Significance
T cells protect the body against pathogens and cancer by recognizing specific foreign peptides on the cell surface. Because antigen recognition occurs at the junction between a migrating T cell and an antigen-presenting cell (APC), it is likely that cellular forces are generated and transmitted through T-cell receptor (TCR)-ligand bonds. Here we develop a DNA-based nanoparticle tension sensor producing the first molecular maps of TCR-ligand forces during T cell activation. We find that TCR forces are orchestrated in space and time, requiring the participation of CD8 coreceptor and adhesion molecules. Loss or damping of TCR forces results in weakened antigen discrimination, showing that T cells harness mechanics to optimize the specificity of response to ligand.
Keywords: T-cell receptor, mechanotransduction, antigen discrimination, cell migration, molecular tension sensor
Abstract
T cells are triggered when the T-cell receptor (TCR) encounters its antigenic ligand, the peptide-major histocompatibility complex (pMHC), on the surface of antigen presenting cells (APCs). Because T cells are highly migratory and antigen recognition occurs at an intermembrane junction where the T cell physically contacts the APC, there are long-standing questions of whether T cells transmit defined forces to their TCR complex and whether chemomechanical coupling influences immune function. Here we develop DNA-based gold nanoparticle tension sensors to provide, to our knowledge, the first pN tension maps of individual TCR-pMHC complexes during T-cell activation. We show that naïve T cells harness cytoskeletal coupling to transmit 12–19 pN of force to their TCRs within seconds of ligand binding and preceding initial calcium signaling. CD8 coreceptor binding and lymphocyte-specific kinase signaling are required for antigen-mediated cell spreading and force generation. Lymphocyte function-associated antigen 1 (LFA-1) mediated adhesion modulates TCR-pMHC tension by intensifying its magnitude to values >19 pN and spatially reorganizes the location of TCR forces to the kinapse, the zone located at the trailing edge of migrating T cells, thus demonstrating chemomechanical crosstalk between TCR and LFA-1 receptor signaling. Finally, T cells display a dampened and poorly specific response to antigen agonists when TCR forces are chemically abolished or physically “filtered” to a level below ∼12 pN using mechanically labile DNA tethers. Therefore, we conclude that T cells tune TCR mechanics with pN resolution to create a checkpoint of agonist quality necessary for specific immune response.
T-cell activation is a crucial step in adaptive immunity, offering defense against pathogens and cancer (1). During activation, the T-cell receptor (TCR) recognizes and binds to its ligand, the antigenic peptide-major histocompatibility complex (pMHC), which is expressed on the surface of antigen-presenting cells (APCs). Because T cells are continuously moving and scanning the surfaces of APCs for evidence of antigens, and TCR-ligand binding occurs at the junction between two cells, it is likely that the TCR experiences mechanical forces during normal T-cell function. Therefore, important questions in this area pertain to whether the TCR-pMHC complex experiences defined forces during T-cell activation, and whether these forces influence immune function (2).
An elegant body of single-molecule experiments further underscores the connection between TCR signaling and mechanics. For example, Lang and Reinherz (3) used optical tweezers to demonstrate that the TCR responds to physical forces applied using an optically trapped bead. This team also showed that the TCR undergoes distinct structural transitions within its FG loop (a region formed by F and G strands) when the pMHC-TCR complex is strained at ∼15 pN (4). Complementary single-molecule force spectroscopy measurements using the biomembrane force probe by Zhu and Evavold (5) showed that the average TCR-pMHC bond lifetime (1/koff rate) is enhanced when ∼10 pN of tension is applied through a specific antigen. Enhancement of bond lifetime under the influence of antigen mechanical strain (∼10 pN) was further demonstrated in CD4+ T cells (6) and for pre–TCR-pMHC interactions (7). These experiments specifically demonstrate an inherent TCR sensitivity to pN forces transmitted through its cognate pMHC ligand (3–8).
The role of forces in modulating bond lifetimes is particularly striking given that the most widely accepted model of TCR activation invokes a kinetic proofreading mechanism, emphasizing the importance of the TCR-pMHC dissociation rate in boosting antigen specificity (2, 9). The implicit model is that a T cell actively regulates forces transmitted to its TCR-pMHC complex to fine-tune bond lifetimes, thereby enhancing selective and differential levels of TCR activation and regulating antigen discrimination and T-cell selection.
The role of mechanics in T-cell activation remains controversial, however. For example, does the T cell itself transmit 10–15 pN of tension to its engaged TCR-pMHC complex during the early stages of antigen proofreading? Although traction force microscopy methods demonstrate that T cells generate contractile forces ∼5 min after activation (10, 11), there is no evidence showing that the TCR-pMHC complex experiences pN forces during initial antigen encounters. Such forces are beyond the spatial and temporal resolution of traction force microscopy. Therefore, new molecular approaches are needed to investigate intrinsic TCR mechanics and to determine the physical basis and physiological consequences of mechanics in immunology.
Results
Fabrication and Characterization of the DNA-Based Nanoparticle Tension Sensors.
