteolytically labile (17, 18). Thus, IGF2 affinity for domain 11 may have reached an optimum that balances both in vivo cellular function (i.e., IGF2 must dissociate in late endosomes) and selectivity for IGF2 over IGF1. Excess IGF2, as occurs in during embryonic development and in tumors, appears to overwhelm the normal clearance function of M6P/IGF2R, to promote growth via IGF1R/IR-A signaling receptor activation. In this instance, alternative mechanisms may limit free IGF2, including increased supply of IGFBPs and release of soluble M6P/IGF2R that may trap the free ligand (13, 19). Full exploitation of selective IGF2 targeting in a wide range of cancers, using either antibodies or soluble receptors, remains an unmet need because of the lack of stable, high-affinity, and high-specificity therapeutic agents (20).
The IGF2-binding site of domain 11 in humans consists of a hydrophobic pocket centered on the CD loop, surrounded by polar and charged residues in the AB, FG, and HI loops that complement surface charge on IGF2 (Fig. 1A) (2). The exception to this rule is an otherwise unfavorable charge–charge interaction between E1544 on the AB loop and D23 on IGF2 (4, 21, 22). Domain 13 indirectly interacts with the AB loop of domain 11, breaking this interaction in the multiple-domain receptor, and contributes to the stability of the complex by decreasing the “off-rate” (koff) of the IGF2 interaction (21). Mutations in the AB loop replace the effect of domain 13, increase the affinity for IGF2 (2, 22), and facilitate subsequent determination of a solution structure of the 24.2-kDa complex of IGF2 with domain 11AB3 that incorporated three mutations of the AB loop (also known as AB3 or E1544K, K1545S, and L1547V; KD = 15 nM) (2). The structural basis of the IGF2:domain 11AB3 interaction occurs with a relatively fixed conformation of the CD loop on complex formation. The AB loop moves to accommodate IGF2 helix 1, which bears the critical recognition residues T16 and F19 (Fig. 1 A and B), and the FG loop repositions between helices 2 and 3 of IGF2 to accommodate the burial of a third important IGF2 residue, L53 (16, 21). The final complex forms a series of complementary hydrophobic surfaces and supports a range of H-bonding and salt-bridging interactions (2).
Fig. 1.
Sequence and structure of the IGF2 binding site of domain 11 and in vitro forward evolution. (A, Left) Ribbon representation of the solution structure of the complex between domain 11AB3 (gray) and IGF2 (pink). Surface representations are shown for the AB (blue), CD (green), FG (yellow), and HI (dark pink) loops. (A, Right) Close-up of the IGF2-binding site and interacting loops of domain 11AB3 and the main three-pronged interaction involving T16, F19, and L53 of IGF2. (B) Sequence of domain 11 secondary structure and binding loops. Mutations of domain 11WT and their effects on affinity for IGF2 are shown below the sequences. (C) Strategy for forward evolution. Mutant library generated by combining random mutagenesis in P. pastoris yeast surface display with flow cytometry screen (green) after validation of biotin-IGF2 binding to Flag tag domain 11 protein on the yeast surface. Mutant clones are selected using a high-throughput SPR screen and, with NMR informed site-directed mutagenesis (purple), further validated (yellow) for IGF2 sensitivity and specificity using high-sensitivity SPR of purified recombinant protein.
Although the IGF1 ligand is highly homologous to IGF2, particularly those residues involved with binding IGFBPs and signaling receptors, T16 of IGF2 confers specificity over IGF1 (where it is replaced by an alanine) for domain 11, most likely through packing interactions that precisely define the spacing of F19 and L53 (2). NMR structures of domain 11 from opossum, echidna, zebrafish, and chicken all share similar β-barrel topology, yet IGF2 does not bind to domain 11 in birds and fish. Binding coevolved with mammalian evolution, as primitive mammals (monotremes) bind IGF2 with 10-fold lower affinity compared with human domain 11WT (KD = 250–400 nM vs. 40–60 nM) (2, 23–25). Thus, acquisition of the IGF2:CD loop hydrophobic interaction appears to be the main structural change during initial binding evolution, a chance event attributed to acquisition of an exon splicing enhancer that altered the amino acid coding of the CD loop (2). As mammalian domain 11 evolved, the surface surrounding the IGF2-binding region provided by AB, FG, and HI loops displayed a loss of charged residues and increased volume and hydrophobicity, combined with the correct shape complementarity (2).
To address the extent to which human IGF2:domain 11 affinity can further evolve, we evaluated a multistep approach to forward evolution. We developed in vitro-directed evolution with yeast cell surface display, combined with structural-directed mutagenesis and surface plasmon resonance (SPR) affinity selection. The directed evolution of proteins to generate adaptive and positive selected functions is a powerful strategy but is known to be dependent on a number of essential factors, including the design and selectivity of the screening strategy, the epistatic effects of permissive existing and coexisting mutations on protein stability, and the practical limitations of generating comprehensive library diversity in bacteria (26–28). We identified previously unidentified and independent gain-of-binding function mutations of all of the binding loops of domain 11, but with retained binding specificity for IGF2. By also introducing structural informed mutations to those selected in the first round of directed evolution of ligand-interacting residues, we iteratively combined individually identified loop mutants to generate domain 11 variants with even higher additive affinities, slower dissociation rates, and improved thermodynamic properties. These previously unidentified high-affinity mutant combinations generate novel structural mechanisms of ligand-receptor binding site interactions, and are the basis of specific soluble ligand traps for IGF2 that antagonize IGF2 function in vivo.
Results
Domain 11 Yeast Surface Display and Selection of High-Affinity AB Loop Mutants.
For the directed evolution screen, we combined domain 11 SPR binding kinetics (22) with an Agα1 yeast surface display system in Pichia pastoris (29). Mutants of an N-terminal Flag domain 11 were expressed, and cell surface expression was quantified with an anti-Flag antibody and an Alexa Fluor 488-labeled secondary antibody (Fig. 1C). After binding of biotinylated human mature IGF21–67 to live yeast expressing domain 11 controls, affinity for domain 11WT, domain 11AB3, and a nonbinding mutant domain 11I1572A were quantified with Alexa Fluor 647-labeled streptavidin (Fig. 1C, Upper Right). In the first round of selection, yeasts expressing mutant domain 11s were flow-sorted on binding to a low concentration of IGF21–67 (5 nM). Gates were based on the binding to domain 11WT and domain 11I1572A (nonbinding mutant), and thresholds were based on domain 11E1544K or domain 11AB3 (higher IGF21–67 affinity; Fig. 1C) (22). After culture of sorted clones, mutants were subcloned into a soluble protein expression vector. Then transfected cells were plated, and single clones were screened in a 96-well format using a Biacore 3000 analyzer (GE Healthcare) with koff (y = y0 ⋅ e−k off ⋅ x) determination (Fig. 1C).
Yeasts expressing soluble mutant his6 domain 11s with slow koff were expanded and protein-purified, and SPR validation was performed using the more sensitive Biacore T200 analyzer (GE Healthcare). The screen was applied to mutagenesis of the AB loop (1542YSEKGL1547), where the E1544K gain-of-function mutation was incorporated (1542YSKKGL1547) (SI Appendix, Table S1). We identified an AB loop with five mutations (domain 11AB5) and with a 10-fold higher IGF21–67 affinity compared with domain 11WT (KD = 5.07 nM vs. 46–64 nM; SI Appendix, Table S2), and no detectable binding to IGF1. The five AB loop mutations (1542YAKGWG1547) in domain 11AB5 generated a very different combination of amino acids than those identified in domain 11AB3 (1542YSKSGV1547) (Fig. 2A).
Fig. 2.
