Significance
B-cell receptor (BCR) signaling promotes the survival of malignant B cells, such as Burkitt’s lymphoma (BL) and the activated B-cell–like subtype of diffuse large B-cell lymphoma (ABC-DLBCL). In contrast to ABC-DLBCL, which depends on chronic activation of the BCR, BL cells rely on tonic BCR signaling that is antigen-independent. Elucidation and systematic comparison of tonic and activated BCR signaling led to the identification of novel signaling effectors, including ACTN4 and ARFGEF2, which were identified as regulators of BL-cell survival. Beyond its relevance to the understanding of BL pathogenesis and the development of targeted therapies, our study complements the general understanding of BCR-induced processes also in physiological settings.
Keywords: lymphoma, B-cell receptor, phosphoproteome, cancer biology
Abstract
Burkitt's lymphoma (BL) is a highly proliferative B-cell neoplasm and is treated with intensive chemotherapy that, because of its toxicity, is often not suitable for the elderly or for patients with endemic BL in developing countries. BL cell survival relies on signals transduced by B-cell antigen receptors (BCRs). However, tonic as well as activated BCR signaling networks and their relevance for targeted therapies in BL remain elusive. We have systematically characterized and compared tonic and activated BCR signaling in BL by quantitative phosphoproteomics to identify novel BCR effectors and potential drug targets. We identified and quantified ∼16,000 phospho-sites in BL cells. Among these sites, 909 were related to tonic BCR signaling, whereas 984 phospho-sites were regulated upon BCR engagement. The majority of the identified BCR signaling effectors have not been described in the context of B cells or lymphomas yet. Most of these newly identified BCR effectors are predicted to be involved in the regulation of kinases, transcription, and cytoskeleton dynamics. Although tonic and activated BCR signaling shared a considerable number of effector proteins, we identified distinct phosphorylation events in tonic BCR signaling. We investigated the functional relevance of some newly identified BCR effectors and show that ACTN4 and ARFGEF2, which have been described as regulators of membrane-trafficking and cytoskeleton-related processes, respectively, are crucial for BL cell survival. Thus, this study provides a comprehensive dataset for tonic and activated BCR signaling and identifies effector proteins that may be relevant for BL cell survival and thus may help to develop new BL treatments.
The B-cell antigen receptors (BCRs) control important cell-fate decisions in the B-lineage—including proliferation, differentiation, and cell survival—by regulating complex intracellular signaling networks (1). The molecular communication between signaling intermediates is achieved mainly through reversible protein phosphorylation of tyrosine, serine, or threonine residues, which ensures regular B-cell function (2). The molecular architecture of this intricate BCR signaling network has so far been studied mostly at the level of individual molecules and signaling complexes.
Dysregulation of BCR signaling is involved in the pathogenesis of immunodeficiency, autoimmune disorders, and B-cell malignancies. For example, chronic lymphocytic leukemia (CLL) and activated B cell-like diffuse large B-cell lymphoma (ABC-DLBCL) depend on chronic active BCR signaling whereas “tonic” BCR signaling, which is thought to provide an antigen-independent constitutive baseline signal, induces cell survival in the majority of Burkitt lymphomas (BLs) (3, 4). Despite the fact that the molecular details remain largely elusive, the essential role of tonic BCR signaling is further emphasized by its importance for pivotal aspects of B-cell biology, such as regular B-cell development (5). In line with the distinct BCR signaling modes found in B-cell malignancies, no relevant antigens have been identified for BL so far whereas self-antigens were reported to trigger oncogenic BCR signaling in CLL and ABC-DLBCL cells (6, 7). Interestingly, different lymphoma entities seem to rely on distinct BCR signaling branches. In ABC-DLBCL, for example, mutated BCR signaling effectors mediate cell survival by inducing chronic active NFκB signaling (8, 9). In contrast, tonic BCR-dependent phosphatidylinositol 3 kinase (PI3K) signals have been reported to promote the survival and proliferation of BL cells (4) whereas NFκB activity seems to be dispensable (10). Despite these initial findings, the functional relevance and molecular basis of BCR signaling in BL remain elusive to date. A more comprehensive knowledge about BCR signaling in BL, however, is necessary to understand the pathogenesis of BL and may lead to the identification of functionally relevant and potentially druggable signaling pathways, which most likely fall into the category of the so far poorly characterized tonic BCR signaling networks.
In this work, we sought to elucidate both tonic and activated BCR signaling in two BL-cell models that, although differing in their genotype, depend on BCR expression. By quantitative phosphoproteomics, we were able to identify, beyond phosphorylation of already known BCR effectors, more than 300 hitherto unknown BCR effectors with their BCR-dependent phosphorylation sites (p-sites). Our study clearly identifies both commonalities and marked differences between the activated and tonic BCR signaling modes. The exemplary examination of the functional relevance of selected identified phosphoproteins for BL-cell survival confirmed that our approach revealed previously unidentified BCR effectors relevant for BL.
Results
Characterization of Activated BCR Signaling in the Human Burkitt’s Lymphoma Models DG75 and Daudi.
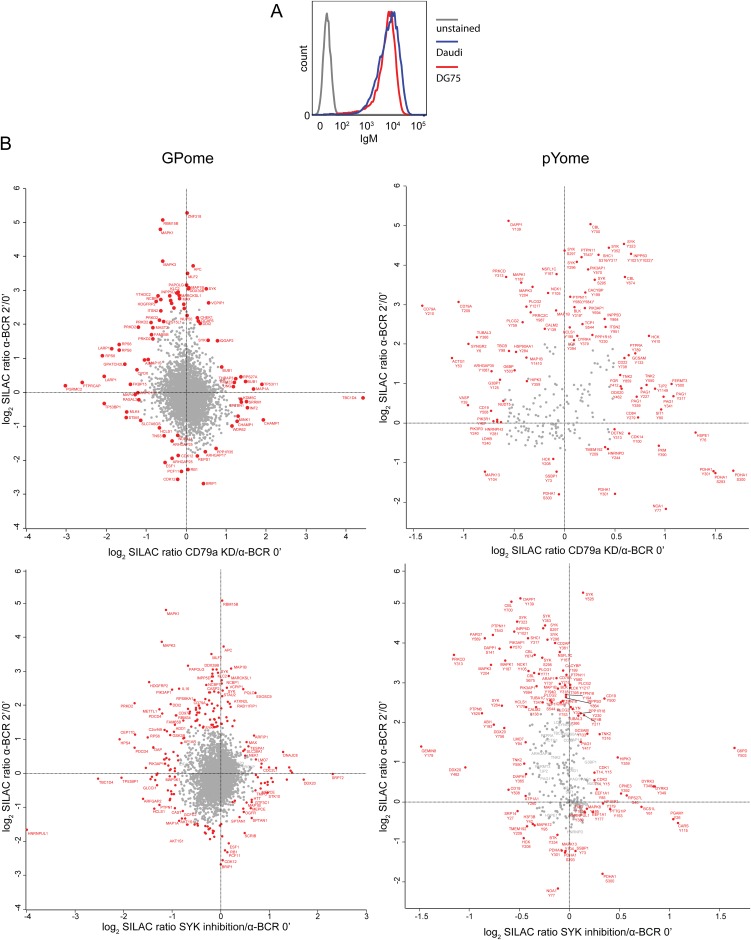
Despite the importance of BCR signaling in the pathogenesis of BL, drug target identification in BL-specific BCR signaling networks is hampered by incomplete understanding of the full spectrum of BCR-mediated events (4, 11). We therefore aimed at comprehensively characterizing BCR signaling in BL by pursuing a quantitative phosphoproteomic approach (Fig. S1 A and B). For this approach, we used the human cell lines DG75 and Daudi because they are frequently used BL models that (i) depend on BCR expression (4) (see also Fig. 2A) and (ii) have functional BCR signaling (Fig. S1C).
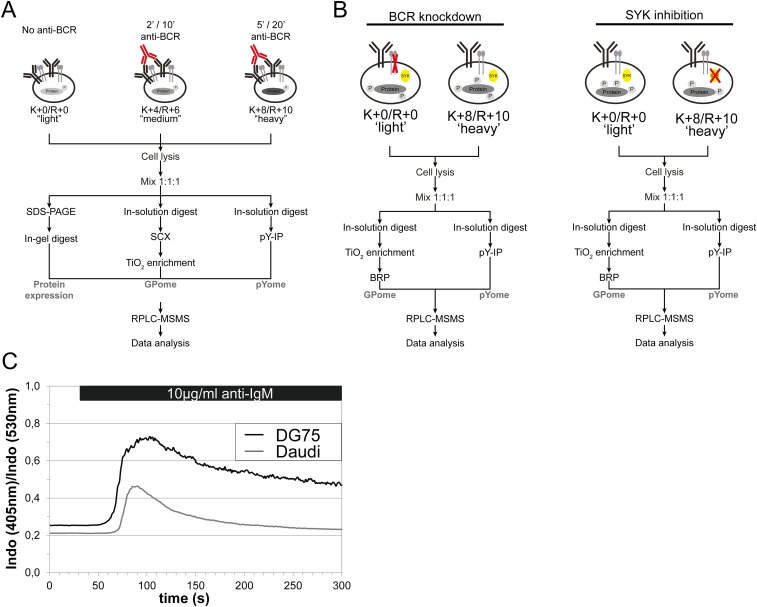
Fig. S1.