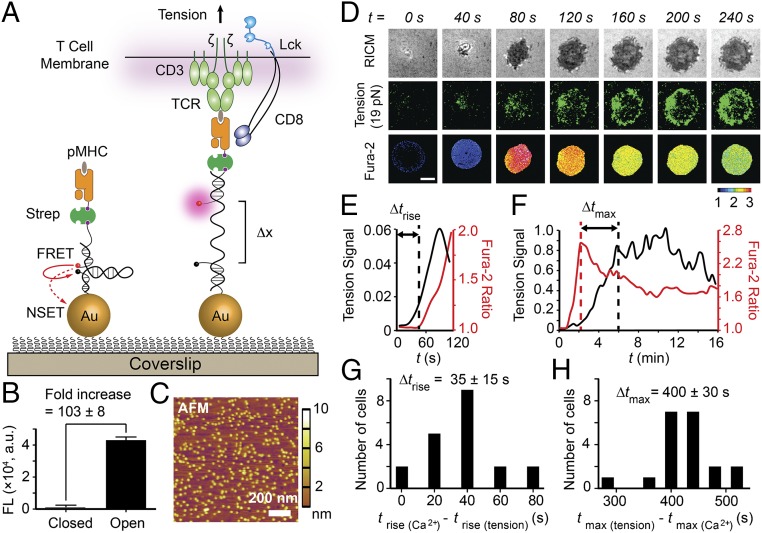
We have developed an improved type of fluorescent tension probe to directly image and quantify the innate forces mediated by individual TCR complexes on ligand binding (Fig. 1A and SI Appendix, Fig. S1). These probes provide unprecedented sensitivity (SI Appendix, Supplementary Note 1), generating a ∼100-fold fluorescence increase in response to pN forces (Fig. 1B and SI Appendix, Fig. S2). This sensitivity is essential given the transient nature and the limited number of TCR-pMHC antigens sufficient to activate T cells (12, 13). In brief, each tension probe consists of a DNA hairpin labeled with a fluorophore-quencher pair, and is immobilized onto a gold nanoparticle (AuNP). The mean interparticle spacing is 48 ± 8 nm (Fig. 1C), which is physiologically optimal for antigen-mediated T-cell adhesion and functional responses (14, 15). Each particle presents an average of 4 ± 1 DNA hairpins (SI Appendix, Fig. S3). The fluorophore is dual-quenched by both the molecular quencher through fluorescence resonance energy transfer (FRET) and the plasmon of the AuNP via nanometal surface energy transfer (NSET) pathway, thus reducing the background signal and boosting sensitivity over conventional molecular tension sensors (16–22). DNA hairpins generate fluorescence when the applied force exceeds the unfolding force, F1/2 (defined as the force at equilibrium for which a hairpin has a 50% probability of unfolding). Note that the probability of mechanical unfolding depends on the loading rate and the duration of the force, and thus the reported F1/2 values serve as a lower bound of the applied force given that the physiological loading rates and bond lifetimes are unknown. Importantly, the F1/2 can be tuned from ∼2.4 to 19.0 pN through the GC content and stem-loop structure of the hairpin (23) (SI Appendix, Table S1). Each probe exclusively responds to forces transmitted through a single TCR molecule (SI Appendix, Fig. S4).
Fig. 1.
T cells apply pN forces to the TCR preceding the rise in Ca2+. (A) Schematic of DNA-based AuNP sensor for mapping TCR-mediated tension. The fluorescence of the Cy3B dye (pink dot) is dequenched on mechanical unfolding of the hairpin, which separates the dye from the black hole quencher 2 (BHQ2, block dot) and AuNP surface (Δx = ∼16.9 nm). (B) Plot showing the fluorescence intensity of the closed and open forms of the hairpin. There is a 103 ± 8-fold increase in fluorescence on the opening of hairpins. n = 3. Error bars indicate SD. (C) AFM image showing the immobilized AuNP sensors on a glass coverslip. (D) Simultaneous imaging of cell spreading (RICM), 19 pN TCR forces (tension), and T-cell activation (fura-2) for a representative OT-1 cell encountering α-CD3 antigen. Unless noted otherwise, all experiments were performed at 23 °C, the temperature at which F1/2 values were determined experimentally. (Scale bar: 3 μm.) (E and F) Representative plots of tension signal and fura-2 ratio showing temporal differences between the initial rise of [Ca2+] and the initial rise in TCR tension (Δtrise) and between the maximum [Ca2+] and the maximum TCR tension signal (Δtmax), and for the cell shown in D. The normalized y-axis applies to E and F. (G and H) Histogram analysis of Δtrise and Δtmax (n = 20 cells).
Piconewton Forces Are Transmitted by Individual TCR Complexes Before T-Cell Activation.
As a proof of concept, we used anti-CD3 antibody (α-CD3)-modified probes (F1/2 = 19 pN) to map TCR forces during the activation of naïve OT-1 cells (Fig. 1D and Movie S1), with F1/2 determined at 23 °C (SI Appendix, Supplementary Note 2). T cells began to spread within seconds of encountering the α-CD3 ligand surface, and then continued to spread up to ∼8–10 μm in diameter. A punctate 19-pN tension signal was generated within seconds of initial surface contact and progressively increased thereafter, becoming enriched at the periphery of the contact area in a ring-like pattern. This result provides, to our knowledge, the first demonstration that the T cell innately transmits pN forces to its TCR complex in a defined spatial and temporal pattern.
Simultaneous tracking of TCR triggering using a ratiometric fura-2 Ca2+ indicator showed that the initial appearance of forces preceded the rise in [Ca2+] by 35 ± 15 s (Fig. 1 D, E, and G; n = 20 cells). The initial Ca2+ flux was observed ∼1 min after initial cell contact, and peaked at ∼2 min (Fig. 1F). Interestingly, the tension signal continued to rise even after the initial Ca2+ flux started to subside, with a 400 ± 30 s delay between the maximum [Ca2+] and the maximum TCR tension signals (Fig. 1 F and H; n = 20 cells). Therefore, initial TCR triggering is tightly associated with its mechanics, and downstream signaling, such as Ca2+ flux, further amplifies TCR forces.