Comparison of domain 11WT, domain 11AB3, and domain 11AB5 high-resolution structures. (A) Sequence comparison of the AB loop from domain 11WT and mutants. (B) High-resolution NMR structural ensembles of the lowest twenty energy models for free domain11AB5 (dark blue; PDB ID code 2M6T) and bound to IGF2 (light blue; PDB ID code 2M68). Structural statistics are provided in in SI Appendix, Table S4. (C) Key residues of the free forms (dark gray) of domain 11AB3 (Left; PDB ID code 2L2A) and domain 11AB5 (dark blue; Right) and corresponding IGF2- bound forms of domain 11AB3 (light gray; PDB ID code 2L29) and domain 11AB5 (light blue). In domain 11AB5 W1546 closes into IGF2 F19 (pink, modeled from 2L29), whereas in domain 11AB3, G1546 (highlighted by red arrow) moves outward and indicates the large conformational change of the AB loop on complexation. (D) Lipari–Szabo model-free S2 parameters showing flexible AB, BC, and CD loops for domain 11WT, shown by S2 values <0.7. These loop regions are more rigid in domain 11AB5, consistent with NOE, R1, and R2 data (SI Appendix, Figs. S1–S4).
In parallel, P. pastoris cultures of nonmutagenized domain 11AB3 controls were screened for IGF21–67 binding, and a surprising number of clones were obtained with slow koff. These selected clones arose as a consequence of random mutation during bulk culture. No mutations were selected in the β-barrel structure, but two spontaneous loop mutants were identified that bound IGF21–67 with high affinity (SI Appendix, Table S3). The first of these was an FG loop mutant (P1597H), with a fourfold reduction in koff associated with a 10-fold improvement in affinity compared with domain 11AB3 (KD = 1.23 nM vs. 15.3 nM). The second mutant was in the hydrophobic CD loop (Q1569R) with enhanced affinity (KD = 2.30 nM). Although mutations within domain 11 that altered IGF2 affinity were expected, a mutant of the CD loop was unexpected because of the importance of hydrophobicity and shape complementarity during evolution of the binding site (2), and because Q1569A had little impact on IGF2 affinity (22).
Structure of the Domain 11AB5 Mutant Informs Selection of Non-AB Loop Mutants.
To determine how the newly identified AB loop mutations in domain 11AB5 altered the IGF2-binding site, high-resolution NMR structures of domain 11AB5 were determined in both the free and IGF21–67-bound forms (Fig. 2B) (2, 30). Domain 11AB5 retained the characteristic domain 11 flattened β-barrel fold (SI Appendix, Tables S4 and S5), which conserves the primary IGF21–67-binding site (Fig. 2A and SI Appendix, Fig. S5A) (2); however, the AB loop adopted a significantly different structure in domain 11AB5. The orientation of the side chain of W1546, which substitutes for G1546, was not well defined in the ensemble of structures, but appeared to reorient adjacent aromatic residues Y1542 and F1567, displacing the AB loop away from the IGF2-binding region. In the domain 11AB3 structure, this portion of the AB loop was hinged around the small G1546 residue, and on complex formation, the AB loop moved to accommodate IGF21–67 (Fig. 2 C and D) (2). In the free form of domain 11AB5, the AB loop more closely resembled the bound form, and no equivalent rearrangement was necessary (Fig. 2 C and D).
Recording of 15N-relaxation data for unbound domain 11WT and domain 11AB5 at two field strengths (600 and 900 MHz) revealed an overall increased rigidity in the AB loop of domain 11AB5 (Fig. 2D and SI Appendix, Figs. S1–S4). The binding of IGF21–67 to domain 11AB5 did draw the W1546 aromatic side chain inward to the binding site, however. This side chain is well defined in the ensemble of NMR structures and, along with aromatic side chains Y1542 and F1567, interacts with the primary binding epitope, F19, from IGF21–67 (2, 21). K1544 was observed to form a salt bridge with D23 of IGF2 in the complex with domain 11AB3 and most likely fulfilled a similar role here. The K1545S mutation in domain 11AB3 was replaced in domain 11AB5 by K1545G, resulting in a loss of hydrogen-bonding capability to Q18 of IGF21–67 from the serine OH. Thus, among the five mutations in the AB loop of domain 11AB5, our data implicate the structural importance of the specific G1546W mutation for the gain in affinity (Fig. 2 C and D).
The Robetta server (robetta.bakerlab.org/alascansubmit.jsp) was used to perform interface alanine scanning mutagenesis on the IGF21–67:domain 11AB3 complex to identify additional “hotspot” loop residues important for complex formation. Mutations of these residues in domain 11AB5 and SPR affinity are shown in Fig. 3A and listed in SI Appendix, Table S2. Compared with the AB loop, the FG loop undergoes a pronounced structural change on complexation in domain 11AB5, suggesting the potential for further optimization of this sequence beyond P1597H. Mutations of FG loop residues K1601R, S1602H, S1602N, and G1603K all exhibited up to a twofold improvement in affinity (koff), suggesting that the AB and FG loops can function in combination. Introduction of structurally predicted site-directed mutations in the HI loop improved affinity only slightly, although a number of single point mutations could be introduced that appeared to slow the koff (e.g., Q1632A and K1631W) (Fig. 4A and SI Appendix, Table S4). Finally, introduction of W1546F into the AB loop reduced the koff by twofold, yet retained a similar overall KD (4.35 nM) as a result of a reduced kon value.
Fig. 3.
Effects of single and combined mutations of the IGF2-binding site in human domain 11AB3 and domain 11AB5. (A) Heat map of IGF2 binding to single loop point mutants on domain 11AB5 (Left) and domain 11AB3 (Right) backgrounds. (Upper) koff. (Lower) KD. The scale is log2 of the domain 11reference/domain 11mutant ratio, where domain 11reference is either domain 11AB5 or domain 11AB3. The reference value is shown in green, the increase in affinity (i.e., decrease in KD or koff) is shown in red, and the decrease in affinity (i.e., increase in KD or koff) is shown in blue. (B) Effect of combinations of loop mutants on IGF2 koff compared with domain 11AB3. (C) Effect of combined loop mutants on IGF2-binding free energy (ΔG°) compared with domain 11AB3. ΔΔG° is calculated as ΔΔG° = ΔG° domain 11mutant − ΔG° domain 11AB3 = RTln (KD domain 11mutant/KD domain 11AB3). Additive and nonadditive contributions of single point mutations are shown with a boxed solid line and dashed line, respectively. Raw data are provided in SI Appendix, Tables S2, S5, and S6. (D) Thermodynamic profiles of the IGF2 binding of domain 11 mutants. Data fitting of the temperature dependence of the dissociation constant according to the van’t Hoff equation is shown. (E) Comparison of koff between domain 11AB3 and domain 11AB5 with combined mutations.
Fig. 4.
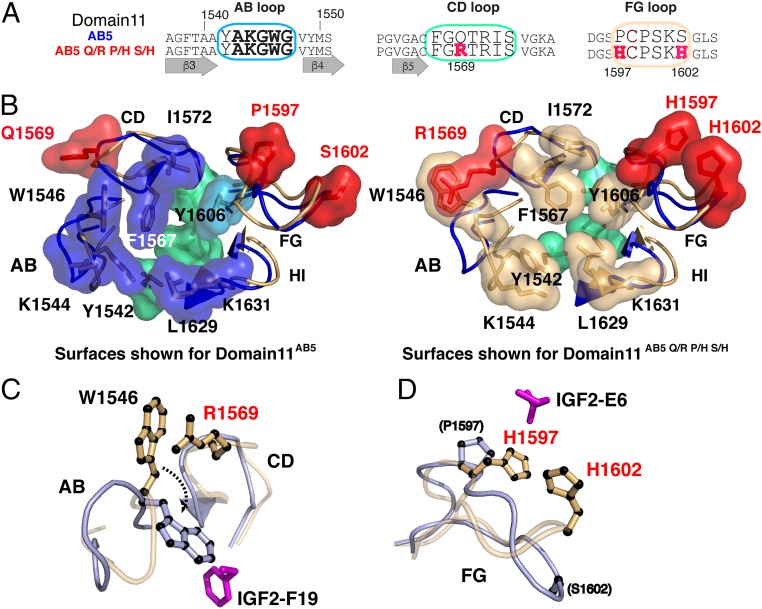
Comparison of domain 11AB5 and domain 11AB5-Q1569R P1597H S1602H high-resolution structures. (A) Sequence comparison of the AB, CD, and FG loop from domain 11AB5-Q1569R P1597H S1602H and domain 11AB5. (B) Superposition of free domain 11AB5 (blue loops) and domain 11AB5-Q1569R P1597H S1602H (gold loops) with residue surfaces shown for domain 11AB5 (Left) and domain 11AB5-Q1569R P1597H S1602H (Right). Hydrophobes within or surrounding the binding site are shown in blue (domain 11AB5) or gold (domain 11AB5-Q1569R P1597H S1602H) with underlying foundation residues in green (V1574, L1629, and L1636). The mutation Q/R 1569, P/H 1597, and S/H 1602 surfaces are shaded in red. (C and D) Comparison of the AB and CD loop orientation (C) and the FG loop (D) between the free form of domain 11AB5-Q1569R P1597H S1602H (orange) and domain11AB5 in the IGF2-bound form (light blue). The reorientation of W1546 required for domain 11AB5-Q1569R P1597H S1602H to achieve the same bound conformation is highlighted by the dashed arrow in C.