(A) Schematic representation of a 3-plex SILAC approach for profiling phosphorylation dynamics in resting and BCR-stimulated DG75 cells. DG75 cells were cultured in SILAC medium as indicated and were left untreated, or were BCR-stimulated, for 2, 5, 10, or 20 min. Daudi cells were stimulated for 2 and 10 min. Lysates were mixed in a 1:1:1 ratio and digested with trypsin. Resulting phosphopeptides were enriched by either SCX/TiO2 chromatography (global phosphoproteome analysis; GPome) or phosphotyrosine immunoprecipitation (pY-IP; pYome analysis), and analyzed by LC-MS/MS. For analysis of protein expression levels, proteins were separated by 1D-PAGE, digested with trypsin, and analyzed by LC-MS/MS (see SI Materials and Methods for details). (B) Schematic representation of a 2-plex SILAC approach for profiling phosphorylation changes upon inducible CD79a knockdown or upon SYK inhibition. DG75 cells were cultured in SILAC medium and treated as indicated. Lysates were processed as described in A. (C) DG75 and Daudi cells were loaded with the ratiometric Ca2+-chelator INDO-1-AM and subjected to BCR-induced Ca2+ flux analysis by flow cytometry.
Fig. 2.
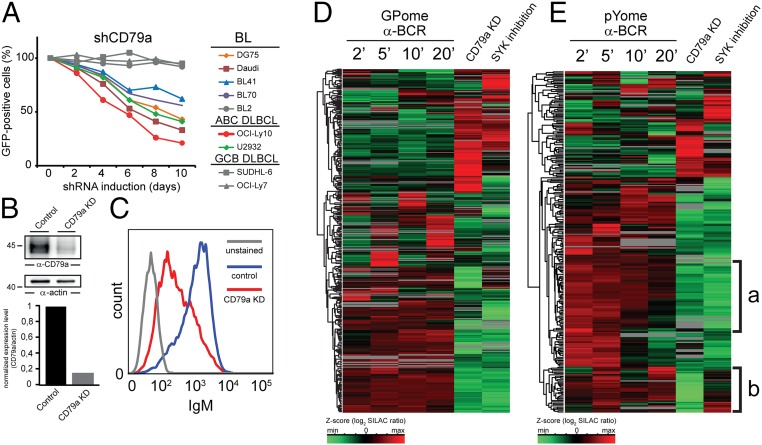
Tonic BCR signaling. (A) CD79a shRNAs are toxic for BL cell lines. The figure shows the fraction of GFP-positive, shRNA-expressing cells relative to the GFP-negative, shRNA-negative fraction at the times indicated (normalized to day 0). Data are representative of three experiments. (B) CD79a and actin immunoblots of lysates derived from DG75 cells that were treated with doxycycline for 18 h to express either unspecific shRNAs (Control) or shRNAs targeting CD79a. (C) BCR cell surface expression in DG75 control cells or CD79a knockdown cells was monitored by flow cytometry 18 h after shRNA induction. (D and E) Unsupervised clustering analysis of all p-sites that were regulated upon BCR stimulation/CD79a knockdown/SYK inhibition. Values for each p-site (row) in all conditions (columns) are colored based on the z-score of the log2-transformed SILAC ratios.
We applied stable-isotope labeling by amino acids in cell culture (SILAC) (12) to DG75 cells and quantified the changes in the phosphorylation of effector proteins after various durations of BCR stimulation (2, 5, 10, and 20 min) by mass spectrometry (MS) (Fig. S1A). Phosphopeptides with pS, pT, and pY sites derived from the corresponding time points of BCR stimulation were enriched by strong-cation-exchange (SCX) chromatography in combination with titanium dioxide (TiO2) solid-phase extraction; we refer to this type of analysis as the “global phosphoproteome” (GPome). Using pY-specific antibodies in an additional analysis, we enriched mainly tyrosine-phosphorylated peptides, which we refer to as the “pYome.” To exclude any possibility that BCR stimulation causes major changes in protein levels that interfere with the quantification of phosphorylation events, we performed large-scale SILAC-based protein expression profiling in parallel. No significant BCR-induced changes in the abundance of proteins relevant for BCR signaling within the DG75 proteome were observed (Fig. S2A).
Fig. S2.
(A) Protein expression profiling: Scatter plots of normalized “protein groups” SILAC ratios (log2) versus intensity (log10) for 2, 5, 10, and 20 min after BCR stimulation. Outlier proteins are marked red, and the respective gene names are shown. (B) Pearson’s correlation analysis of normalized SILAC ratios (log2) of all p-sites quantified in four biological replicates (indicated by 1, 2, 3, and 4) from the global phosphoproteome experiment of DG75 cells. Pearson’s correlation coefficient is indicated in blue in the upper left corner of each scatter plot. The pink box highlights the correlation of the biological replicates of 2 min, blue 5 min, yellow 10 min, and green 20 min BCR stimulation. (C) Pearson’s correlation analysis of normalized SILAC ratios (log2) of biological replicates (indicated by 1 and 2) of all p-sites derived from the pYome experiment. Pearson’s correlation coefficient is shown in blue in the upper left corner of each scatter plot. The pink box highlights the correlation of the biological replicates of 2 min, blue 5 min, yellow 10 min, and green 20 min BCR stimulation.
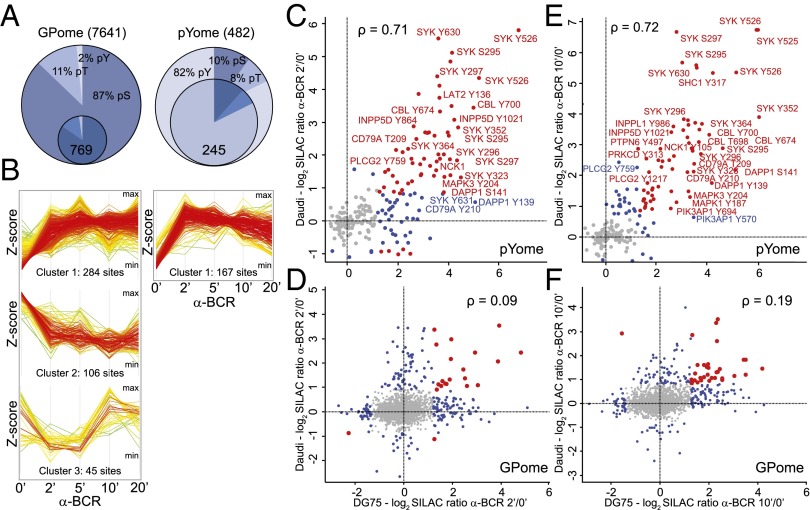
In the GPome of DG75 cells, we detected around 9,000 class I p-sites (p-sites with a localization probability of >75%), of which 7,641 were quantifiable for all four durations of BCR stimulation (2, 5, 10, and 20 min) (Fig. 1A, Fig. S2B, and Dataset S1). The majority of the p-sites identified and quantified were identified as serine (∼87%), followed by threonine (∼11%) and tyrosine (∼2%) residues (Fig. 1A). The pYome analysis revealed 482 quantifiable class I p-sites (including pS and pT sites) for all stimulation durations. Confirming the validity of our phospho-tyrosine enrichment approach, 80% of these p-sites were found on tyrosine residues (Fig. 1A, Fig. S2C, and Dataset S1). A total of 769 unique class I p-sites in the GPome and 245 p-sites in the pYome in a total of 577 proteins showed significant phosphorylation changes upon BCR stimulation (Fig. 1A and Dataset S1).
Fig. 1.
Activated BCR signaling in BL cells. (A) Numbers of quantified and regulated p-sites in the GPome and the pYome of DG75 cells. Outer circles represent the number of quantified class I p-sites at all stimulation durations. Inner circles represent the relative abundance of regulated class I p-sites. (B) Unsupervised clustering analysis of all regulated p-sites quantified at all BCR stimulation durations in DG75 cells. Line graphs illustrate the phosphorylation dynamics upon BCR stimulation for each identified cluster for the indicated stimulation time course (y axis, z-score of the log2 SILAC ratios; x axis, minutes). (C–F) Scatter plots of fold-change of p-sites on tyrosines (C and E) and serines/threonines (D and F) in Daudi (y axis) versus DG75 (x axis) cells as determined by quantitative MS upon 2-min (C and D) and 10-min (E and F) BCR stimulation. Selected phosphorylated proteins are shown. Colored dots indicate p-sites that are significantly regulated in both cell lines (red) or just in one cell line (blue). The Spearman’s correlation coefficient (ρ) is indicated.