Although several lines of evidence demonstrate that actin-mediated cytoskeleton dynamics are crucial in T-cell biomechanics (24–26), the cytoskeletal factors that specifically initiate the rise of TCR-ligand forces remain unclear. To address this question, we treated naïve OT-1 cells with a library of cytoskeletal inhibitors and measured cell spreading and TCR-ligand forces (SI Appendix, Fig. S5). The GTPase Cdc42 was identified as critical to initiating the rise of TCR-ligand forces. Myosin light chain kinase and Arp2/3 were found to be important in maintaining TCR-ligand forces, and their inhibition caused drastic cell retraction and ring-shaped condensation of tension signals (Movie S2). This result mirrors the disruptive reorganization of F-actin after blebbistatin and jasplakinolide treatment of Jurkat cells, where actin retrograde flow is hypothesized to exert pushing and pulling forces (25). Furthermore, immunostaining showed that TCR forces highly colocalized with F-actin and were further surrounded by a slightly inward ring pattern of myosin light chain kinase (SI Appendix, Fig. S6). This is consistent with the actomyosin-enriched contraction arc in the lamella (25). Therefore, TCR-ligand forces are highly regulated by precise coordination between actin dynamics and actomyosin contractility.
TCR-Ligand Tension Requires Coreceptor Engagement and Is Modulated by Adhesion Molecules.
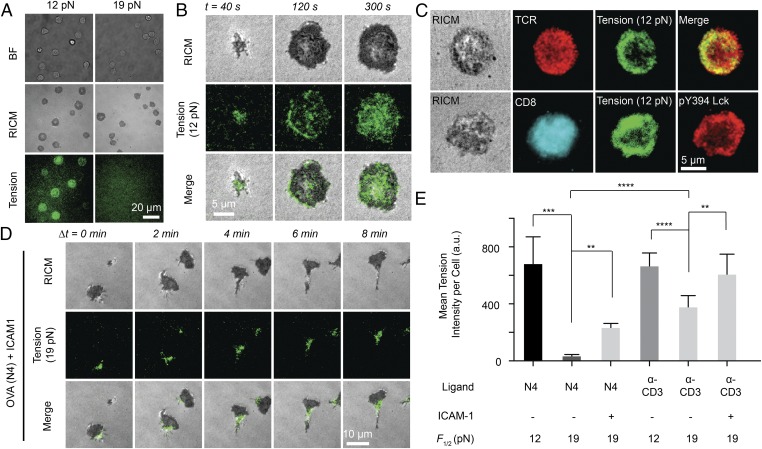
We next aimed to define the magnitude of innate T-cell forces transmitted through the TCR-pMHC bond. For this, we used two DNA tension probes with F1/2 = 12 pN or 19 pN and monitored hairpin unfolding in response to the wild-type N4 ligand, the most potent of the ovalbumin (OVA) antigenic ligands (Fig. 2A). After naïve OT-1 T lymphocytes engaged the surface for 15 min, the cells generated a strong tension signal on the 12-pN probe surface. In contrast, OT-1 T cells failed to unfold the 19-pN sensor (N4 antigen) despite fully spreading on this surface, as determined by reflection interference contrast microscopy (RICM). Because shear flow is known to generate TCR forces (27), we applied external flow to cells immobilized through the 19-pN probe (with N4 antigen), and observed a rapid and robust increase in tension signal (SI Appendix, Fig. S7). This experiment demonstrates that the pMHC-TCR bond can withstand forces >19 pN in the presence of CD8 coreceptor, but the innate TCR forces fall within the 12–19 pN range. Thus, TCR mechanics are precisely tuned with an accuracy of a few ∼kcal/mol.
Fig. 2.
Magnitude and spatial organization of TCR-antigen forces are highly dependent on antigen and adhesion receptor binding. (A) Representative bright-field, RICM, and tension (12 pN and 19 pN) images of OT-1 cells cultured on tension probe surfaces modified with N4 pMHC. (B) Representative RICM and tension (12 pN) images taken from a time-lapse movie for an OT-1 cell on N4 ligand stimulation (Movie S3). (C) Representative immunostaining images showing colocalization between TCR and 12 pN tension (Upper) and colocalization among active Lck (pY 394), CD8, and 12 pN TCR tension (Lower) at t = 5 min. (D) Representative RICM and 19 pN TCR tension images taken from a time-lapse movie for an OT-1 cell on N4 and ICAM-1 stimulation (Movie S4). (E) Bar graph showing the TCR tension intensity on ligand stimulation with N4, α-CD3, and ICAM-1. n = 20 cells. Error bars represent SD. **P < 0.01; ***P < 0.001; ****P < 0.0001.
To determine the characteristic time course of initiation of TCR-pMHC forces, we next imaged the surface immediately after culture of naïve OT-1 cells onto 12-pN tension probes. The T cells generated punctate fluorescence tension signal within 40 s of cell-surface contact (Fig. 2B and Movie S3). This tension signal increased in intensity and spread until it reached a steady state, coinciding with cell spreading as determined by the RICM channel. Spatial analysis at t = 2 min showed that forces were generally concentrated in a ring-like structure 1–2 μm wide at the cell periphery (SI Appendix, Fig. S8). This ring-like distribution was short-lived, however, becoming diffuse across the junction at 2–5 min after cell-surface contact. Subsequently (>5 min), tension was enriched at the center of the contact zone, reminiscent of the central supramolecular activating cluster (28).