Directed Evolution of Domain 11 HI Loop Ligand Interacting Residues Selects a Higher-Affinity Mutant.
The affinity of IGF2 for domain 11AB5 was at least twofold higher than that of the highest-affinity mutant detected previously. Because we had already discovered single point mutations of the FG and CD loops that enhanced domain 11AB5 affinity, we systematically determined the potential for the remaining interacting HI loop in domain 11AB5 to specifically stabilize the IGF2-binding interaction. We first introduced a library of mutants for four ligand proximal residues of the HI loop (1630DKQT1633) into domain 11WT, so that we could discriminate clear affinity gains compared with domain 11AB5, and screened >1 × 105 fully representative clones representing every possible amino acid combination (SI Appendix, Table S1). After flow sorting and subcloning of 2.2 × 105 mutants into the soluble expression system, >1,000 clones were screened by SPR (Fig. 1C and SI Appendix, Table S3). Only one mutant among several false-positives was identified (1630HFQS1633) as having a slower koff than domain 11WT.
Combinations of Domain 11 Loop Mutants Improves Affinity by 100-Fold with Retained IGF2 Specificity.
The first round of directed evolution combined with structural predictions had evolved domain 11 with respect to higher IGF21–67 affinity in vitro with previously unidentified and independent AB, CD, FG, and HI loop mutants. Moreover, the specific mutations in the CD and FG loops on AB loop background of either of domain 11AB5 or domain 11AB3 modified the koff and overall affinity by at least twofold (Fig. 3A and SI Appendix, Tables S2 and S3). We determined whether the individual loop mutants identified by either the screens or by structural predictions could be combined to generate even higher binding affinity, but with retained IGF21–67 specificity. In particular, CD loop (Q1596Rscreen), FG loop (P1597Hscreen/Kstructure, S1602Hstructure, G1603Kstructure), and HI loop (K1631Wstructure, 1630HFQS1633/screen) mutations were selected as candidates. Successive addition of the majority of mutations resulted in additive at best, rather than synergistic, affinity effects, with improved binding kinetics on both domain 11AB3 and domain 11AB5 backgrounds (Fig. 3 B and C and SI Appendix, Table S6). The addition of the HI loop mutants K1631W and 1630HFQS1633 often failed to result in stable expressed protein. The introduction of multiple mutations (up to 11) into domain 11AB5 also appeared less well tolerated in terms of protein yields than similar mutations in domain 11AB3, indicating potential AB loop-dependent epistatic effects on protein folding and stability (SI Appendix, Table S6).
Thermodynamic parameters reflected the changes in affinity for IGF21–67 between domain 11WT, AB loop mutants domain 11AB3 and domain 11AB5, double-loop mutants domain 11AB5-Q1569R (AB + CD loops) and domain 11AB5-P1597H S1602H (AB + FG loops), and the triple-loop mutant domain 11AB5-Q1569R P1597H S1602H (AB + CD + FG loops, also referred to as domain 11AB5 Q/R P/H S/H, and later abbreviated as domain 11AB5-RHH) (22, 31) (Fig. 3D and SI Appendix, Fig. S6). The temperature dependence of the dissociation constant according to the van’t Hoff equation showed that the interaction of IGF21–67 with domain 11WT appeared to be enthalpically driven (ΔH <0), with a relatively small and unfavorable entropic contribution (−TΔS >0) (Fig. 3D). Mutations in the AB loop appeared to have different effects. Domain 11AB5 retained a similar unfavorable thermodynamic profile as domain 11WT, with an unfavorable entropic contribution. The entropic contribution appeared to be more favorable in domain 11AB3, as we had previously shown for the E1544K AB loop mutant (22). The addition of mutations to domain 11AB5 via CD or FG loops introduced more favorable changes in the entropic contribution and koff, similar to domain 11AB3 (Fig. 3 D and E). Further mutations in the CD and FG loops of domain 11AB5 introduced overall improvement in the free energy of binding (ΔGo), with varied entropic and enthalpic compensation, consistent with the affinity improvement of the triple-loop mutant domain 11AB5-Q1569R P1597H S1602H (Fig. 3 D and E).
Four higher-affinity and -specificity IGF21–67-binding versions of domain 11 were identified with at least a 100-fold higher affinity compared with domain 11WT and 5- to 10-fold higher affinity compared with the full-length M6P/IGF2R. Within the domain 11AB5 background, domain 11AB5-Q1569R P1597H S1602H (KD = 0.65 nM, pH 7.4; KD = 0.71 nM, pH 6.5) and domain 11AB5-Q1569R P1597H G1603K (KD = 0.87 nM, pH 7.4; KD = 0.69 nM, pH 6.5) were identified, independent of pH between 6.5 and 7.4 (SI Appendix, Tables S6 and S7). For the domain 11AB3 background, domain 11AB3-Q1569R P1597H S1602H (KD = 0.74 nM, pH 7.4; KD = 0.40 nM, pH 6.5) and domain 11AB3-Q1569R P1597H S1602H G1603K (KD = 0.78 nM, pH 7.4; KD = 0.43 nM, pH 6.5) were selected because the affinity (i.e., slower koff) was observed to increase at lower pH.
X-Ray Crystallographic Structure of Domain 11AB5-Q1569R P1597H S1602H That Combines Eight Loop Mutations.
The X-ray crystallographic structure of one of the highest-affinity domains, domain 11AB5-Q1569R P1597H S1602H, was solved to 2.8-Å resolution. This showed several unexpected structural rearrangements of the hydrophobic core compared with domain 11AB5 (SI Appendix, Table S8). In domain 11AB5, W1546 adopted multiple conformations, but these were bounded by a neighboring surface, defined by Q1569 (Fig. 4 A and B). This was reversed in domain 11AB5-Q1569R P1597H S1602H, where R1569 lined the IGF2-binding site and engaged W1546 in a cation–π interaction, directing the indole ring away from the interface (Fig. 4B). R1569 and W1546 were separated by 3.5 Å, consistent with the expected range for this type of interaction (2–6.0 Å) (32, 33). Similarly, K1544 was directed away from the IGF2-binding site in domain 11AB5, but rotated to point toward it in domain 11AB5-Q1569R P1597H S1602H. Y1542 (AB loop) and F1567 (CD loop) showed minor conformational changes that opened a deeper pocket at the F19-binding site than that observed in the IGF2:domain 11AB3 complex. Other residues in these loops adopted comparable conformations.
The FG loop of domain 11AB5-Q1569R P1597H S1602H is well defined, and the side chains of both mutated histidine residues were visible, revealing a significant rearrangement of this portion of the loop compared with domain 11AB5 (Fig. 4B). Whereas P1597 and S1602 did not interact in either the free form or the bound form of domain 11AB5, the histidine residues associated closely in domain 11AB5-Q1569R P1597H S1602H to produce a positively charged patch. Finally, the HI loop backbone and side chains superimposed well between domain 11AB5 and domain 11AB5-Q1569R P1597H S1602H. In terms of the complex with IGF21–67, we speculate that packing of W1546 into the hydrophobic core as seen with domain 11AB5 would require breaking the cation–π interaction with R1569. This would allow W1546 to interact with F19 and R1569 to form new electrostatic interactions with D15 and D23 of IGF2 (Fig. 4C). Second, the positively charged patch formed from H1597 and H1602 on the FG loop may interact with the N terminus of IGF21–67 (e.g., E6) (Fig. 4D); however, S1600 and K1601 are also reoriented favorably for interaction with IGF2 D52, another key residue for binding on IGF2 (16). The positions of I1572 and Y1606 do not occlude binding of T16 based on the domain 11AB3:IGF21–67 complex and may contribute to the retained specificity for IGF21–67 over IGF1. These observations will require confirmation in future X-ray crystallography studies of the IGF21–67:domain 11AB5-Q1569R P1597H S1602H complex (Fig. 4D).