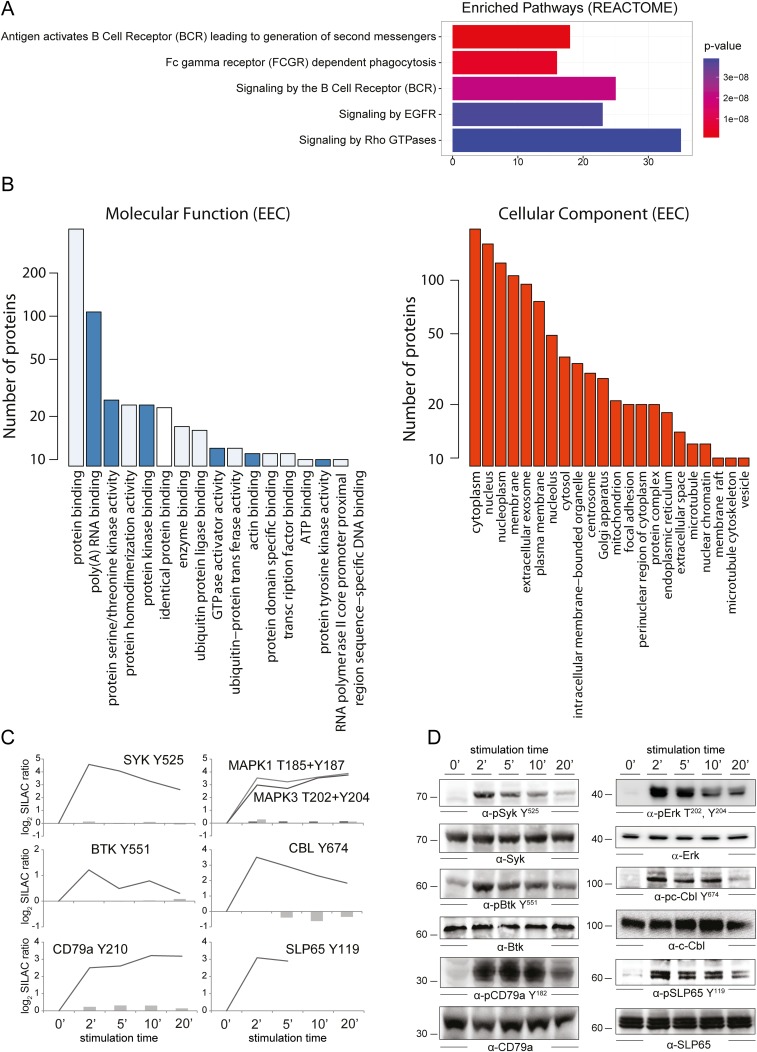
To assign BCR-dependent p-sites with distinct phosphorylation dynamics to signaling clusters, we performed an unsupervised clustering analysis, with p-sites quantified at all stimulation durations using stringent filtering parameters (SI Materials and Methods). For the GPome, we identified three separate clusters, with the largest cluster containing phosphorylation events early after BCR engagement (284 p-sites), followed by down-regulated p-sites (106 p-sites) and late p-sites (45 p-sites) (Fig. 1B, Left). In marked contrast, we found only one cluster for the pYome data containing p-sites that are up-regulated early upon BCR stimulation (Fig. 1B, Right), a finding that is consistent with the pivotal role of tyrosine phosphorylation in BCR-proximal signaling processes. These data represent the largest resource for BCR-induced phosphorylation events in human lymphoma cells to date. The identified p-sites directly link to BCR effector proteins. Pathway enrichment analysis confirmed a strong representation of known BCR effector proteins in our dataset (Fig. S3A). Because most annotation databases [e.g., Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology (GO), and Ingenuity] do not contain the latest published information, we performed an additional extensive literature (PubMed) search to determine how many novel BCR effectors were identified. This approach revealed that around 60% of the identified BCR effectors have not yet been described in the context of BCR signal transduction per se (Dataset S1).
Fig. S3.
(A) Pathway enrichment analysis for BCR effectors identified upon BCR stimulation of DG75 cells. (B) Experimentally validated molecular function and cellular component ontology terms annotated to BCR effector proteins identified in DG75 cells. Bars representing the numbers of cytoskeleton and transcriptional regulators, kinases, and RNA-binding proteins are marked in dark blue. See SI Materials and Methods for details. (C) Mass-spectrometric quantification of expression levels (grey bars) and p-sites (line graphs) of the indicated BCR signaling effectors. (D) Phosphorylation dynamics and respective expression levels of proteins analyzed in C were monitored by immunoblotting.
A bioinformatic annotation of putative protein functions revealed that, apart from kinases, transcriptional regulators, and RNA-binding proteins, cytoskeletal regulators are among the most prominent functional groups of BCR effectors (Fig. S3B, highlighted in dark blue). This finding emphasizes that, beyond kinase activity and transcriptional regulation, cytoskeletal rearrangements are important downstream effects of BCR engagement as well (13).
We validated our quantitative MS results by immunoblot analyses using phosphosite-specific antibodies directed against some of the quantified p-sites. As depicted in Fig. S3 C and D, phosphorylation of the BCR-proximal effectors Igα (CD79a) (Y210), SYK (Y525), SLP65 (also known as BLNK) (Y119), BTK (Y551), c-CBL (Y674), and the more BCR-distal MAPK1/3 (ERK1/2, T185, Y187, T202/Y204) showed phosphorylation patterns very similar to those found by mass spectrometry.
The BCR Signaling Response Is in Part Genotype-Specific.
BL is a genetically heterogeneous disease characterized by certain recurrently mutated (onco)genes that cooperate with Myc, the defining oncogene of this cancer (4, 14). In contrast to ABC-DLBCL, where mutations in BCR signaling effectors cause oncogenic “chronic active” BCR signaling, no mutations in BCR proximal signaling effectors were identified in BL (4). To investigate whether the BL-genotype with the corresponding mutations in the BL-specific (onco)genes still affects BCR signaling, we characterized BCR signaling in the BL cell line Daudi. Daudi cells genetically differ from DG75 cells because they harbor an inactivating ID3 mutation (4, 15), but have in common with DG75 cells that they (i) depend on BCR signaling (Fig. 2A) and (ii) express similar amounts of BCR on their surface (Fig. S4A).
Fig. S4.
(A) BCR cell surface expression was monitored by flow cytometry (red line, DG75; blue line, Daudi). (B) Scatter plots showing the fold-change of p-sites on mainly serines/threonines (GPome, Left) and mainly tyrosines (pYome, Right) as determined by quantitative MS upon BCR stimulation versus CD79a knockdown and SYK inhibition. Selected phosphorylated proteins and p-sites are highlighted.
By phosphoproteomics, we identified and quantified 8,171 pS/pT-sites and 319 pY-sites in Daudi cells. Of these sites, 500 (pS/pT) and 145 (pY) were significantly regulated upon BCR activation (Dataset S1). We found that most pY-sites were concordantly regulated in DG75 and Daudi cells upon BCR stimulation (indicated in red in Fig. 1 C and E); these pY-sites were mostly found in BCR-proximal effector proteins, such as the BCR subunit CD79a, spleen tyrosine kinase (Syk), the E3 ubiquitin protein ligase CBL, the cytoplasmic adaptor protein NCK, and others that are involved in early BCR signaling events (Fig. 1 C and E). In contrast, a large proportion of pS- and pT-sites were differentially phosphorylated among the two cell types (indicated in blue in Fig. 1 D and F), in particular those that act further downstream of the BCR [e.g., p-sites in transcription factors, such as the general transcription factor II-I (GTF2I) and POU domain class 2 transcription factor 1 (POU2F1)] (Fig. 1 D and F). The concordant regulation of tyrosine phosphorylation in BL cells of different genotypes is promising with respect to the identification of potential drug targets, given the fact that targeted therapies in other B-cell malignancies have been reported to interfere with proximal BCR signaling (16, 17).
Elucidation of Tonic BCR Signaling.
Recent studies suggest that BL cells depend on tonic antigen-independent BCR signaling because (i) they lack activation-inducing mutations in BCR effectors, which is in contrast to ABC-DLBCL (4, 9), (ii) no autoantigens were reported for their BCR, and (iii) the majority of BL cells depend on BCR expression (4). In line with these findings, we observed a profound reduction of viable BL cells for most of the BL cell lines analyzed upon induction of shRNAs targeting the BCR component CD79a, confirming previously published data (Fig. 2A). ABC and germinal center B-cell–like (GCB) DLBCL cell lines served as positive and negative controls, respectively. Because tonic BCR signaling is essential for BL cell survival, knowledge about the pathways activated by such tonic BCR signaling would foster the identification of novel drug targets and improve our understanding of BL pathogenesis.
To elucidate tonic BCR signaling in BL, we analyzed phosphoproteomic changes that occurred either upon inducible CD79a knockdown or upon pharmacological SYK inhibition, using the small molecule inhibitor PRT-062607 (Fig. S1B). This SYK inhibitor has been validated in previous studies, where it showed suppression of BCR-dependent SYK activity in B cells (18, 19). Upon induction of shRNAs targeting CD79a in DG75 cells, we observed a profound reduction of CD79a expression leading to diminished expression of BCR on the cell surface (Fig. 2 B and C). An shRNA induction duration of 18 h was identified as the optimum condition for the analysis of tonic BCR signaling because the BCR expression level was already significantly reduced (Fig. 2C) whereas, at the same time, more than 95% of the cells were still viable. We identified and quantified ∼16,000 class I p-sites upon BCR stimulation, inducible CD79a knockdown, and SYK inhibition. Of these p-sites, 4,524 sites were identified under all conditions and were subsequently subjected to comparative analysis.
A total of 515 and 441 p-sites were significantly regulated upon CD79a knockdown and SYK inhibition, respectively (Dataset S2). Most p-sites that were up-regulated upon BCR stimulation were found to be down-regulated upon CD79a knockdown and SYK inhibition (Fig. 2 D and E, cluster a). However, we also identified specific p-sites that were regulated upon disruption of tonic BCR signaling whereas they were not affected upon BCR activation and vice versa (Fig. S4B). This result points toward specific qualitative differences between the activated and tonic BCR signaling modes.