We further analyzed the spatial colocalization between tension, TCR, CD8 (coreceptor), and lymphocyte-specific kinase (Lck), which is essential for T-cell activation (29, 30). In contrast to the tension signal near the cell edge, TCRs were distributed uniformly across the cell–substrate interface (Fig. 2C). Therefore, the T cell applies force only to a subset of its TCRs. Notably, CD8 also was distributed homogeneously across the cell membrane, whereas phosphorylated (active) Lck (pY394) was highly colocalized with TCR tension (Pearson correlation = 0.84 ± 0.04; n = 20 cells) (Fig. 2C and SI Appendix, Fig. S9), further supporting the relationship between mechanics and early signaling. To probe this relationship, we treated T cells with a Lck-specific inhibitor (31) before and after cell plating (SI Appendix, Fig. S10). Inhibition of Lck after cell plating led to an ∼20–30% reduction in tension signals, whereas pretreatment with Lck inhibitor abolished tension and cell spreading. This abolishment was also achieved by antibody blocking of the CD8 coreceptor or with a mutant pMHC that inhibits coreceptor binding (SI Appendix, Fig. S10). Therefore, the generation of TCR forces >12 pN requires CD8 coreceptor and its associated Lck.
TCR-pMHC binding is the first step in the TCR triggering cascade that includes a variety of ligand-receptor pairs during T cell–APC contact. For example, adhesion receptors such as LFA-1 and other costimulatory receptors also bind to their ligands at the intercellular junction, which has been proposed to modulate TCR-pMHC forces (32). A binary surface copresenting ICAM-1 ligands and the N4 ligands was engineered using orthogonal chemistry such that TCR forces were exclusively visualized and quantified through the N4 antigen and without convolution from adhesion-mediated forces (Fig. 2D and SI Appendix, Fig. S11). Our results show that the incorporation of ICAM-1 not only led to enhanced TCR forces beyond 19 pN, but also triggered a profound change in T-cell morphology and motility. Shortly after engaging the surface (1–5 min), OT-1 cells polarized, forming a kinapse structure that coincided with cell migration at velocities of ∼1–2 μm/min (Movie S4). TCR forces were observed primarily at the trailing edge (focal zone) of the T cell, in agreement with a recent model integrating cell motility (surveillance) and TCR signaling (33, 34). Fig. 2E summarizes the average TCR forces with N4 and α-CD3 ligands, as well as the role of ICAM-1 in modulating this force (n = 20 cells per group). These data unambiguously show that TCR forces are regulated by antigen and adhesion ligand engagement.
T Cells Harness Mechanical Forces as a Checkpoint of Antigen Quality.
A key property of T cells is their ability to differentiate nearly identical pMHC ligands with distinct levels of response (35, 36). We asked whether TCR mechanics contribute to the specificity of its response to antigen. To answer this question, we used the less potent ligands Q4 and V4, differing by single amino acid mutations at the fourth position (36) and compared tension signals with that of the OVA N4 antigen. As an initial test, time-lapse imaging showed that the TCR mechanically interrogates the less potent V4 ligand with >12 pN forces, albeit at differing time scales (SI Appendix, Fig. S12A and Movie S5). TCR-pMHC forces were more transient and punctate for V4, in contrast to the greater mechanical response to N4 ligand (SI Appendix, Fig. S12B and Movie S6). Moreover, the delay between the rise in [Ca2+] and the rise in tension exceeded 5 min for V4, further confirming that TCR-pMHC mechanics are associated with early antigen discrimination.
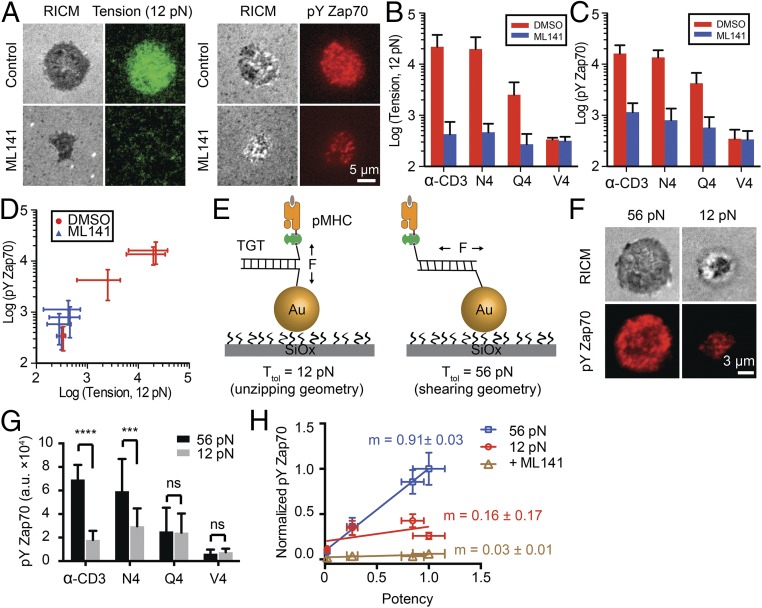
To relate TCR mechanics with T-cell functional response, we plated naïve OT-1 cells onto 12-pN tension sensors displaying N4, Q4, and V4 OVA pMHCs as well as α-CD3. Simultaneously, we measured T-cell activation by quantifying the immunofluorescence of Zap70 phosphorylation (pY319) when Cdc42-mediated tension was chemically inhibited and compared it with the value in the DMSO control (Fig. 3A). The tension signal and ligand potency decreased from α-CD3 to N4, Q4, and V4 (Fig. 3 B and C, red bars). Ligand potency was consistent with literature values determined using IFN-γ production (36). When the Cdc42 inhibitor (ML141) was used, there were marked reductions in TCR tension and T-cell activation (Fig. 3 B and C, blue bar). Plotting the total TCR tension against Zap70 phosphorylation showed a strong correlation (Fig. 3D; Pearson correlation = 0.99), further supporting a strong relationship between TCR tension and T-cell activation. Importantly, the range of T-cell responses are dampened on Cdc42 inhibition, indicating that TCR tension and ligand discrimination are related.