Development of Specific and Soluble IGF2 Antagonists Based on Domain 11AB5 Mutants.
Because domain 11AB5 and the combined variant domain 11AB5-Q1569R P1597H S1602H had affinities exceeding those of endogenous M6P/IGF2R, IGF1R, and IR-A, we evaluated whether these domains could be translated into the basis of improved IGF2 antagonists or ligand traps that function in vivo (14). Human IgG1 Fc fusion domain 11AB5 and human optimized IgG2 Fc domain 11AB5-Q1569R P1597H S1602H (domain 11AB5-RHH hereinafter) were bulk-produced as initial optimized IGF2 antagonists, along with an IgG1 Fc domain 11I1572A control with mutation of the CD loop known to impair IGF2 binding (SI Appendix, Fig. S7).
SPR binding kinetics revealed an affinity and specificity for IGF21–67 close to that of the single soluble domain 11AB5 when IgG1 Fc domain 11AB5 was immobilized using an anti-human Fc antibody (KD = 3.89 nM) (Fig. 5A and SI Appendix, Table S9). IGF21–67 binding by IgG2 Fc fusion domain 11AB5-RHH also showed a significantly reduced koff (0.0014 s−1) and slower kon, to yield a similar overall KD as that of the respective single domain (Fig. 5A and SI Appendix, Table S9).
Fig. 5.
Fc domain 11AB5/AB5-RHH binds IGF2 isoforms with high affinity and selectivity. (A) SPR sensorgrams of the interactions among Fc domain 11AB5, Fc domain 11AB5-RHH, and Fc domain 11I1572A (control non-IGF2 binding) with IGF1 (red), IGF21–67, IGF21–104, and IGF21–156. Recombinant IGF1 and the different IGF2 forms were injected at concentrations ranging from 64 nM to 0.25 nM over Fc domain 11AB5 immobilized on a CM5 surface by antibody capture. (B) Fc domain 11AB5 or Fc domain 11I1572A pull-down assay of different recombinant IGF2 isoforms. (C) Fc domain 11 pull-down assay as in B of the different IGF2 isoforms produced by tumor cell lines. Supernatants of the HCC cell lines Hep3B and Huh7 and of the NIH 3T3 control cell line expressing pro-IGF2R104A were incubated with Fc domain 11AB5 or with Fc domain 11I1572A as a control.
IGF2 in serum, and commonly in tumors, occurs as a range of pro-IGF2 isoforms attributed to incomplete processing of the E-domain by proprotein convertases (34). Binding kinetics of recombinant pro-IGF2 isoforms (mature IGF21–67, 7.5 kDa; pro-IGF21–104, 11 kDa; and pro-IGF21–156, 17 kDa) to Fc domain 11AB5 and Fc domain 11AB5-RHH, including specificity of binding relative to IGF1 were determined. We observed a higher relative affinity of pro-IGF21–104 to Fc domain 11AB5-RHH than to Fc domain 11AB5, and, surprisingly, binding of Fc domain 11I1572A to both pro-IGF21–156 and to a lesser extent pro-IGF21–104 (Fig. 5A and SI Appendix, Table S9). Binding of domain 11AB5 and domain 11AB5-RHH was independent of the Fc fragment, given that the same affinity to pro-IGF2 isoforms was obtained when immobilized human Fc alone was used in the control flow cell. No binding to IGF1 was observed (Fig. 5A). Solution pull-downs confirmed the specific capture of recombinant IGF21–67, pro-IGF21–104, and pro-IGF21–156 (note the multiple forms) by Fc domain 11AB5, and binding of Fc domain 11I1572A to pro-IGF21–156 (Fig. 5B).
To further test the functional activity of IGF binding, we obtained culture supernatants from human hepatocellular carcinoma cell lines (HuH7 and Hep3B) and a Tet-inducible NIH 3T3 line that expressed big IGF2 isoforms and IGF2R104A, a mutation that impairs processing of IGF21–156 (34), respectively (Fig. 5C). Solution pull-downs from cell line supernatants confirmed Fc domain 11AB5 binding to larger pro-IGF2 glycosylated isoforms, and domain 11I1572A particularly to IGF21–156, indicating the potential for additional binding interactions of the E-domain of IGF2 to domain 11 independent of the main IGF2-binding surface.
Trapping IGF2 with Fc Domain 11AB5-RHH (IGF2-TRAP) Inhibits IGF2- Dependent Signaling in Vivo.
To test whether the domain 11AB5 backbone could function as an IGF2 antagonist or ligand trap in vivo, we tested the acute metabolic signaling effects of exogenous IGF21–67-induced hypoglycemia in mice. Coinjection of IGF2 and Fc domain 11AB5 or Fc domain 11AB5-RHH at a 1:1 or 1:0.5 molar ratio, respectively, resulted in abrogation of the IGF21–67-induced hypoglycemia, an effect not detected with the Fc domain 11I1572A non–IGF2-binding control (Fig. 6A and SI Appendix, Fig. S8). When the IGF2:Fc domain 11 ratio was increased, the advantages of a higher-affinity mutant were evident. A molar ratio of 1:0.23 significantly counteracted IGF2-induced hypoglycemia in the case of domain 11AB5-RHH, but not with Fc domain 11AB5. Moreover, preloading or postloading of Fc domain 11AB5 also abrogated IGF21–67-induced hypoglycemia (SI Appendix, Fig. S9).
Fig. 6.
Fc domain 11AB5 and Fc domain 11AB5-RHH (IGF2-TRAP) inhibit IGF2 signaling in vivo. (A) Fc domain 11AB5 and Fc domain 11AB5-RHH (IGF2-TRAP) abrogate an IGF21–67-induced hypoglycemia in a mouse model. Mice were anesthetized (t = −30 min), and blood glucose levels were allowed to stabilize for 30 min (expressed relative to this blood glucose level). Subsequently (t = 0 min), the mice received 1 mg kg-1 IGF21–67 alone (n = 4), or premixed with Fc domain 11AB5 or Fc domain 11AB5 RHH at a molar ratio of 1:1 (n = 3) (P = 0.0133, two-way ANOVA with Bonferroni post-test) or 1:0.5 (n = 3) (P = 0.0023, two-way ANOVA with Bonferroni post-test), respectively. With a molar ratio of 1:0.23, Fc domain 11AB5 RHH is a more efficient IGF2 antagonist than Fc domain 11AB5 (P = 0.0026, two-way ANOVA with Bonferroni post-test). (B) IGF2-TRAP reduces IGF2-dependent xenograft growth (SKNMC-IGF21–67). There were 5 × 106 cells per injection site in CD-1 nude mice, with a single infused concentration of IGF2-TRAP (40 mg kg−1 per week) (green; n = 7; n = 2 injection error, n = 1 unexplained death) or PBS control (blue; n = 10). (P = 0.002, Wilcoxon test across all time points). (C) IGF2-TRAP administration resulted in reduced levels of serum IGF2 independent of IGF1, GH, and IGFBP level (day 28; control PBS, n = 10 out of 10; IGF2-TRAP, n = 6 out of 7). (D) Oncology drug synergistic screen in SKNMC-FLAG-IGF2104 with IGF2-TRAP. Heat maps show top-ranked drugs (30 shown; blue for synergy, red for antagonism) with low P values (RP method) of the interaction score at 1 µM and the IGF2-TRAP (three replicates, R1–R3). (E) Validation dose–response curves for PI3kinase inhibitors (PF-04691502 and pictilisib) in the presence (green) and absence (blue) of the IGF2-TRAP. IC50 values are shown. A leftward shift indicates synergism (P < 0.0001 comparing the IC50 of drug alone vs. drug + IGF2-TRAP, F test). Asterisks indicate the concentrations at which synergistic interactions occur (Q >1.15). (F) IGF2-TRAP modifies the molecular distribution of IGF2 in human serum. Normal (Left) and NICTH (Right) serum samples were fractionated in a gel filtration column at neutral pH, alongside a molecular weight calibration marker, before and after incubation and depletion with IGF2-TRAP–loaded protein G beads. Elution fractions were evaluated by Western blot analysis.