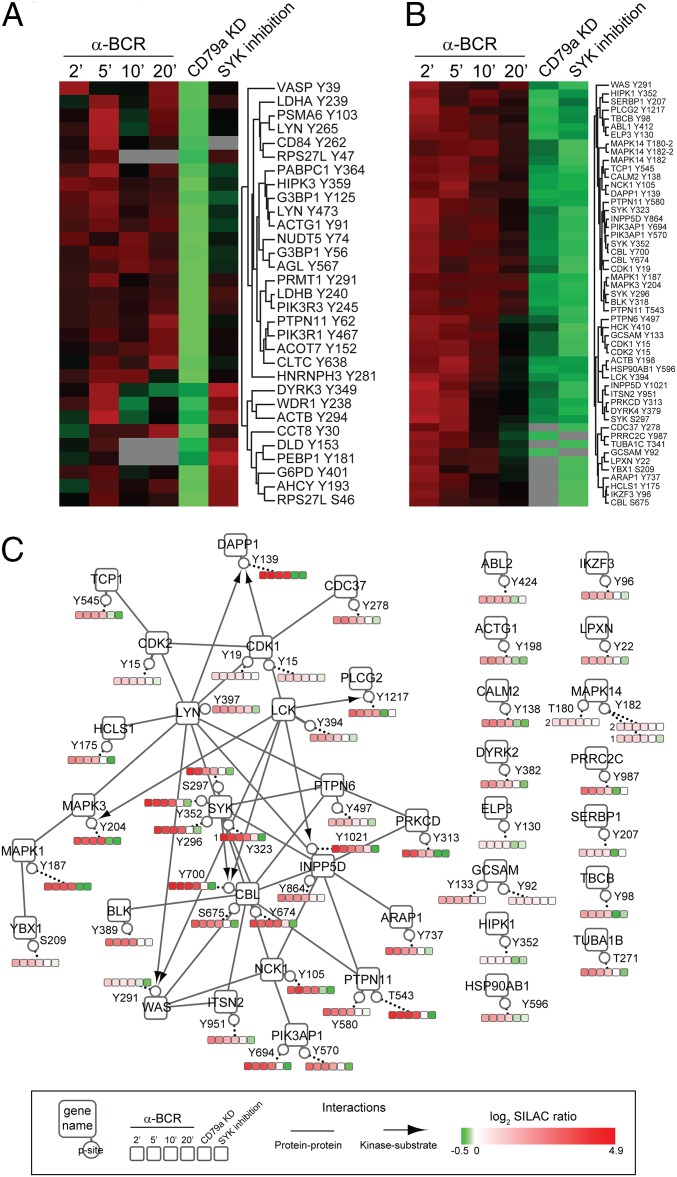
Notably, we found p-sites that were down-regulated upon CD79a knockdown but were not affected by SYK inhibition (Fig. 2E, cluster b, and Fig. 3A). Such p-sites were found on proteins such as tyrosine kinase LYN and regulatory subunits of PI3K, both of which are known to act upstream of SYK in BCR signaling (3, 20). Thus, these proteins are likely to represent effectors that act either in close proximity to the BCR and upstream of SYK or just independently of SYK.
Fig. 3.
BCR signaling networks. (A and B) Enlarged versions of cluster “b” (A) and cluster “a” (B) of the heatmap shown in Fig. 2E including protein names and p-sites. (C) Cytoscape derived signaling network showing selected BCR effectors (derived from cluster “a” of Fig. 2E) that contain regulated phospho-sites upon BCR stimulation and upon interference with tonic BCR signaling. Proteins were grouped according to their known protein–protein interaction status (BioGRID database). Kinase–substrate relations were derived from the PhosphoSitePlus database. The phosphorylation status of individual p-sites is shown in color-coded rectangles.
We also investigated tonic BCR signaling in Daudi cells in the presence of SYK inhibitor (Dataset S3). However, investigation of phosphoproteomic changes in Daudi cells upon CD79a knockdown was not possible, owing to the fact that apoptosis occurred very early under these conditions. Nevertheless, Daudi cells share a considerable amount of phosphorylation changes under SYK inhibitor treatment.
BCR Signaling Networks in BL.
To systematically integrate the identified effectors and, more specifically, tyrosine phosphorylation events into BCR signaling cascades, we generated a pYome signaling network based on our data from the pYome analyses of tonic and activated BCR signaling. We first focused on some selected proteins that were identified as being phosphorylated upon BCR stimulation, while dephosphorylated upon CD79a knockdown and SYK inhibition [Fig. 3B (cluster “a” from Fig. 2E)]. Such proteins are supposedly involved in both tonic and activated BCR signaling. To generate a signaling network, we combined our information about differentially phosphorylated proteins with known protein–protein interactions derived from the BioGRID database and kinase-substrate relations derived from the PhosphoSitePlus database. Fig. 3C shows the pYome network thus generated, in which pivotal and well-studied BCR-proximal signaling effectors, including Src kinases, SYK, phospholipase C-gamma-2 (PLCγ2), CBL, and mitogen-activated protein kinases (MAPK) like ERK, are found in a highly interconnected module. Previously published data showed an important role of PI3K function in tonic BCR signaling in BL (4). In accordance with these data, we found that the B-cell–specific PI3K activating complex consisting of LYN, NCK, and phosphoinositide-3-kinase adaptor protein (PIK3AP1) (also known as BCAP) (20), as well as downstream effectors of PI3K signaling like dual adaptor protein of phosphotyrosine and 3-phosphoinositides (DAPP1) (also known as BAM32) (21), are phosphorylated in tonic BCR signaling. Notably, effector proteins, which were also shown to be phosphorylated in tonic as well as activated BCR signaling, are not yet linked to the main BCR signaling hub and may point to hitherto unknown BCR-signaling complexes. These effector proteins include components of the cytoskeleton, such as γ-actin (ACTG1) and α-tubulin (TUBA1B), as well as putative cytoskeleton regulators like Abelson protein tyrosine kinase 2 (ABL2) (22) and Leupaxin (LPXN) (23). The latter has also been described as a negative regulator of BCR signaling (24). We also identified significantly regulated phosphorylation of the Ikaros transcription factor family member Aiolos (IKZF3), which is known to be important for B-cell activation (25) and to be up-regulated in CLL (26). Ikaros proteins are pivotal regulators of hematopoiesis and immunity (27) and have been reported to be essential for B-cell development (28). Interestingly, we identified tyrosine residue 96 of Aiolos to be phosphorylated in tonic and activated BCR signaling. Although serine phosphorylation of IKZF1-encoded Ikaros has been shown to control its cellular localization (29), a regulation of Ikaros proteins by tyrosine phosphorylation is hitherto unknown. Therefore, our data might help to understand how BCR-proximal processes are linked to the regulation of this protein family.
Identification of BCR Effectors Involved in Regulation of BL Cell Survival.
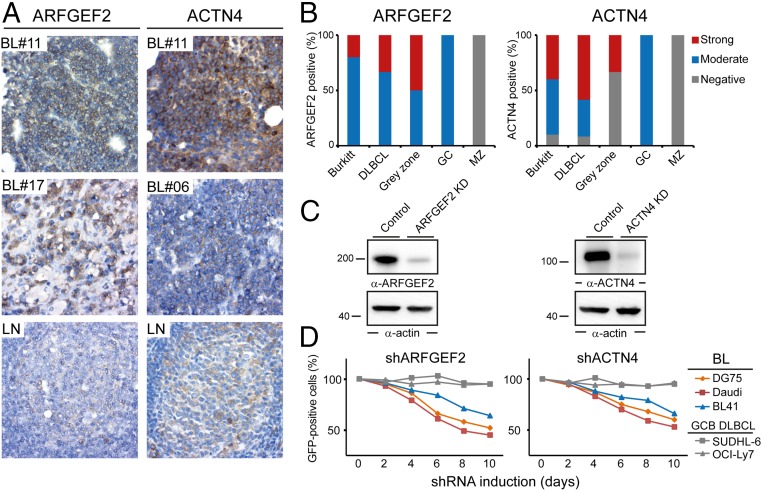
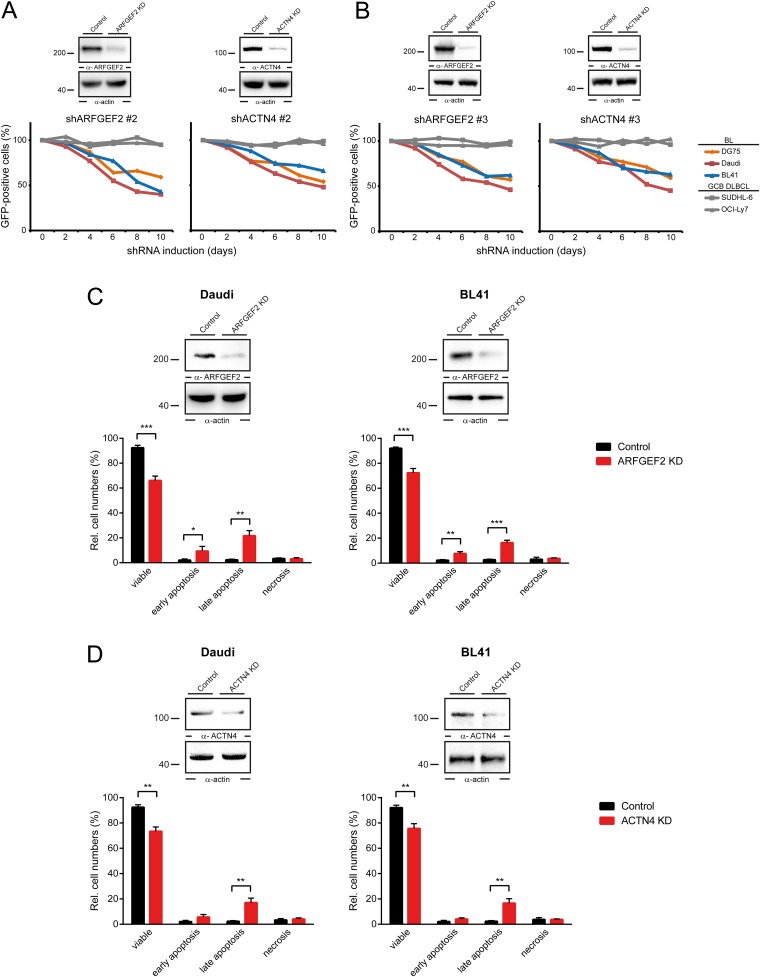
Based on the identification of regulated p-sites in BCR signaling, we next investigated, in an exemplary manner, whether the newly identified BCR effectors are relevant for BL-cell fitness and survival. Therefore, we targeted a subset of selected genes that encode proteins that were identified as being phosphorylated in a BCR-dependent manner by an shRNA-based approach. Among these genes were several that have not yet been described as relevant for BL pathophysiology, including ADP ribosylation factor guanine nucleotide-exchange factor 2 (ARFGEF2) and actinin-4 (ACTN4). In other cell types, ARFGEF2 and ACTN4 have been described as regulators of membrane-trafficking and cytoskeleton-related processes, respectively (30, 31). We first confirmed the expression of ARFGEF2 and ACTN4 in patient-derived Burkitt’s lymphoma samples by immunohistochemical analysis (Fig. 4 A and B). Subsequently, we interfered with the expression of ARFGEF2 and ACTN4 by using specific shRNAs (Fig. 4C), allowing us to investigate their impact on the regulation of BL-cell fitness and survival. We monitored the relative proportions of GFP-positive cells that express shRNAs for ARFGEF2 or ACTN4, respectively (Dataset S4). We found their abundance in DG75, Daudi, and BL41 cell cultures to be markedly decreased over time whereas cells derived from GCB-DLBCL were not affected (Fig. 4D). To exclude off-target effects of the shRNAs, the same type of analysis was repeated with two additional shRNAs per gene, which confirmed a role of ARFGEF2 and ACTN4 in the regulation of BL-cell survival (Fig. S5 A and B). Finally, we performed Annexin V-based apoptosis assays to see whether the reduction of ARFGEF2 or ACTN4 expression directly affects cell survival. This analysis revealed an augmented apoptosis rate in the ARFGEF2 and ACTN4 knockdown cells (Fig. S5 C and D). See Dataset S4 for shRNA sequences.