Fig. 3.
TCR forces enhance antigen specificity. (A) Representative images showing TCR tension and pYZap70 on N4 stimulation with or without ML141 treatment for 30 min. (B and C) Bar graphs quantifying TCR tension signals (B) and pYZap70 staining levels (C) for single OT-1 cells on ligand stimulation with α-CD3, N4, Q4, and V4 antigens. n = 20 cells for each group. Error bars represent SD. (D) Plot showing the correlation between TCR tension signal and pYZap70 levels for different ligands. (E) Schematic showing TGTs for modulating TCR forces and T-cell activation. TGTs in an unzipping mode (12 pN) or in a shearing mode of TGT (56 pN) were immobilized onto the 9-nm AuNP through an Au–thiol interaction. Different ligands were conjugated to the TGTs through biotin-streptavidin binding. (F) Representative images showing differential T-cell activation on N4 pMHC-modified 12 pN TGT compared with the 56 pN TGT. (G) Bar graph showing pYZap70 levels on stimulation with pMHCs anchored through the 12 or 56 pN TGT at 37 °C. n = 20 cells for each group. Error bars represent SD. ***P < 0.001; ****P < 0.0001. (H) Plot of pYZap70 levels in response to ligands with increasing potency. The slope (m) indicates the T-cell specificity to different ligands. n = 20 cells from triplicate measurements. Error bars represent SEM.
We next adapted the recently reported DNA tension gauge tether (TGT) (37, 38) into our AuNP platform to physically modulate TCR forces (Fig. 3E). The TGT is a DNA duplex tailored to dissociate at force levels that exceed its mechanical tolerance, Ttol (defined as the rupture force when a constant force is applied for 2 s). In this way, the chemical recognition (affinity) between the TCR and antigenic ligand is maintained; however, the TGT sets the upper limit of forces experienced between a cell-surface receptor and its ligand. Given that TCR forces are in the range of 12–19 pN for the N4 antigen (Fig. 2E), we designed ligands anchored through previously validated TGTs with Ttol values of 56 pN and 12 pN (SI Appendix, Table S1), which provide the largest possible difference in mechanical resistance (Ttol determined at 37 °C) (SI Appendix, Supplementary Note 2). Indeed, the 12-pN TGT presenting α-CD3 displayed greater dissociation compared with the 56-pN TGT (SI Appendix, Fig. S13). After culturing naïve OT-1 cells for 30 min onto the 12 pN and 56 pN TGT surfaces decorated with N4, Q4, and V4 OVA pMHCs as well as α-CD3, T-cell activation was quantified using pYZap70 (pY319) (Fig. 3 F and G). Note that all TGT experiments were performed at 37 °C, the temperature at which Ttol values were determined.
Interestingly, the most potent ligands, α-CD3 and N4, displayed the greatest pYZap70 levels when tethered through the 56-pN TGT, with a significant reduction in activation for the 12-pN TGT. Thus, more potent ligands display enhanced T-cell triggering by a pulling force >12 pN. The less potent Q4 and V4 antigens did not show a differential response to force (Fig. 3G). Therefore, T-cell activation levels are sensitive to the amount of self-generated force transmitted to the different TCR-pMHC bonds, allowing for the optimal (i.e., more potent) ligands to selectively benefit from intensified mechanical tension. With the 12-pN TGT, ligand potency is statistically similar across the different antigens. Capping the TCR-pMHC force to 12 pN likely leads to dampening differences in TCR-pMHC dwell times (39). When the cell is allowed to intensify TCR-pMHC tension before it dissociates, the tested ligands further resolve with different levels of pYZap70 and functional responses. This is shown graphically by plotting the pYZap70 levels against ligand potency (Fig. 3H). Taken together, these experiments demonstrate that cell-generated forces transmitted to the TCR-pMHC bond amplify the specificity of antigen recognition, which is a required feature of T-cell function.
Discussion
We imaged TCR forces with the highest spatial and temporal resolution reported to date using improved molecular tension sensors. We reveal that naïve T cells generate forces in the 12–19 pN range during initial antigen recognition (Fig. 2 and SI Appendix, Fig. S12). Importantly, our results complement and further validate the physiological relevance of a significant body of single-molecule measurements showing that external forces, in the ∼10–20 pN range, drive structural transitions in the TCR and enhance the TCR-pMHC bond lifetime for strong agonists (4, 5, 7). It is no coincidence that the T cell strains its TCR-pMHC complex to tension values similar to those that achieve maximal “catch bond” behavior (a counterintuitive enhancement in bond lifetime that occurs when a bond experiences mechanical strain), as determined in single-molecule experiments.