Fc domain 11AB5 inhibited cell growth of Hep3B and Huh7 cells in culture, but growth of these cell lines as xenografts failed in CD-1 nude mice in vivo (SI Appendix, Figs. 10 and 11). An alternative autocrine-IGF21–67 tumor model was developed using the Ewing sarcoma cell line (SKNMC) selected for IGF2 dependency (35). Growth of retroviral-transformed SKNMC cells expressing constitutive IGF21–67 and a luciferase reporter was inhibited by Fc domain 11AB5-RHH in culture, and inhibited IGF2 downstream signaling (SI Appendix, Fig. S12). SKNMC-IGF21–67 xenografts are predicted to be dependent on autocrine-IGF21–67 supply, given that adult mice do not express IGF2. IgG2 Fc domain 11AB5-RHH purified from CHO cells was infused using an osmotic minipump before SKNMC-IGF21–67 cell line injection. Reduced xenograft growth rates in mice were observed with Fc domain 11AB5-RHH infusion (Fig. 6B and SI Appendix, Fig. S13A), with induced regions of tumor cell death (SI Appendix, Fig. S13B) that resulted in a reduced bioluminescence signal:volume ratio (SI Appendix, Fig. S13 C and D). Circulating IGF21–67 was readily detected in serum after 28 d of SKNMC-IGF21–67 tumor growth, but was less evident in treated Fc domain 11AB5-RHH mice, and without alteration in either total serum IGF1, GH, or IGFBP levels (Fig. 6C).
The genomic complexity of cancer reduces any supposition that a drug targeting IGF2 alone in established tumors will lead to durable consequences, yet there may be specific circumstances in which dual targeting with other agents may unmask a synergistic or conditional dependency on IGF2. A number of candidate additive and synergistic lethality interactions were observed in SKNMC-IGF2104 treated with Fc domain 11AB5-RHH and an extended oncology compound library (222 agents plus 77 additional agents from the DTP Approved Oncology Drug Set containing oncology-specific compounds that have been tested in man; www.tdi.ox.ac.uk/home). The high-throughput screen compared the effects of the oncology drugs in the presence and absence of a fixed concentration of Fc domain 11AB5-RHH (IGF2-TRAP) that resulted in partial reduction in cell viability (10–20%) at the assay endpoint of 72 h (SI Appendix, Fig. S14 A and B). Evaluation of the metrics of raw data (Pearson’s r scores) indicated good replicate correlation, with average r ≥0.92 for all plates with an average inhibitory effect of the IGF2-TRAP alone of ∼10%. Drugs showing P < 0.05 in the rank product method applied to their interaction score were selected. A number of hits were identified, including two independent PI3 kinase inhibitors, PF-04691502 (P = 0.011) and pictilisib (P = 0.017) (Fig. 6D). These were further validated by comparison of the dose–response curve in both the presence and absence of IGF2-TRAP (Fc domain 11AB5-RHH) (Fig. 6E). The IC50 for PF-04691502 shifted from 0.42 µM to 92 nM in the presence of the IGF2-TRAP, and from 1.1 µM to 0.14 µM in the case of pictilisib, both of which are significant based on an F-test. Such 10-fold differences following combination with IGF2-TRAP suggest that antagonizing IGF2 signaling can reveal synergistic activity that is likely to be clinically advantageous, at least in reducing the dosage of these PI3 kinase inhibitors.
Changes in the molecular distribution of circulating IGF2 on IGF2-TRAP treatment were analyzed by neutral size-fractionation of human serum (Fig. 6F). In adult human serum, Western blot analysis of the fractions reveals that IGF2 is mainly the mature lower molecular mass isoform and is present predominantly in a ternary complex with IGFBP3 and the acid labile subunit (ALS) (∼150 kDa), and to a lesser extent in a binary complex with IGFBP3 (∼40 kDa), with free IGF2 below the limit of detection. In this sample, IGF2-TRAP depleted IGF2 in the binary complex, and also partially decreased the amount of IGF2 in the ternary complex (Fig. 6F, Lower Left). Pull-downs confirmed that the depleted IGF2 specifically bound IGF2-TRAP. In a second experiment, serum from a patient with non–islet cell tumor-associated hypoglycemia (NICTH) secondary to a GIST tumor was used. Western blot analysis of the fractions showed that IGF2 was present with additional higher molecular weight isoforms, predominantly in a binary complex. IGF2-TRAP depleted both lower and higher molecular weight isoforms of IGF2, and pull-downs confirmed specific binding to both isoforms (Fig. 6F, Lower Right). These data suggest that IGF2-TRAP can target higher molecular weight pro-IGF2 isoforms associated with human cancer and NICTH (IGF2 syndrome).
Discussion
Functional Evolution of Affinity Without Negative Consequences on Specificity.
A comparison of binding kinetics in the context of high-resolution NMR structures of domain 11WT, domain 11AB3, and the new structure domain 11AB5 showed that structural perturbations within the binding site are localized mainly to the AB loop (2). Although the G1546W mutation in domain 11AB5 introduces a bulky side chain into the AB loop and the binding site, residues in the neighboring CD and FG loops appear to be only minimally perturbed in this structure; however, the exciting structural and functional observation is that affinity for IGF2 can be increased by further ligand-interacting surface and rim (non-AB loop) mutations that confer reconfiguration of the binding surface complementarity, rather than acting indirectly through non–ligand-interacting surface sites remote (40 Å) from the interface (36). The predominant polar and electrostatic mutations are consistent with the potential for both ligand-loop stabilization of existing hotspot interactions through second sphere residues, because the specific effects are selected based mainly on koff and the structural complementarity surrounding F19, L53, and T16 of IGF2. Importantly, this supports the maintained selectivity through T16 of IGF2, thus retaining specificity over IGF1.
The thermodynamic payoffs between entropy (predominantly solvent exclusion) and enthalpy (charge attraction) of the interaction between IGF2 and domain 11 mutants showed some consistent differences, including the effects of CD and FG loop mutations. Mutation of the AB loop in the domain 11AB3 would be consistent with rigidification and improved solvent exclusion, but this effect was unmasked only in domain 11AB5 with respect to thermodynamic terms after the introduction of additional mutations in the CD and FG loops (domain 11AB5-RHH). Thus, the overall improvement in Gibbs free energy of the interaction (ΔGo) appeared to require combinations of mutations that reduced the entropic barrier (i.e., improved solvent exclusion), offsetting the negative cooperativity of the AB loop mutations. Ultimately, the combinatorial gain-of-function mutations in the AB, CD, and FG loops in domain 11AB5-RHH yield a subnanomolar IGF2 affinity that is exceptional for non-Ig scaffolds (37). This scaffold includes the unexpected abstraction of AB loop W1546 via R1569 from the IGF2-binding site that significantly modifies the IGF2 interaction, as well as the E6 IGF2 interaction with the FG intraloop histidine–histidine bridge. These molecular mechanisms never could have been predicted even with previous structural knowledge, but were ultimately discovered only through integration of mutagenesis, selection, and structure-based reengineering. Whether changing the permissive background of domain 11 binding site amino acids would have ultimately altered the specific selection of high-affinity mutations is not known but is likely, and has implications for the modification of other ligand–receptor interactions (38).