Fig. 4.
(A and B) Tissues derived from lymph nodes of either patients with Burkitt lymphoma (n = 11) (A, Top and Middle), DLBCL (n = 13), and Grey zone lymphoma (n = 6) or healthy donors (n = 4) (A, Bottom) were immunohistochemically stained with antibodies against ARFGEF2 and ACTN4. Two independent pathologists evaluated all tissue sections on the basis of staining intensity using a three-stage staining score. A 40-fold magnification was used. GC, Germinal center; MZ, marginal zone. (C) Immunoblots with antibodies against ARFGEF2 and ACTN4 of cleared cellular lysates derived from Daudi cells that were treated with unspecific shRNAs and shRNAs specific for either ARFGEF2 or ACTN4 (Upper). Protein loading was monitored by anti-actin immunoblotting (Lower). (D) ARFGEF2 and ACTN4 shRNAs are toxic for BL. The figure shows the fraction of GFP-positive, shRNA-expressing cells relative to the GFP-negative, shRNA-negative fraction at the times indicated (normalized to the day 0 values). Data are representative of three experiments.
Fig. S5.
(A and B) Immunoblots of cleared cellular lysates derived from control cells that were treated with unspecific shRNAs and the respective knockdown cells with antibodies against ARFGEF2 and ACTN4 (Upper). Protein loading was monitored by anti-actin immunoblotting (Lower). ARFGEF2 and ACTN4 shRNAs are toxic for BL. The figure shows the fraction of GFP-positive, shRNA-expressing cells relative to the GFP-negative, shRNA-negative fraction at the indicated times (normalized to the day 0 values). Data represent three experiments. (C and D) Immunoblots showing reduction of ARFGEF2 (C) and ACTN4 (D) expression levels in Daudi and BL41 cells upon 24 h doxycycline treatment whereas induction of nonspecific control shRNAs showed no reduction of ARFGEF2 and ACTN4 expression levels. Actin is shown as a loading control (Upper). Flow cytometric Annexin V/7-AAD–based apoptosis analysis of Daudi and BL41 cells 4 d after induction of ARFGEF2 and ACTN4-specific shRNAs or unspecific control shRNAs (Lower). Statistical significance: *P < 0.05, **P < 0.01 using Student’s t test, ***P < 0.001.
SI Materials and Methods
Cell Culture, BCR Stimulation, and Cell Lysis.
All cell lines were cultured in RPMI medium (Invitrogen) supplemented with 10–20% (vol/vol) heat-inactivated FBS (Invitrogen), penicillin/streptomycin (Invitrogen), and l-glutamine (Invitrogen) at 37 °C and 5% CO2.
SILAC labeling of DG75 and Daudi cells was performed by culturing the cells in SILAC-RPMI medium 1640 devoid of arginine and lysine (Pierce) supplemented with 10% (vol/vol) dialyzed FCS (PAA) and the respective SILAC amino acids (all from Cambridge Isotopes). “Light” (L) SILAC medium contained [12C614N4]-l-arginine and [12C614N2]-l-lysine; “intermediate” (M) SILAC medium contained [13C614N4]-l-arginine and 4,4,5,5-d4-l-lysine; “heavy” (H) SILAC medium contained [13C615N4]-l-arginine and [13C615N2]-l-lysine.
For BCR stimulation, DG75 and Daudi cells were starved in RPMI without supplements for 15 min, and subsequently BCR were stimulated for the indicated durations with 10 µg/mL F(ab)2 anti-human IgM antibodies (Dianova) at 37 °C. For phosphoproteomic and protein expression profiling of DG75 cells, the L-labeled batches were left untreated whereas the M-labeled batches were BCR-stimulated for 2 min and the H-labeled batches for 5 min. In a second experimental part, the L-batches were again left untreated whereas the M- and H-batches were stimulated for 10 and 20 min, respectively. L-labeled Daudi batches were left unstimulated (0 min) whereas M-labeled cells were BCR-stimulated for 2 min and the H-labeled cells for 10 min. For analysis of tonic BCR signaling, L-batches were treated with shRNAs targeting CD79a or DMSO and H-batches were treated with either unspecific shRNAs or the SYK inhibitor PRT-062607.
Cell lysis for SILAC-labeled and label-free cells was performed as described in Oellerich et al. (41), except that the phosphatase inhibitor mixtures 2 and 3 (Sigma) were added according to the manufacturer’s instructions. Cell lysis for proteomic profiling of tyrosine phosphorylation was performed according to the manufacturer’s instructions by using a urea-based lysis buffer (PTMScan Kit; Cell Signaling Technology).
Antibodies, Compounds, Plasmids, and Reagents.
Antibodies against the following proteins were used: SLP65, pSLP65, PLCγ2, pPLCγ2, ERK, pERK, SYK, pSYK, BTK, pBTK, CD79a, pCD79a, CBL, pCBL, ACTN4, actin (all from Cell Signaling Technology), pTyr (4G10; Millipore) and ARFGEF2 (Abcam). Tyrosine-phosphorylated peptides were enriched by using the PTMscan Phosphotyrosine Rabbit mAb P-Tyr-1000 kit (Cell Signaling Technology).
PRT-062607 was obtained from Selleck Chemicals, dissolved in DMSO at 10 mM, used in a final concentration of 250 nM, and stored at –80 °C.
For lentiviral knockdown of CD79a, ARFGEF2, and ACTN4, we used the inducible retroviral vector described by Fellmann et al. (43).
shRNA-Mediated Knockdown Experiments.
Cells were transduced with retroviral vectors coexpressing inducible unspecific shRNAs and GFP or shRNAs targeting the 3′ UTR of CD79a and GFP. Transduced cells were enriched by FACS sorting so that ∼50% of cells were GFP-positive. At this point, shRNA expression was induced using 1 μg/mL doxycycline, and the cell fraction positive for GFP was monitored over time by flow cytometry as described in Schmitz et al. (4). The same experimental approach was performed with shRNAs targeting ARFGEF2 and ACTN4.
Immunohistochemistry.
Each tissue sample was fixed in 4% (vol/vol) buffered formalin and embedded in paraffin. Informed consent from all patients at the University Medical Center Göttingen and a corresponding ethics vote was obtained, which allows for the utilization of biomaterial for biobanking and scientific purposes.
For the immunohistochemical analysis, 2-μm tissue sections were incubated in EnVision Flex Target Retrieval Solution, pH low for ACTN4 and ARFEF2 staining (both from Dako), followed by incubation with primary antibodies against ACTN4 diluted at 1:1,000 (Atlas Antibodies) or against ARFGEF2 (Abcam) for 40 min at room temperature. Polymeric secondary antibodies coupled to horseradish peroxidase (EnVision Flex+; Dako) and DAB (Dako) were applied to visualize the sites of immunoprecipitations. Tissue samples were analyzed by light microscopy after counterstaining with Meyer’s hematoxylin.
Two independent pathologists evaluated all tissue sections using a three-stage staining score: 0, negative; 1, moderate intensity of staining; 2, strong intensity of staining.
Apoptosis Studies.