Our results also suggest that agonists, both strong and weak, experience a pN mechanical stringency test, but strong agonists selectively display enhanced activation, presumably owing to enhanced bond lifetime. T cells harness mechanical forces to aid discrimination between strong and weak agonists by inducing differential phosphorylation levels of Zap70 (Fig. 3), further supporting the force-dependent mechanisms of T-cell activation (40). Given that LFA-1 engagement is not required to generate TCR-pMHC tension, mechanical testing of antigen seems to be independent of T-cell migration processes.
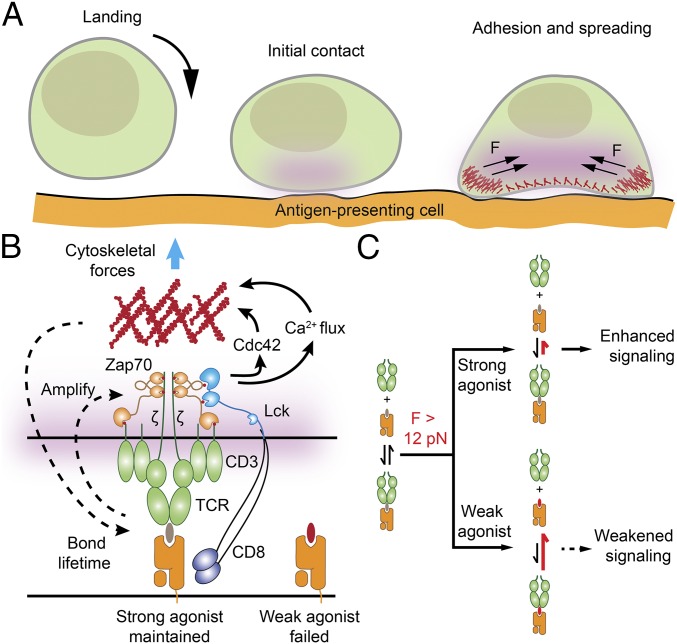
Our data also point to a dual role for mechanics in T-cell function (Fig. 4). First, TCR-pMHC forces are involved in initial ligand discrimination as a fidelity checkpoint. As discussed above, T cells likely harness mechanics to maximize the TCR-pMHC bond lifetime for cognate ligands. Second, following T-cell triggering and Ca2+ flux, myosin contractility is enhanced, further mounting TCR forces and leading to cell spreading. This in turn increases the number of TCR-pMHC engagements at the T cell–APC junction by simply flattening the cell–cell interface (Fig. 4A). This chemomechanical feedback maximally amplifies the distinct TCR signaling levels between strong and weak agonists, demonstrating a mechanically regulated model for antigen discrimination (Fig. 4 B and C).
Fig. 4.
Proposed model for T-cell force generation and antigen discrimination. (A) Initial ligand-receptor engagement occurs when a T cell physically encounters an antigen-presenting cell. If strong pMHC agonists are encountered, the TCR is triggered and the initial signal is amplified, leading to cell adhesion, spreading, and force enhancement. (B) Speculative model depicting how T cells harness a chemomechanical feedback mechanism to increase the specificity of TCR signaling and distinguish between strong and weak agonists. The chemical triggering of TCR activates cytoskeletal processes that further enhance mechanical testing of the TCR–pMHC bond. (C) Data showing that TCR-pMHC complexes experience tension >12 pN during initial ligand-receptor sampling and engagement. Moreover, TCR-pMHC forces >12 pN lead to greater levels of downstream signaling for strong agonists compared with the levels achieved through weaker agonists. The mechanism of differential response to force is likely through catch bond behavior, as shown by Liu et al. (5).
By decorating the tension sensor with different ligands, we have shown that the magnitude of TCR forces are significantly lower in response to cognate N4 antigen compared with the α-CD3 ligand (Fig. 2). This conclusion is not simply a result of selective bond rupture; rather, T cells transmit defined forces to a subset of engaged receptors (SI Appendix, Fig. S7). These data suggest the existence of a self-regulatory mechanism for the fine-tuning of force generation. The observations that CD8 binding and Lck kinase activity are essential in mediating force generation (SI Appendix, Fig. S10) suggest chemical mechanisms of force regulation.
LFA-1/ICAM-1 binding results in a migratory phenotype of OT-1 cells and also generates a higher level of TCR-pMHC tension within the focal zone (Fig. 2 D and E and Movie S4), where adhesion molecules including CD2 (41), talin (42) and Rho-associated kinase (43) are enriched. Our data support the emerging motile synapse model in migratory OT-1 cells (33) and further demonstrate active crosstalk between TCR signaling and LFA-1 activation. Because T-cell migration relies on LFA-1 mediated detachment of the trailing edge (focal zone), our observation points to an idea that TCR signaling is coupled to and modulated by mechanics in the kinapse during lymphocyte surveillance and immune function.
Finally, our method provides, to our knowledge, the first platform for decoupling the specific forces transmitted through the TCR from those forces mediated by LFA-1/ICAM-1 interactions (Fig. 2D, SI Appendix, Fig. S11, and Movie S4). In principle, the high modularity of the method should permit a generalization to investigate the mechanics of any specific surface receptors in the context of other intercellular interactions (e.g., receptor–ligand and glycan–glycan interactions), which normally show synergistic effects at the cellular level. This design of a molecular tension sensor better resembles the complex nature of cell–cell junctions and provides a readout of mechanics with molecular specificity that is beyond the capabilities of conventional traction force microscopy and single-molecule force spectroscopy methods.