Similar to mutations introduced into other protein–protein interaction sites, our data suggest that single point mutations in loops CD, HI, and FG can result in relatively small (less than twofold) incremental gains in affinity in a domain isolated from the natural context of the receptor. It is also evident that although such small gains are possible, they have not been selected in the current mammalian evolutionary context, indicating that they have had little consequence on IGF2 and M6P/IGF2R function. The larger incremental gains in affinity of 8- to 10-fold observed during the evolution of early mammals (ancestors of monotremes) do, however, suggest that loop mutations have had a very significant impact on gain in fitness in the past (2). Combinatorial mutations in separate loops that result in much higher affinities would have a very low stochastic likelihood of occurring simultaneously in vivo; thus, the five mutations of domain 11AB5 may have been sufficient to generate a significant incremental gain of function, and may be selected for potential improvement in fitness over time if they could be introduced.
A Functional High-Affinity and -Specificity IGF2-Soluble Antagonist.
Activation of the insulin-like growth factor signaling pathway in cancer relates predominantly to gain of function of IGF2 ligand (expression and bioavailability) and the subsequent activation of both IGF1 and IR-A signaling receptors (39). Multiple mechanisms result in increased IGF2 expression and ligand supply, from genomic amplification to proteolysis of IGF-binding proteins (40–46). Despite intensive efforts, the sole targeting of IGF1R downstream of IGF2 has had only limited success in clinical trials (47). One problem appears to be the underestimated effects of IGF1(R) blockade on activation of a central nervous system negative feedback loop, resulting in increased systemic GH driving total IGF1 production from the liver, GH-mediated insulin resistance, and increased insulin supply, resulting in antagonism of the IGF1R inhibitor and systemic metabolic toxicity (20). Similar compensatory feedback responses are also observed with anti-IGF ligand antibodies when administered at therapeutic levels, likely related to the lack of selectivity and subsequent sequestration of free IGF1 (48, 49). Anti-ligand antibodies appear to bind IGF2 with a cross-reaction with IGF1, e.g., Dx-2647 (49 pmol L−1 for IGF2 vs. 9.6 nmol L−1 for IGF1), Medi-573 (2 pmol L−1 for IGF2 vs. 294 pmol L−1 for IGF1), and m660 chimeric antibody (700 pmol L−1 for IGF2 vs. 8 nmol L−1 for IGF1) (14, 50–52).
It is well established that for ligand trap molecules to be functional, a >10-fold higher affinity than that for the signaling receptor appears to be necessary to achieve maximal antagonistic activity (53). A specific IGF2 antagonist, such as Fc domain 11AB5-RHH (IGF2-TRAP), may have this significant advantage, because its higher specific affinity means that it can inhibit IGF2 activation of both IGF1R and IR-A signaling at the level of the cell, as well as the potential advantage of not perturbing the endocrine hypothalamic IGF1-GH-insulin feedback that occurs when targeting IGF1. The latter possibility requires evaluation in human trials, however, because humans have an adult liver IGF2 promoter, resulting in gene expression and circulating ligand.
The synergistic effects with PI3 kinase inhibitors reported here are also in keeping with reports of the intrinsic resistance conferred by the IGF pathway following PI3 kinase inhibition (ZSTK474) in gastric carcinoma cell lines, suggesting that conditional effects acting directly on the intracellular signaling pathway may unmask such IGF1R/IR-A and IGF2 ligand dependency (54). Similar intracellular feedback mechanisms may extend to regulation of ligand bioavailability, given that EGFR pathway resistance may be conferred by reduced IGFBP levels, and resistance to IGF pathway inhibition may be improved by downstream MEK inhibition (55, 56). Our data support the concept that specific IGF2 targeting also might address unmet needs for IGF2-dependent clinical phenotypes, such as the mosaic overgrowth Beckwith–Weidemann syndrome (GCID: GC11M002113), and as stated above, the systemic excess of higher molecular weight or big IGF2 (e.g., IGF287, IGF2104) in NICTH (IGF2 syndrome) (57, 58). Our functional data suggest that IGF2 signaling, with or without other targeted cell signaling agents, can be inhibited by the high affinity and specificity generated by a combinatorial mutation in Fc domain 11AB5-RHH (IGF2-TRAP) in vivo.
In summary, we have rapidly evolved the domain 11 IGF2-binding site and identified previously unidentified combinatorial molecular and structural mechanisms of enhanced affinity that are confined to the surface of the binding interface. Specifically, these include interdomain (AB-CD) and intradomain (FG-FG) amino acid side chain interactions that alter the complementary interactions with IGF2 but retain IGF2–T16 interactions that confer specificity over IGF1. The 100-fold improvement in IGF2:domain 11 affinity and our functional assays of ligand trap activity, including binding to pathological pro-IGF2 isoforms, accelerate the impetus to apply the advantages of these domains as IGF2 ligand traps for cancer therapy.
Materials and Methods
Further details are provided in SI Appendix, Materials and Methods.
Yeast Surface Display of Domain 11 and IGF2-Binding Screens.
The yeast surface display of an N-terminal Flag-tagged fusion of domain 11 and the C-terminal domain of α-agglutinin was developed in P. pastoris for loop-specific mutant library selection using binding with biotinylated IGF2 and flow sorting (SI Appendix, Table S1), as described previously (59). Selected mutant proteins were purified by an Ni-NTA column before validation by SPR using a Biacore T200 system. Thermodynamic data were obtained across the temperature range of 10–30 °C. van’t Hoff and Erying plots were generated, and ΔH, −TΔS, and ΔG values were determined as described previously (22).
Structure-Informed Loop-Specific Site-Directed Mutagenesis.
The domain 11AB3:IGF2 structure (PDB ID code 2L29) was used to identify residues in the binding loops that could interact with IGF2. These were manually mutated in Pymol v1.5 to determine whether they might stabilize the complex (www.pymol.org/). In addition, the Robetta server (robetta.bakerlab.org/alascansubmit.jsp) was used to interface alanine scanning mutagenesis on the AB3:IGF2 complex.
Expression and Purification of Domain 11WT, Domain 11AB5, and IGF2 Domain 11AB5 Complexes for NMR and 15N Relaxation Studies of Domain 11WT and Domain 11AB5.
His6-tagged domain 11WT and domain 11AB5 were isotopically 15N- or dual 15N- and 13C-labeled and purified from E. coli (2). Structural calculations of the domain 11AB5, both free and complexed with IGF2, were as described previously (2). NOE, 15N-T1, and 15N-T2 NMR relaxation data were acquired in two fields (600 and 900 MHz) at 25 °C. T1 and T2 experiments were run in duplicate (SI Appendix, Fig. S2).
Expression, Purification, and Crystal Structure of Domain 11AB5-Q1569R P1597H S1602H.
Domain 11AB5-Q1569R P1597H S1602H protein (160 µM) was screened for crystals, and data collection was done using a PILATUS detector system (Dectris) at station I04 of the Diamond Synchrotron (SI Appendix, Table S8).
Fc Fusion Protein Expression and Purification.
IgG1 Fc domain 11AB5 fusion protein was produced by transient transfection of HEK293T cells and IgG2 Fc domain 11AB5-RHH in Chinese hamster ovary GS knockout cells, and purified by protein A affinity (SI Appendix, Fig. S7).
IGF2-Induced Hypoglycemia.
All animal experiments were approved by the United Kingdom Home Office (PPL 30/2695). Blood glucose levels were allowed to normalize for 30 min after induction of anesthesia before IGF2 injection either alone or premixed with different molar ratios, Fc domain 11AB5, or Fc domain 11I1572A.
IGF2-Dependent Human Cancer Cell Xenografts.
SKNMC-IGF21–67 cells mixed with Matrigel were injected into CD-1 (Crl:NU-1 Foxn1nu) at 24 h after placement of an ALZET osmotic minipump (Durect) delivering 40 mg kg−1/week of either IGF2-TRAP or PBS control. Tumor growth was monitored with both caliper measurements and bioluminescence.
IGF2-TRAP Synergistic Lethality Screens.
SKNMC-IGF2104-IRES-luc cells were exposed to a 307 oncology drug library that has been tested in humans (Target Discovery Institute Expanded Oncology Drug Set) using 10 µM, 1 µM, 100 nM, and 10 nM concentrations, in the presence and absence of 250 nM IGF2-TRAP.
Neutral Size-Fractionation of Human Serum and IGF2 Depletion with IGF2-TRAP.