Daudi and BL41 cells were treated with doxycycline for 4 d to express shRNAs specific for (i) ARFGEF2 or (ii) ACTN4 or unspecific shRNAs. Annexin V APC/7-AAD staining was performed with the Annexin V-APC/7-AAD Apoptosis Detection Kit (Biolegend). Cells were analyzed by flow cytometry with a FACSCanto II flow cytometer (Becton Dickinson).
Flow Cytometry.
For detection of BCR cell surface expression, 106 B cells were fixed in 2% (vol/vol) formaldehyde. Afterward, Fc receptor blocking reagent (Miltenyi Biotec) was applied according to the manufacturer’s instructions before staining of IgM-BCR by APC-labeled anti-IgM (eBioscience). The APC signal was detected by flow cytometry (LSR Fortessa; BD Biosciences).
Pathway Enrichment Analysis and Functional Annotation.
Pathway enrichment analysis was performed using R 3.2.1 and the R/Bioconductor package ReactomePA v1.14.4 [Yu and He (44); bioconductor.org/packages/release/bioc/html/ReactomePA.html]. P values were adjusted for multiple comparisons using the Benjamini–Hochberg procedure.
Gene ontology functional annotations were retrieved using the mygene package (version 1.6.0). Only terms supported by experimental evidence codes (EXP, IDA, IPI, IMP, IGI, and IEP) were considered for the analysis.
Full Proteome Analysis.
For protein expression analysis, light-, medium-, and heavy-labeled DG75 cell lysates were mixed in a 1:1:1 ratio. A total of 150 µg of protein was separated by SDS/PAGE using precast Bis-Tris minigels (NuPAGE Novex 4–12%; Life Technologies) and visualized by staining with Coomassie Brilliant Blue (Serva). Each lane was cut into 23 slices, reduced with DTT (Sigma-Aldrich), and alkylated with iodoacetamide (IAM; Sigma-Aldrich), digested in-gel with trypsin (Serva), extracted, and analyzed by mass spectrometry.
Protein Digestion and Phosphopeptide Enrichment for SILAC-Based Quantitative Global Phosphoproteomic Analysis.
For investigation of phosphorylation dynamics, equal amounts of SILAC-labeled cell lysates were mixed, treated with Benzonase (Novagen) for 1 h at 37 °C, and precipitated with acetone. The precipitate was dissolved in 1% RapiGest Surfactant (Waters) in 25 mM ammonium bicarbonate (Sigma-Aldrich), reduced with 10 mM DTT for 1 h at 65 °C, and alkylated by IAM at a final concentration of 20 mM for 1 h at 37 °C. Proteins were digested with trypsin (Promega) at a 1:20–1:50 (wt/wt) trypsin:protein ratio in the presence of 0.1% RapiGest at 37 °C overnight. The digest was acidified to 1% formic acid and cleared of precipitated material by centrifugation at maximum speed for 30 min; the supernatant was then evaporated to dryness in a SpeedVac concentrator (Thermo Scientific). For samples of DG75 BCR stimulations, peptides were subsequently fractionated by strong cation exchange (SCX) chromatography (BioBasic SCX 50 × 2.1 mm; Thermo Fisher) on an FPLC system (SMART; Pharmacia) with a salt concentration gradient. Buffer A contained 10 mM ammonium formate (Sigma-Aldrich) and buffer B 500 mM ammonium formate, both at pH 2.65 and containing 30% (vol/vol) acetonitrile (ACN). Twenty fractions were collected over a 50-min gradient at a flow rate of 100 µL⋅min–1. Fractions 1–12 were dried, resuspended in a solution of 200 mg/mL 2,5-dihydroxybenzoic acid (DHB; Sigma-Aldrich) in 80% (vol/vol) ACN and 5% (vol/vol) TFA, and loaded on in-house made TiO2 (GL Science) spin columns as described previously (1). TiO2 beads were washed three times with 200 mg⋅mL–1 DHB in 80% (vol/vol) ACN, 5% (vol/vol) TFA, and five times with 80% (vol/vol) ACN, 5% (vol/vol) TFA. Phosphopeptides were eluted with 0.3 M NH4OH (pH ≥ 10.5), dried in a SpeedVac concentrator, and analyzed by mass spectrometry.
For all other samples, phosphopeptides were enriched directly from cell lysates before prefractionation. In brief, digested peptides were resuspended in incubation buffer [80% (vol/vol) ACN, 5% (vol/vol) TFA, 5% (vol/vol) glycerol] and incubated with TiO2 beads (10 µm; GL Science) at a 1:8 peptide:bead ratio (wt/wt) with end-over-end rotation at 37 °C for 20 min. The peptide concentration was maintained at around 2 mg/mL during the incubation. After incubation, all TiO2 beads were loaded onto an empty spin column (5 µm frit; Hoefer Inc.) and washed three times each with incubation buffer, 80% (vol/vol) ACN, 5% (vol/vol) TFA and 60% (vol/vol) ACN, 0.1% FA. Phosphopeptides were then eluted with 1 M 2,2'-(propane-1,3-diyldiimino)bis[2-(hydroxymethyl)propane-1,3-diol] in 0.5 M NH4OH (pH ≥10.5) and acidified immediately with 10% (vol/vol) FA. The eluate was desalted on a C18 spin column and fractionated using basic pH reverse phase chromatography (XBridge C18 3.5 µm, 150 × 1.0 mm; Waters).
Phosphopeptide Enrichment for pYome Analysis.
Antibody-based enrichment for tyrosine-phosphorylated peptides was done with the Phospho-Tyrosine (P-Tyr-1000) Rabbit mAb Kit (Cell Signaling Technology). SILAC-labeled DG75 and Daudi cells were lysed in 8 M urea (2 M thiourea) buffer, mixed 1:1:1 (or 1:1) according to cell number/protein concentration and prepared as described in the manufacturer’s instructions (Cell Signaling Technology).
LC-MS/MS Analysis and Data Processing.
Peptide fractions of the global phospho-proteome and technical replicates of the pYome of BCR-stimulated DG75 cells were analyzed on a hybrid linear ion trap-Orbitrap mass spectrometer (LTQ Orbitrap Velos; Thermo Scientific) coupled to a nanoflow liquid chromatography system (Agilent 1100 series; Agilent). Samples were preconcentrated and desalted on a trap column (ReproSil-Pur 120 C18-AQ, 5 µm; 20 × 0.100 mm, packed in-house; Dr. Maisch GmbH) at 5 μL/min–1 in loading buffer [2% (vol/vol) acetonitrile, 0.1% formic acid in water]. Peptides were separated on an analytical column [ReproSil-Pur 120 C18-AQ, 3 µm, (Dr. Maisch GmbH); 200 × 0.075 mm, packed in-house into a PF360-75-15-N picofrit capillary, (New Objective)] using a 105-min linear gradient from 5% to 40% (vol/vol) acetonitrile containing 0.1% formic acid at a flow rate of 300 nL⋅min–1. The mass spectrometer was operated in data-dependent acquisition (DDA) mode, automatically switching between MS and MS/MS acquisitions. Survey spectra from m/z 350–1,600 were acquired in the Orbitrap at a resolution setting of 60,000 FWHM at m/z 400. Product-ion spectra of the 10 most abundant precursors with charge states 2+ to 5+ per cycle were acquired using collision-induced dissociation (CID) in the linear ion trap at a normalized collision energy (NCE) of 35% and an isolation width of 2 m/z. Automatic gain control (AGC) target values and maximum injection times for MS and MS/MS were 1 × 106 in 500 ms and 5 × 104 in 100 ms, respectively.
DG75 pYome samples were measured on an EASY n-LC 1000 (Thermo Scientific) coupled to a hybrid quadrupole-Orbitrap mass spectrometer (Q Exactive; Thermo Scientific). The samples were preconcentrated and desalted on a trap column (20 × 0.1 mm, ReproSil-Pur 120 C18-AQ, 5 µm, packed in-house; Dr. Maisch GmbH) at 5 μL⋅min−1 in loading buffer [2% (vol/vol) ACN, 0.1% FA]. Peptides were separated on an analytical column (200 × 0.075 mm, ReproSil-Pur 120 C18-AQ, 3 µm, packed in-house; Dr. Maisch GmbH) using an 80-min linear gradient from 4% to 34% buffer B [95% (vol/vol) ACN, 0.1% FA] versus a decreasing concentration of buffer A (0.1% FA) at a flow rate of 300 nL/min. The Q Exactive was operated in a DDA selecting the top 12 most abundant precursors for higher energy collisional dissociation (HCD) in the collision cell with an isolation width of 2 m/z and an NCE setting of 28%. Survey spectra from m/z 350–1600 were acquired with an MS resolution setting of 70,000 FWHM at m/z 200 and product ion spectra with a MS/MS resolution of 17,500 in the Orbitrap. AGC target values and maximum injection times for MS and MS/MS were set to 1 × 106 in 60 ms and 2 × 105 in 60 ms, respectively.
For all other samples, phosphopeptides were analyzed on a Q Exactive HF mass spectrometer (Thermo Fisher) coupled with an Ultimate 3000 RSLC (Dionex). Phosphopeptides were separated on a self-made capillary column (ReproSil-Pur 120 C18-AQ, 3 µm, 350 × 0.075 mm; Dr. Maisch GmbH) with a 70-min linear gradient of 2–40% buffer B [80% (vol/vol) ACN, 0.1% FA] and versus buffer A (0.1% FA in water) at a constant flow rate of 300 nL⋅min–1. The mass spectrometer was operated in DDA mode using a top 20 method with a survey scan resolution setting of 120,000 FWHM and an MS/MS resolution setting of 35,000 FWHM at 200 m/z. HCD was performed with an NCE setting of 28% and an isolation width of 1.4 m/z. AGC target values and maximum ion injection times for MS and MS/MS were set 1 × 106 in 40 ms and 1 × 105 in 64 ms, respectively.