Materials and Methods
DNA Labeling.
A mixture of A21B (10 nmol) and excess Cy3B NHS ester in 0.1 M sodium bicarbonate solution was allowed to react at room temperature overnight. The mixture was then subjected to P2 gel filtration to remove salts, organic solvent, and unreacted reactants, and further purified by reverse-phase HPLC (solvent A: 0.1 M TEAA; solvent B: 100% MeCN); the initial condition was 10% B with a gradient of 1%/min and a flow rate of 1 mL/min. The desired product was characterized by MALDI-TOF mass spectrometry.
Optical Microscopy.
Live cells were imaged in Hank’s balanced salt imaging buffer at 23 °C. In the TGT experiments, cells were incubated in the imaging buffer at 37 °C for 30 min before fixation. The microscope was a Nikon Eclipse Ti driven by the Elements software package. The microscope featured an Evolve electron multiplying charge coupled device (Photometrics), an Intensilight epifluorescence source (Nikon), a CFI Apo 100× NA 1.49 objective (Nikon), and a TIRF launcher with three laser lines: 488 nm (10 mW), 561 nm (50 mW), and 638 nm (20 mW). This microscope also included the Nikon Perfect Focus System, an interferometry-based focus lock that allowed the capture of multipoint and time-lapse images without loss of focus. All of the reported experiments were performed using the following Chroma filter cubes: TIRF 488, TIRF 640, TRITC, and RICM.
More detailed information about the materials and methods used in this study is provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health (NIH) Tetramer Facility at Emory University for providing the biotinylated pMHC monomers. K.S. was supported by NIH Grant R01 GM097399, an Alfred P. Sloan Research Fellowship, a Camille Dreyfus Teacher-Scholar Award, and National Science Foundation CAREER Award 1350829. L.B. was supported by a postdoctoral fellowship from the National Multiple Sclerosis Society (FG1963A1/1). R.A. was supported by NIH Grant T32 AI007610. B.E. and R.A. were supported by NIH Grant R01 AI096879.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600163113/-/DCSupplemental.
References
- 1.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chakraborty AK, Weiss A. Insights into the initiation of TCR signaling. Nat Immunol. 2014;15(9):798–807. doi: 10.1038/ni.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim ST, et al. The alphabeta T cell receptor is an anisotropic mechanosensor. J Biol Chem. 2009;284(45):31028–31037. doi: 10.1074/jbc.M109.052712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das DK, et al. Force-dependent transition in the T-cell receptor β-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc Natl Acad Sci USA. 2015;112(5):1517–1522. doi: 10.1073/pnas.1424829112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu B, Chen W, Evavold BD, Zhu C. Accumulation of dynamic catch bonds between TCR and agonist peptide MHC triggers T cell signaling. Cell. 2014;157(2):357–368. doi: 10.1016/j.cell.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong J, et al. Force-regulated in situ TCR peptide-bound MHC class II kinetics determine functions of CD4+ T cells. J Immunol. 2015;195(8):3557–3564. doi: 10.4049/jimmunol.1501407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mallis RJ, et al. Pre-TCR ligand binding impacts thymocyte development before αβTCR expression. Proc Natl Acad Sci USA. 2015;112(27):8373–8378. doi: 10.1073/pnas.1504971112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z, et al. Nanoscale optomechanical actuators for controlling mechanotransduction in living cells. Nat Methods. 2016;13(2):143–146. doi: 10.1038/nmeth.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKeithan TW. Kinetic proofreading in T-cell receptor signal transduction. Proc Natl Acad Sci USA. 1995;92(11):5042–5046. doi: 10.1073/pnas.92.11.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bashour KT, et al. CD28 and CD3 have complementary roles in T-cell traction forces. Proc Natl Acad Sci USA. 2014;111(6):2241–2246. doi: 10.1073/pnas.1315606111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hui KL, Balagopalan L, Samelson LE, Upadhyaya A. Cytoskeletal forces during signaling activation in Jurkat T-cells. Mol Biol Cell. 2015;26(4):685–695. doi: 10.1091/mbc.E14-03-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huppa JB, et al. TCR-peptide MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 2010;463(7283):963–967. doi: 10.1038/nature08746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Donoghue GP, Pielak RM, Smoligovets AA, Lin JJ, Groves JT. Direct single molecule measurement of TCR triggering by agonist pMHC in living primary T cells. eLife. 2013;2:e00778. doi: 10.7554/eLife.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deeg J, et al. T cell activation is determined by the number of presented antigens. Nano Lett. 2013;13(11):5619–5626. doi: 10.1021/nl403266t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delcassian D, et al. Nanoscale ligand spacing influences receptor triggering in T cells and NK cells. Nano Lett. 2013;13(11):5608–5614. doi: 10.1021/nl403252x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Yehl K, Narui Y, Salaita K. Tension sensing nanoparticles for mechano-imaging at the living/nonliving interface. J Am Chem Soc. 2013;135(14):5320–5323. doi: 10.1021/ja401494e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, et al. Nanoparticle tension probes patterned at the nanoscale: Impact of integrin clustering on force transmission. Nano Lett. 2014;14(10):5539–5546. doi: 10.1021/nl501912g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Ge C, Zhu C, Salaita K. DNA-based digital tension probes reveal integrin forces during early cell adhesion. Nat Commun. 