Human serum was size-fractionated by gel filtration and assayed for the presence of ALS, IGFBP3, and IGF2 by Western blot analysis. Pull-downs were performed using protein G magnetic beads (SI Appendix, Materials and Methods).
Supplementary Material
Acknowledgments
We thank Emily Richards, Leanne Minnall, and Sara Whittaker for the NMR data and Hans-Jürgen Hoppe, Sermet Can, Claudia Bühnemann, Nigel Rust, Martin Glennie, Anton van der Merwe, and Neil Barclay for discussions. We also thank the Wellcome Trust for the use of 900-MHz NMR spectrometer at the Henry Wellcome Building for NMR in Birmingham, United Kingdom. The research described in this manuscript is the subject of a worldwide patent application (WO2007020402A), granted in Europe as EP1945663B and in the United States as US8293875 and US8658423. Support for this work was provided by Cancer Research UK (ABH C429), the Engineering and Physical Sciences Research Council, the Algerian Government (D.R.), the National Institute for Health Research Biomedical Research Centre and Unit, the Clarendon Fund, and Agilent Technologies (formerly Varian).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The ensembles of NMR structures and associated NMR chemical shifts have been deposited in the Protein Data Bank (PDB) and BioMagResBank [free domain 11AB5 (PDB ID code 2M6T; 19153) and IGF2-bound domain 11AB5 (PDB ID code 2M68; 19117)]. The crystal structure of domain 11AB5-RHH has been deposited in the PDB (PDB ID code 5IEI).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1513023113/-/DCSupplemental.
References
- 1.Harms MJ, Thornton JW. Evolutionary biochemistry: Revealing the historical and physical causes of protein properties. Nat Rev Genet. 2013;14(8):559–571. doi: 10.1038/nrg3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams C, et al. An exon splice enhancer primes IGF2:IGF2R binding site structure and function evolution. Science. 2012;338(6111):1209–1213. doi: 10.1126/science.1228633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghosh P, Dahms NM, Kornfeld S. Mannose 6-phosphate receptors: New twists in the tale. Nat Rev Mol Cell Biol. 2003;4(3):202–212. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- 4.Linnell J, Groeger G, Hassan AB. Real time kinetics of insulin-like growth factor II (IGF-II) interaction with the IGF-II/mannose 6-phosphate receptor: The effects of domain 13 and pH. J Biol Chem. 2001;276(26):23986–23991. doi: 10.1074/jbc.M100700200. [DOI] [PubMed] [Google Scholar]
- 5.Morgan DO, et al. Insulin-like growth factor II receptor as a multifunctional binding protein. Nature. 1987;329(6137):301–307. doi: 10.1038/329301a0. [DOI] [PubMed] [Google Scholar]
- 6.Wilkins JF, Haig D. What good is genomic imprinting: The function of parent-specific gene expression. Nat Rev Genet. 2003;4(5):359–368. doi: 10.1038/nrg1062. [DOI] [PubMed] [Google Scholar]
- 7.Hughes J, Frago S, Bühnemann C, Carter EJ, Hassan AB. Maternal transmission of a humanised Igf2r allele results in an Igf2 dependent hypomorphic and non-viable growth phenotype. PLoS One. 2013;8(2):e57270. doi: 10.1371/journal.pone.0057270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau MM, et al. Loss of the imprinted IGF2/cation-independent mannose 6-phosphate receptor results in fetal overgrowth and perinatal lethality. Genes Dev. 1994;8(24):2953–2963. doi: 10.1101/gad.8.24.2953. [DOI] [PubMed] [Google Scholar]
- 9.Wang ZQ, Fung MR, Barlow DP, Wagner EF. Regulation of embryonic growth and lysosomal targeting by the imprinted Igf2/Mpr gene. Nature. 1994;372(6505):464–467. doi: 10.1038/372464a0. [DOI] [PubMed] [Google Scholar]
- 10.Wylie AA, et al. Tissue-specific inactivation of murine M6P/IGF2R. Am J Pathol. 2003;162(1):321–328. doi: 10.1016/S0002-9440(10)63823-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Souza AT, Hankins GR, Washington MK, Orton TC, Jirtle RL. M6P/IGF2R gene is mutated in human hepatocellular carcinomas with loss of heterozygosity. Nat Genet. 1995;11(4):447–449. doi: 10.1038/ng1295-447. [DOI] [PubMed] [Google Scholar]
- 12.Hankins GR, et al. M6P/IGF2 receptor: A candidate breast tumor suppressor gene. Oncogene. 1996;12(9):2003–2009. [PubMed] [Google Scholar]
- 13.Harper J, et al. Soluble IGF2 receptor rescues Apc(Min/+) intestinal adenoma progression induced by Igf2 loss of imprinting. Cancer Res. 2006;66(4):1940–1948. doi: 10.1158/0008-5472.CAN-05-2036. [DOI] [PubMed] [Google Scholar]
- 14.Prince SN, Foulstone EJ, Zaccheo OJ, Williams C, Hassan AB. Functional evaluation of novel soluble insulin-like growth factor (IGF)-II–specific ligand traps based on modified domain 11 of the human IGF2 receptor. Mol Cancer Ther. 2007;6(2):607–617. doi: 10.1158/1535-7163.MCT-06-0509. [DOI] [PubMed] [Google Scholar]
- 15.Surinya KH, et al. An investigation of the ligand binding properties and negative cooperativity of soluble insulin-like growth factor receptors. J Biol Chem. 2008;283(9):5355–5363. doi: 10.1074/jbc.M707054200. [DOI] [PubMed] [Google Scholar]
- 16.Delaine C, et al. A novel binding site for the human insulin-like growth factor-II (IGF-II)/mannose 6-phosphate receptor on IGF-II. J Biol Chem. 2007;282(26):18886–18894. doi: 10.1074/jbc.M700531200. [DOI] [PubMed] [Google Scholar]
- 17.Bach LA, Headey SJ, Norton RS. IGF-binding proteins: The pieces are falling into place. Trends Endocrinol Metab. 2005;16(5):228–234. doi: 10.1016/j.tem.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Greenall SA, et al. Biochemical characterization of individual human glycosylated proinsulin-like growth factor (IGF)-II and big-IGF-II isoforms associated with cancer. J Biol Chem. 2013;288(1):59–68. doi: 10.1074/jbc.M112.432013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaina S, Squire S. The soluble type 2 insulin-like growth factor (IGF-II) receptor reduces organ size by IGF-II–mediated and IGF-II–independent mechanisms. J Biol Chem. 1998;273(44):28610–28616. doi: 10.1074/jbc.273.44.28610. [DOI] [PubMed] [Google Scholar]
- 20.Yee D. Insulin-like growth factor receptor inhibitors: Baby or the bathwater? J Natl Cancer Inst. 2012;104(13):975–981. doi: 10.1093/jnci/djs258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown J, et al. Structure and functional analysis of the IGF-II/IGF2R interaction. EMBO J. 2008;27(1):265–276. doi: 10.1038/sj.emboj.7601938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaccheo OJ, et al. Kinetics of insulin-like growth factor II (IGF-II) interaction with domain 11 of the human IGF-II/mannose 6-phosphate receptor: Function of CD and AB loop solvent-exposed residues. J Mol Biol. 2006;359(2):403–421. doi: 10.1016/j.jmb.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 23.Canfield WM, Kornfeld S. The chicken liver cation-independent mannose 6-phosphate receptor lacks the high affinity binding site for insulin-like growth factor II. J Biol Chem. 1989;264(13):7100–7103. [PubMed] [Google Scholar]
- 24.Clairmont KB, Czech MP. Chicken and Xenopus mannose 6-phosphate receptors fail to bind insulin-like growth factor II. J Biol Chem. 1989;264(28):16390–16392. [PubMed] [Google Scholar]
- 25.Yandell CA, Dunbar AJ, Wheldrake JF, Upton Z. The kangaroo cation-independent mannose 6-phosphate receptor binds insulin-like growth factor II with low affinity. J Biol Chem. 1999;274(38):27076–27082. doi: 10.1074/jbc.274.38.27076. [DOI] [PubMed] [Google Scholar]
- 26.Bloom JD, Arnold FH. In the light of directed evolution: Pathways of adaptive protein evolution. Proc Natl Acad Sci USA. 2009;106(Suppl 1):9995–10000. doi: 10.1073/pnas.0901522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levin KB, et al. Following evolutionary paths to protein–protein interactions with high affinity and selectivity. Nat Struct Mol Biol. 2009;16(10):1049–1055. doi: 10.1038/nsmb.1670. [DOI] [PubMed] [Google Scholar]