All raw files were processed using MaxQuant software (v1.5.2.8, MPI for Biochemistry) (45). MS/MS spectra were searched against a UniProtKB/Swiss-Prot human database containing 88,993 protein entries (downloaded July 2014) supplemented with 245 frequently observed contaminants via the Andromeda search engine (46). Precursor and fragment ion mass tolerances were set to 6 and 20 ppm after initial recalibration, respectively. STY phosphorylation, protein N-terminal acetylation, and methionine oxidation were allowed as variable modifications. Cysteine carbamidomethylation was defined as a fixed modification. Minimal peptide length was set to seven amino acids, with a maximum of two missed cleavages. The false discovery rate (FDR) was set to 1% on both the peptide and the protein level using a forward-and-reverse concatenated decoy database approach.
For SILAC quantitation, multiplicity was set to two or three for double (Lys+0/Arg+0, Lys+8/Arg+10) or triple (Lys+0/Arg+0, Lys+4/Arg+6, Lys+8/Arg+10) labeling, respectively. At least two ratio counts were required for peptide quantitation. Both the “match between runs” and “re-quantify” options of MaxQuant were enabled.
All of the raw files and MaxQuant search results have been deposited to the ProteomeXchange Consortium (proteomecentral.proteomexchange.org/cgi/GetDataset) via the PRIDE partner repository (47) with the dataset identifier PXD003492.
Data Evaluation and Bioinformatics.
Data analysis was conducted with Perseus software (v1.5.2.4, MPI for Biochemistry). After removing decoy and contaminant entries, identified phosphosites with a localization probability lower than <0.75 were filtered out. SILAC ratios and peptide intensities were logarithmized (log2 and log10, respectively). For quality control, multiscatter plots displaying Pearson’s correlation coefficients were generated (Fig. S2). P-sites with SILAC ratios showing Z scores >2 or <−2 in at least one time point in each dataset were defined as significantly regulated. Unsupervised clustering analysis (row clustering with Euclidean distance, average linkage, preprocessed with k-means, number of clusters 300) was performed in Perseus using only regulated common p-sites after normalization as described by Deshmukh et al. (48).
Additional network visualization of quantitative phosphoproteomic datasets was performed with the Cytoscape (v3.2.1) application PhosphoPath (v1.1) (cytoscape.org) [Shannon et al. (49)]. Protein–protein interaction information was retrieved from the BioGRID database (www.thebiogrid.org), and kinase–substrate information from PhosphoSitePlus (www.phosphosite.org/homeAction.action). The quantitative networks were prepared as described by Raaijmakers et al. (50).
Discussion
BCR signaling critically regulates the development, proliferation, and activation of B cells. Further emphasizing this importance, several lymphoid neoplasias, such as CLL, ABC-DLBCL, and BL, are addicted to dysregulated BCR signaling. Although, in CLL and ABC-DLBCL, a mechanistic understanding of altered BCR signaling has already led to development of efficient targeted therapies (16, 17), little is known so far about BCR signaling and its functional impact in BL (3). In contrast to CLL and ABC-DLBCL, in BL, no mutations were found in BCR signaling effectors, such as CD79a/b and CARD11, which implies the existence of different pathomechanisms and makes the identification of potential drug targets within the BL-specific BCR-signaling network more difficult (4, 14).
Contrary to CLL and ABC-DLBCL cells, which depend on chronic active BCR signaling, BL cells are believed to rely on tonic antigen-independent BCR signaling (4). The first biochemical evidence for tonic BCR signals was reported by Wienands et al. (32). In their study, the inhibition of protein tyrosine phosphatases resulted in a phosphotyrosine “signaling signature” that was similar to that present in BCR-activated cells. This finding implied that BCR complexes constitutively generate basal signals that were later shown to be functionally relevant for various aspects of B-cell biology (33–35), as well as BL pathophysiology (4). Our comparative proteomic study of tonic and antigen-induced BCR signaling is in line with these previous studies and revealed, beyond commonly used signaling patterns, distinct effectors for activated and tonic conditions, thus strongly implying a certain degree of specificity for both modes of BCR signaling.
Recently, Satpathy et al. (36) described phosphoproteomic changes that occurred upon stimulation of BCR in the murine B-cell line A20 (36). In addition to a common set of BCR signaling effectors that were identified in both studies, we identified more than 200 so far unreported BCR effectors. There are several possible explanations for the differences observed in BCR signaling between BL and A20 cells. First of all, the BL cells we analyzed are derived from humans whereas the A20 cells are of murine origin. Second, we investigated activated and tonic BCR signaling whereas Satpathy et al. focused only on activated BCR signaling. Third, the BL models that we analyzed express IgM-BCR on the cell surface whereas A20 cells express IgG-BCR. Fourth, A20 cells do not harbor any BL-specific mutations.
Our analyses represent a substantial complement to recent genetic studies in which the mutational landscape in BL was elucidated and the mutational pattern in BL was shown to differ markedly from those in DLBCL and CLL (4, 14, 37). We included in our study genetically distinct BL; therefore, our comprehensive datasets, which include numerous time-resolved BCR signaling events, may represent a valuable resource for identifying potential drug targets. This finding is of particular importance because targeted therapies for BL are largely lacking at present and because they would be needed to improve outcomes—both in elderly BL patients, who still have a poor prognosis (38), and in patients with endemic BL in developing countries, where the administration of intensive chemotherapy is still a challenge.
Our resource datasets will be of importance for further elucidating BCR-mediated processes, in general, and BL-specific signaling, in particular. For example, ARFGEF2 and ACTN4—proteins that have not yet been described in the context of lymphoma pathophysiology—were identified as differentially phosphorylated upon knockdown of CD79a and SYK inhibition in BL cells in our study. We found that both newly identified BCR effectors are crucial for BL cell survival whereas they did not affect cell survival of other lymphoma subtypes. It is known that both proteins play a role in cytoskeleton reorganization and vesicle trafficking (30, 31); thus, it is tempting to speculate that these processes might contribute to the survival of BL cells. Several studies have indicated that the initiation of BCR signaling itself requires defined reorganization of the actin- and microtubule-dependent cytoskeleton, but the underlying molecular mechanisms remain largely elusive (13). We have shown here that cytoskeleton regulators represent one of the largest groups of phosphoacceptor proteins and have identified putative candidates that might control these processes, including ACTN4. Moreover, a recent study indicates that vesicles deliver preassembled SH2–domain-containing leukocyte protein of 65 kD (SLP65) signaling cargos to sites of BCR activation (39). Thus, the down-regulation of ARFGEF2 expression might interfere with vesicle trafficking and thereby disturb tonic BCR signaling.
The presented resource for BL-specific BCR signaling will help in gaining an understanding of fundamental BCR-induced processes. It also provides a useful basis for further biochemical and functional studies aimed at identifying and validating further potential therapeutic options for Burkitt’s lymphoma.
Materials and Methods
Cell Culture, BCR Stimulation, and Cell Lysis.
The lymphoma cell lines DG75, Daudi, BL-2, BL-41, BL-70, U-2932, and SU-DHL-6 were obtained from DSMZ or ATCC. OCI-Ly10 and OCI-Ly7 cells were kindly provided by A. Rosenwald, Institute of Pathology, University of Würzburg, Wuerzburg, Germany. Cell lines were authenticated by using Multiplex Cell Authentication (Multiplexion) (40). Further information is provided in SI Materials and Methods.
Mass Spectrometry and Data Analysis.
Mass spectrometry and data analysis are described in SI Materials and Methods.
Flow Cytometry.
Flow-cytometric calcium measurements were performed as described in Oellerich et al. (41). Further details for flow cytometric analyses are provided in SI Materials and Methods.
Western Blotting.
Western blotting was performed as described in Oellerich et al. (42).
Immunohistochemistry.
Lymphoma and normal lymph node tissue for immunohistochemistry (IHC) analyses was acquired from 32 patients from the University Medical Center Göttingen. Approval for using the human patient material in this study was obtained from the Ethics Committee of the University Medical Center Göttingen and informed consent was obtained. Investigated cases included 11 Burkitt’s lymphoma, 13 DLBCL, 6 Grey zone lymphoma, and 4 healthy lymph nodes. Further details for IHC analyses are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Silvia Münch, Monika Raabe, Annika Kühn, and Uwe Plessmann for their technical support. T.O. and M.E. were supported by Deutsche Krebshilfe (Grant 111399); M.A.R. and H.S. were supported by the Hessian Ministry of Higher Education, Research and the Arts (LOEWE Center for Cell and Gene Therapy Frankfurt, Funding Reference III L4 518/17.004). Parts of this work were supported by a grant from the Deutsche Technico-Gesellschaft e.V. (to H.U. and J.W.). M.E. and J.W. were supported by the Deutsche Forschungsgemeinschaft through TRR130. Work in the Department of Haematology in Cambridge/Green laboratory is supported by Bloodwise (Grant 13003), the Wellcome Trust (Grant 104710/Z/14/Z), the Medical Research Council, the Kay Kendall Leukaemia Fund, the Cambridge NIHR Biomedical Research Center, the Cambridge Experimental Cancer Medicine Centre, the Leukemia and Lymphoma Society of America (Grant 07037), and a core support grant from the Wellcome Trust and MRC to the Wellcome Trust‐Medical Research Council Cambridge Stem Cell Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The raw and processed mass spectrometry data of our study have been deposited in the PRIDE proteomics data repository, www.ebi.ac.uk/pride/archive/ [project no. PXD003492 (username: reviewer60796@ebi.ac.uk; password: TwMpaYmy)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601053113/-/DCSupplemental.