2014;5:5167. doi: 10.1038/ncomms6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blakely BL, et al. A DNA-based molecular probe for optically reporting cellular traction forces. Nat Methods. 2014;11(12):1229–1232. doi: 10.1038/nmeth.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galior K, Liu Y, Yehl K, Vivek S, Salaita K. Titin-based nanoparticle tension sensors map high-magnitude integrin forces within focal adhesions. Nano Lett. 2016;16(1):341–348. doi: 10.1021/acs.nanolett.5b03888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jurchenko C, Chang Y, Narui Y, Zhang Y, Salaita KS. Integrin-generated forces lead to streptavidin-biotin unbinding in cellular adhesions. Biophys J. 2014;106(7):1436–1446. doi: 10.1016/j.bpj.2014.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang Y, et al. A general approach for generating fluorescent probes to visualize piconewton forces at the cell surface. J Am Chem Soc. 2016;138(9):2901–2904. doi: 10.1021/jacs.5b11602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woodside MT, et al. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc Natl Acad Sci USA. 2006;103(16):6190–6195. doi: 10.1073/pnas.0511048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu Y, Fay NC, Smoligovets AA, Wu HJ, Groves JT. Myosin IIA modulates T cell receptor transport and CasL phosphorylation during early immunological synapse formation. PLoS One. 2012;7(2):e30704. doi: 10.1371/journal.pone.0030704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi J, Wu XS, Crites T, Hammer JA., 3rd Actin retrograde flow and actomyosin II arc contraction drive receptor cluster dynamics at the immunological synapse in Jurkat T cells. Mol Biol Cell. 2012;23(5):834–852. doi: 10.1091/mbc.E11-08-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Babich A, et al. F-actin polymerization and retrograde flow drive sustained PLCγ1 signaling during T cell activation. J Cell Biol. 2012;197(6):775–787. doi: 10.1083/jcb.201201018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li YC, et al. Cutting edge: Mechanical forces acting on T cells immobilized via the TCR complex can trigger TCR signaling. J Immunol. 2010;184(11):5959–5963. doi: 10.4049/jimmunol.0900775. [DOI] [PubMed] [Google Scholar]
- 28.Grakoui A, et al. The immunological synapse: A molecular machine controlling T cell activation. Science. 1999;285(5425):221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 29.Lee KH, et al. T cell receptor signaling precedes immunological synapse formation. Science. 2002;295(5559):1539–1542. doi: 10.1126/science.1067710. [DOI] [PubMed] [Google Scholar]
- 30.Casas J, et al. Ligand-engaged TCR is triggered by Lck not associated with CD8 coreceptor. Nat Commun. 2014;5:5624. doi: 10.1038/ncomms6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hui E, Vale RD. In vitro membrane reconstitution of the T-cell receptor proximal signaling network. Nat Struct Mol Biol. 2014;21(2):133–142. doi: 10.1038/nsmb.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beemiller P, Krummel MF. Mediation of T-cell activation by actin meshworks. Cold Spring Harb Perspect Biol. 2010;2(9):a002444. doi: 10.1101/cshperspect.a002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beemiller P, Jacobelli J, Krummel MF. Integration of the movement of signaling microclusters with cellular motility in immunological synapses. Nat Immunol. 2012;13(8):787–795. doi: 10.1038/ni.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choudhuri K, et al. Polarized release of T-cell receptor-enriched microvesicles at the immunological synapse. Nature. 2014;507(7490):118–123. doi: 10.1038/nature12951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evavold BD, Allen PM. Separation of IL-4 production from Th cell proliferation by an altered T cell receptor ligand. Science. 1991;252(5010):1308–1310. doi: 10.1126/science.1833816. [DOI] [PubMed] [Google Scholar]
- 36.Zehn D, Lee SY, Bevan MJ. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009;458(7235):211–214. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Ha T. Defining single molecular forces required to activate integrin and notch signaling. Science. 2013;340(6135):991–994. doi: 10.1126/science.1231041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma PV, et al. The mechanically-induced catalytic amplification reaction for readout of receptor-mediated cellular forces. Angew Chem Int Ed. 2016 doi: 10.1002/anie.201600351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klammt C, et al. T cell receptor dwell times control the kinase activity of Zap70. Nat Immunol. 2015;16(9):961–969. doi: 10.1038/ni.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Depoil D, Dustin ML. Force and affinity in ligand discrimination by the TCR. Trends Immunol. 2014;35(12):597–603. doi: 10.1016/j.it.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tibaldi EV, Salgia R, Reinherz EL. CD2 molecules redistribute to the uropod during T cell scanning: Implications for cellular activation and immune surveillance. Proc Natl Acad Sci USA. 2002;99(11):7582–7587. doi: 10.1073/pnas.112212699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith A, et al. A talin-dependent LFA-1 focal zone is formed by rapidly migrating T lymphocytes. J Cell Biol. 2005;170(1):141–151. doi: 10.1083/jcb.200412032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith A, Bracke M, Leitinger B, Porter JC, Hogg N. LFA-1-induced T cell migration on ICAM-1 involves regulation of MLCK-mediated attachment and ROCK-dependent detachment. J Cell Sci. 2003;116(Pt 15):3123–3133. doi: 10.1242/jcs.00606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.