- 28.Yuen CM, Liu DR. Dissecting protein structure and function using directed evolution. Nat Methods. 2007;4(12):995–997. doi: 10.1038/nmeth1207-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mergler M, Wolf K, Zimmermann M. Development of a bisphenol A-adsorbing yeast by surface display of the Kluyveromyces yellow enzyme on Pichia pastoris. Appl Microbiol Biotechnol. 2004;63(4):418–421. doi: 10.1007/s00253-003-1361-0. [DOI] [PubMed] [Google Scholar]
- 30.Williams C, et al. Structural insights into the interaction of insulin-like growth factor 2 with IGF2R domain 11. Structure. 2007;15(9):1065–1078. doi: 10.1016/j.str.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 31.Tinoco I, Sauer K, Wang JC. Physical Chemistry: Prinicples and Applications in Biological Sciences. Prentice-Hall; Englewood Cliffs, NJ: 1978. [Google Scholar]
- 32.Dougherty DA. Cation-pi interactions involving aromatic amino acids. J Nutr. 2007;137(6) Suppl 1:1504S–1508S, discussion 1516S–1517S. doi: 10.1093/jn/137.6.1504S. [DOI] [PubMed] [Google Scholar]
- 33.Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc Natl Acad Sci USA. 1999;96(17):9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duguay SJ, et al. Post-translational processing of the insulin-like growth factor-2 precursor. Analysis of O-glycosylation and endoproteolysis. J Biol Chem. 1998;273(29):18443–18451. doi: 10.1074/jbc.273.29.18443. [DOI] [PubMed] [Google Scholar]
- 35.Bühnemann C, et al. Quantification of the heterogeneity of prognostic cellular biomarkers in Ewing sarcoma using automated image and random survival forest analysis. PLoS One. 2014;9(9):e107105. doi: 10.1371/journal.pone.0107105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kastritis PL, Rodrigues JP, Folkers GE, Boelens R, Bonvin AM. Proteins feel more than they see: Fine-tuning of binding affinity by properties of the non-interacting surface. J Mol Biol. 2014;426(14):2632–2652. doi: 10.1016/j.jmb.2014.04.017. [DOI] [PubMed] [Google Scholar]
- 37.Škrlec K, Štrukelj B, Berlec A. Non-immunoglobulin scaffolds: A focus on their targets. Trends Biotechnol. 2015;33(7):408–418. doi: 10.1016/j.tibtech.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 38.Harms MJ, Thornton JW. Historical contingency and its biophysical basis in glucocorticoid receptor evolution. Nature. 2014;512(7513):203–207. doi: 10.1038/nature13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brouwer-Visser J, Huang GS. IGF2 signaling and regulation in cancer. Cytokine Growth Factor Rev. 2015;26(3):371–377. doi: 10.1016/j.cytogfr.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 40.Chess A. Mechanisms and consequences of widespread random monoallelic expression. Nat Rev Genet. 2012;13(6):421–428. doi: 10.1038/nrg3239. [DOI] [PubMed] [Google Scholar]
- 41.Gaudet F, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300(5618):489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 42.Lui JC, Baron J. Evidence that Igf2 down-regulation in postnatal tissues and up-regulation in malignancies is driven by transcription factor E2f3. Proc Natl Acad Sci USA. 2013;110(15):6181–6186. doi: 10.1073/pnas.1219079110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyamoto S, et al. Matrix metalloproteinase-7 triggers the matricrine action of insulin-like growth factor-II via proteinase activity on insulin-like growth factor binding protein 2 in the extracellular matrix. Cancer Sci. 2007;98(5):685–691. doi: 10.1111/j.1349-7006.2007.00448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seshagiri S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488(7413):660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun Y, et al. IGF2 is critical for tumorigenesis by synovial sarcoma oncoprotein SYT-SSX1. Oncogene. 2006;25(7):1042–1052. doi: 10.1038/sj.onc.1209143. [DOI] [PubMed] [Google Scholar]
- 46.Vidal SJ, et al. A targetable GATA2-IGF2 axis confers aggressiveness in lethal prostate cancer. Cancer Cell. 2015;27(2):223–239. doi: 10.1016/j.ccell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yee D. A tale of two receptors: Insulin and insulin-like growth factor signaling in cancer. Clin Cancer Res. 2015;21(4):667–669. doi: 10.1158/1078-0432.CCR-14-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haluska P, et al. Phase I dose-escalation study of MEDI-573, a bispecific, antiligand monoclonal antibody against IGFI and IGFII, in patients with advanced solid tumors. Clin Cancer Res. 2014;20(18):4747–4757. doi: 10.1158/1078-0432.CCR-14-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mireuta M, Birman E, Barmash M, Pollak M. Quantification of binding of IGF-1 to BI 836845, a candidate therapeutic antibody against IGF-1 and IGF-2, and effects of this antibody on IGF-1:IGFBP-3 complexes in vitro and in male C57BL/6 mice. Endocrinology. 2014;155(3):703–715. doi: 10.1210/en.2013-1791. [DOI] [PubMed] [Google Scholar]
- 50.Gao J, et al. Dual IGF-I/II-neutralizing antibody MEDI-573 potently inhibits IGF signaling and tumor growth. Cancer Res. 2011;71(3):1029–1040. doi: 10.1158/0008-5472.CAN-10-2274. [DOI] [PubMed] [Google Scholar]
- 51.Chen W, Feng Y, Zhao Q, Zhu Z, Dimitrov DS. Human monoclonal antibodies targeting nonoverlapping epitopes on insulin-like growth factor II as a novel type of candidate cancer therapeutics. Mol Cancer Ther. 2012;11(7):1400–1410. doi: 10.1158/1535-7163.MCT-12-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dransfield DT, et al. A human monoclonal antibody against insulin-like growth factor-II blocks the growth of human hepatocellular carcinoma cell lines in vitro and in vivo. Mol Cancer Ther. 2010;9(6):1809–1819. doi: 10.1158/1535-7163.MCT-09-1134. [DOI] [PubMed] [Google Scholar]
- 53.Forsten KE, Lauffenburger DA. Interrupting autocrine ligand-receptor binding: Comparison between receptor blockers and ligand decoys. Biophys J. 1992;63(3):857–861. doi: 10.1016/S0006-3495(92)81661-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Isoyama S, et al. Basal expression of insulin-like growth factor 1 receptor determines intrinsic resistance of cancer cells to a phosphatidylinositol 3-kinase inhibitor ZSTK474. Cancer Sci. 2015;106(2):171–178. doi: 10.1111/cas.12582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flanigan SA, et al. Overcoming IGF1R/IR resistance through inhibition of MEK signaling in colorectal cancer models. Clin Cancer Res. 2013;19(22):6219–6229. doi: 10.1158/1078-0432.CCR-13-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guix M, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest. 2008;118(7):2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fukuda I, et al. Clinical features of insulin-like growth factor-II producing non-islet-cell tumor hypoglycemia. Growth Horm IGF Res. 2006;16(4):211–216. doi: 10.1016/j.ghir.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 58.Rikhof B, et al. Insulin-like growth factors and insulin-like growth factor-binding proteins in relation to disease status and incidence of hypoglycaemia in patients with a gastrointestinal stromal tumour. Ann Oncol. 2009;20(9):1582–1588. doi: 10.1093/annonc/mdp038. [DOI] [PubMed] [Google Scholar]
- 59.Wu S, Letchworth GJ. High efficiency transformation by electroporation of Pichia pastoris pretreated with lithium acetate and dithiothreitol. Biotechniques. 2004;36(1):152–154. doi: 10.2144/04361DD02. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.