References
- 1.Kurosaki T, Shinohara H, Baba Y. B cell signaling and fate decision. Annu Rev Immunol. 2010;28:21–55. doi: 10.1146/annurev.immunol.021908.132541. [DOI] [PubMed] [Google Scholar]
- 2.Kurosaki T. Regulation of BCR signaling. Mol Immunol. 2011;48(11):1287–1291. doi: 10.1016/j.molimm.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Young RM, Staudt LM. Targeting pathological B cell receptor signalling in lymphoid malignancies. Nat Rev Drug Discov. 2013;12(3):229–243. doi: 10.1038/nrd3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmitz R, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490(7418):116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monroe JG. Ligand-independent tonic signaling in B-cell receptor function. Curr Opin Immunol. 2004;16(3):288–295. doi: 10.1016/j.coi.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Dühren-von Minden M, et al. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature. 2012;489(7415):309–312. doi: 10.1038/nature11309. [DOI] [PubMed] [Google Scholar]
- 7.Young RM, et al. Survival of human lymphoma cells requires B-cell receptor engagement by self-antigens. Proc Natl Acad Sci USA. 2015;112(44):13447–13454. doi: 10.1073/pnas.1514944112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lenz G, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319(5870):1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 9.Davis RE, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keller U, Nilsson JA, Maclean KH, Old JB, Cleveland JL. Nfkb 1 is dispensable for Myc-induced lymphomagenesis. Oncogene. 2005;24(41):6231–6240. doi: 10.1038/sj.onc.1208779. [DOI] [PubMed] [Google Scholar]
- 11.Refaeli Y, et al. The B cell antigen receptor and overexpression of MYC can cooperate in the genesis of B cell lymphomas. PLoS Biol. 2008;6(6):e152. doi: 10.1371/journal.pbio.0060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ong SE, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat Protoc. 2006;1(6):2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 13.Harwood NE, Batista FD. The cytoskeleton coordinates the early events of B-cell activation. Cold Spring Harb Perspect Biol. 2011;3(2):a002360. doi: 10.1101/cshperspect.a002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richter J, et al. ICGC MMML-Seq Project Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat Genet. 2012;44(12):1316–1320. doi: 10.1038/ng.2469. [DOI] [PubMed] [Google Scholar]
- 15.Kreck B, et al. Base-pair resolution DNA methylome of the EBV-positive endemic Burkitt lymphoma cell line DAUDI determined by SOLiD bisulfite-sequencing. Leukemia. 2013;27(8):1751–1753. doi: 10.1038/leu.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byrd JC, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369(1):32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson WH, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med. 2015;21(8):922–926. doi: 10.1038/nm.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coffey G, et al. Specific inhibition of spleen tyrosine kinase suppresses leukocyte immune function and inflammation in animal models of rheumatoid arthritis. J Pharmacol Exp Ther. 2012;340(2):350–359. doi: 10.1124/jpet.111.188441. [DOI] [PubMed] [Google Scholar]
- 19.Hoellenriegel J, et al. Selective, novel spleen tyrosine kinase (Syk) inhibitors suppress chronic lymphocytic leukemia B-cell activation and migration. Leukemia. 2012;26(7):1576–1583. doi: 10.1038/leu.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castello A, et al. Nck-mediated recruitment of BCAP to the BCR regulates the PI(3)K-Akt pathway in B cells. Nat Immunol. 2013;14(9):966–975. doi: 10.1038/ni.2685. [DOI] [PubMed] [Google Scholar]
- 21.Marshall AJ, et al. A novel B lymphocyte-associated adaptor protein, Bam32, regulates antigen receptor signaling downstream of phosphatidylinositol 3-kinase. J Exp Med. 2000;191(8):1319–1332. doi: 10.1084/jem.191.8.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Courtemanche N, Gifford SM, Simpson MA, Pollard TD, Koleske AJ. Abl2/Abl-related gene stabilizes actin filaments, stimulates actin branching by actin-related protein 2/3 complex, and promotes actin filament severing by cofilin. J Biol Chem. 2015;290(7):4038–4046. doi: 10.1074/jbc.M114.608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipsky BP, Beals CR, Staunton DE. Leupaxin is a novel LIM domain protein that forms a complex with PYK2. J Biol Chem. 1998;273(19):11709–11713. doi: 10.1074/jbc.273.19.11709. [DOI] [PubMed] [Google Scholar]
- 24.Chew V, Lam KP. Leupaxin negatively regulates B cell receptor signaling. J Biol Chem. 2007;282(37):27181–27191. doi: 10.1074/jbc.M704625200. [DOI] [PubMed] [Google Scholar]
- 25.Wang JH, et al. Aiolos regulates B cell activation and maturation to effector state. Immunity. 1998;9(4):543–553. doi: 10.1016/s1074-7613(00)80637-8. [DOI] [PubMed] [Google Scholar]
- 26.Duhamel M, Arrouss I, Merle-Béral H, Rebollo A. The Aiolos transcription factor is up-regulated in chronic lymphocytic leukemia. Blood. 2008;111(6):3225–3228. doi: 10.1182/blood-2007-09-113191. [DOI] [PubMed] [Google Scholar]
- 27.John LB, Ward AC. The Ikaros gene family: Transcriptional regulators of hematopoiesis and immunity. Mol Immunol. 2011;48(9-10):1272–1278. doi: 10.1016/j.molimm.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 28.Schwickert TA, et al. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol. 2014;15(3):283–293. doi: 10.1038/ni.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uckun FM, et al. Serine phosphorylation by SYK is critical for nuclear localization and transcription factor function of Ikaros. Proc Natl Acad Sci USA. 2012;109(44):18072–18077. doi: 10.1073/pnas.1209828109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Honda K. The biological role of actinin-4 (ACTN4) in malignant phenotypes of cancer. Cell Biosci. 2015;5:41. doi: 10.1186/s13578-015-0031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheen VL, et al. Mutations in ARFGEF2 implicate vesicle trafficking in neural progenitor proliferation and migration in the human cerebral cortex. Nat Genet. 2004;36(1):69–76. doi: 10.1038/ng1276. [DOI] [PubMed] [Google Scholar]
- 32.Wienands J, Larbolette O, Reth M. Evidence for a preformed transducer complex organized by the B cell antigen receptor. Proc Natl Acad Sci USA. 1996;93(15):7865–7870. doi: 10.1073/pnas.93.15.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kraus M, Alimzhanov MB, Rajewsky N, Rajewsky K. Survival of resting mature B lymphocytes depends on BCR signaling via the Igalpha/beta heterodimer. Cell. 2004;117(6):787–800. doi: 10.1016/j.cell.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 34.Lam KP, Kühn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90(6):1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- 35.Monroe JG. ITAM-mediated tonic signalling through pre-BCR and BCR complexes. Nat Rev Immunol. 2006;6(4):283–294. doi: 10.1038/nri1808. [DOI] [PubMed] [Google Scholar]
- 36.Satpathy S, et al. Systems-wide analysis of BCR signalosomes and downstream phosphorylation and ubiquitylation. Mol Syst Biol. 2015;11(6):810. doi: 10.15252/msb.20145880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puente XS, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475(7354):101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoelzer D, et al. German Multicenter Study Group for Adult Acute Lymphoblastic Leukemia Improved outcome of adult Burkitt lymphoma/leukemia with rituximab and chemotherapy: Report of a large prospective multicenter trial. Blood. 2014;124(26):3870–3879. doi: 10.1182/blood-2014-03-563627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engelke M, et al. Macromolecular assembly of the adaptor SLP-65 at intracellular vesicles in resting B cells. Sci Signal. 2014;7(339):ra79. doi: 10.1126/scitranslmed.2005104. [DOI] [PubMed] [Google Scholar]
- 40.Castro F, et al. High-throughput SNP-based authentication of human cell lines. Int J Cancer. 2013;132(2):308–314. doi: 10.1002/ijc.27675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oellerich T, et al. SLP-65 phosphorylation dynamics reveals a functional basis for signal integration by receptor-proximal adaptor proteins. Mol Cell Proteomics. 2009;8(7):1738–1750. doi: 10.1074/mcp.M800567-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oellerich T, et al. β2 integrin-derived signals induce cell survival and proliferation of AML blasts by activating a Syk/STAT signaling axis. Blood. 2013;121(19):3889–3899, S1–S66. doi: 10.1182/blood-2012-09-457887. [DOI] [PubMed] [Google Scholar]
- 43.Fellmann C, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Reports. 2013;5(6):1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 44.Yu G, He QY. ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst. 2015;12(2):477–479. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
- 45.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 46.Cox J, et al. Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 47.Vizcaíno JA, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32(3):223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deshmukh AS, et al. Deep proteomics of mouse skeletal muscle enables quantitation of protein isoforms, metabolic pathways, and transcription factors. Mol Cell Proteomics. 2015;14(4):841–853. doi: 10.1074/mcp.M114.044222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raaijmakers LM, et al. PhosphoPath: Visualization of phosphosite-centric dynamics in temporal molecular networks. J Proteome Res. 2015;14(10):4332–4341. doi: 10.1021/acs.jproteome.5b00529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.