Significance
Predicting the arrival of alien species remains a big challenge, which is assumed to be a consequence of the complexity of the invasion process. Here, we demonstrate that spreading of alien marine species can be predicted by a simple model using only global shipping intensities, environmental variables, and species occurrence data. We provide species lists of the next potentially invading species in a local habitat or species causing harmful algal blooms with their associated probability of invasion. This will help to improve mitigation strategies to reduce the further introduction of alien species. Although this study focuses on marine algae, the model approach can be easily adopted to other taxonomic groups and their respective drivers of invasion.
Keywords: alien species, model predictions, species identity, marine ecoregion, climate change
Abstract
The human-mediated translocation of species poses a distinct threat to nature, human health, and economy. Although existing models calculate the invasion probability of any species, frameworks for species-specific forecasts are still missing. Here, we developed a model approach using global ship movements and environmental conditions to simulate the successive global spread of marine alien species that allows predicting the identity of those species likely to arrive next in a given habitat. In a first step, we simulated the historical stepping-stone spreading dynamics of 40 marine alien species and compared predicted and observed alien species ranges. With an accuracy of 77%, the model correctly predicted the presence/absence of an alien species in an ecoregion. Spreading dynamics followed a common pattern with an initial invasion of most suitable habitats worldwide and a subsequent spread into neighboring habitats. In a second step, we used the reported distribution of 97 marine algal species with a known invasion history, and six species causing harmful algal blooms, to determine the ecoregions most likely to be invaded next under climate warming. Cluster analysis revealed that species can be classified according to three characteristic spreading profiles: emerging species, high-risk species, and widespread species. For the North Sea, the model predictions could be confirmed because two of the predicted high-risk species have recently invaded the North Sea. This study highlights that even simple models considering only shipping intensities and habitat matches are able to correctly predict the identity of the next invading marine species.
The number of alien species transported by human assistance has increased rapidly during the last decades with serious consequences for native flora and fauna (1–3). These biological invasions are considered to be one of the major drivers of biodiversity changes (4–6). Once an unwanted alien species has naturalized in the new environment, it is nearly impossible to eradicate the species, and thus the mitigation of further introduction is the most efficient way of combating biological invasions (6, 7). However, a targeted monitoring and an efficient adaptive management requires knowledge about spreading dynamics of the next potential invaders and thus about the distribution of species, their invasiveness, and the likelihood of new introductions. Although all of these topics have been analyzed on their own, the potential to predict the spreading of alien species while combining these components remains to be tested.
A large amount of recent introductions can be attributed to the intensified global trade and transport as many species were accidentally or deliberately translocated through the exchange of commodities or the movements of transportation means (8, 9). The amount of exchanged commodities and the intensity of global traffic have therefore been found to be a good predictor to model the global spread of alien species (10–12). In most cases, predictions of alien species introductions are difficult to assess as model results could not be validated (i.e., quantitatively assessed using observed data) thoroughly due to the paucity of high-quality distributional data of alien species. Without any model validation, however, it is nearly impossible to assess the quality and the reliability of model predictions, which hampers the application of models for the management of alien species. In recent years, appropriate high-quality data have been made accessible by various online databases, but testing model predictions with these data has still been lacking.
Model frameworks to predict the likelihood of new invasions have already been developed (10, 11, 13). However, these were not able to predict the identity of new invaders, but only the likelihood that any new species arrives from a certain source region on Earth. Here, we combined such a model, a slightly modified version of the vector-based model of marine invasion adopted from ref. 10, with datasets about the global distribution of marine alien species, which enabled us to predict the identity of the next species to arrive in a given local habitat. The model is a statistical model that describes how the probability a given species successfully invades a specific location depends on the shipping traffic and the environmental differences (temperature and salinity) between locations.
In a first step, to test the accuracy of the model, we used native ranges of 40 marine species from various taxonomic groups, ranging from algae to fish, as initial condition. For each species, we simulated the global spread outside its native range and compared the predicted alien range at each simulation time step with the observed one. This procedure allows the assessment of the quality of model predictions, although the degree of expansion distinctly varied among species, as some species are already widespread, whereas others occupy only a few alien regions either because they just started to spread or there are only a limited number of suitable habitats available.
In a second step, we used the reported distribution of 97 marine algal species with a known invasion history and six harmful algal species obtained from AlgaeBase (www.algaebase.org) and determined the species-specific invasion probabilities for each marine ecoregion not occupied by that species. Algae are particularly well suited for such an analysis because they are easily translocated by the exchange of ballast water, and especially invasive seaweeds are of global concerns because over 400 introductions have been reported worldwide (14). Furthermore, seaweeds deeply shape marine ecosystems and they can have strong detrimental ecological and economic impacts (14, 15). Using our model, we identified the likely hot spots of future invasions among 90 marine ecoregions of the world and algal species with the highest probability to arrive next.
Results
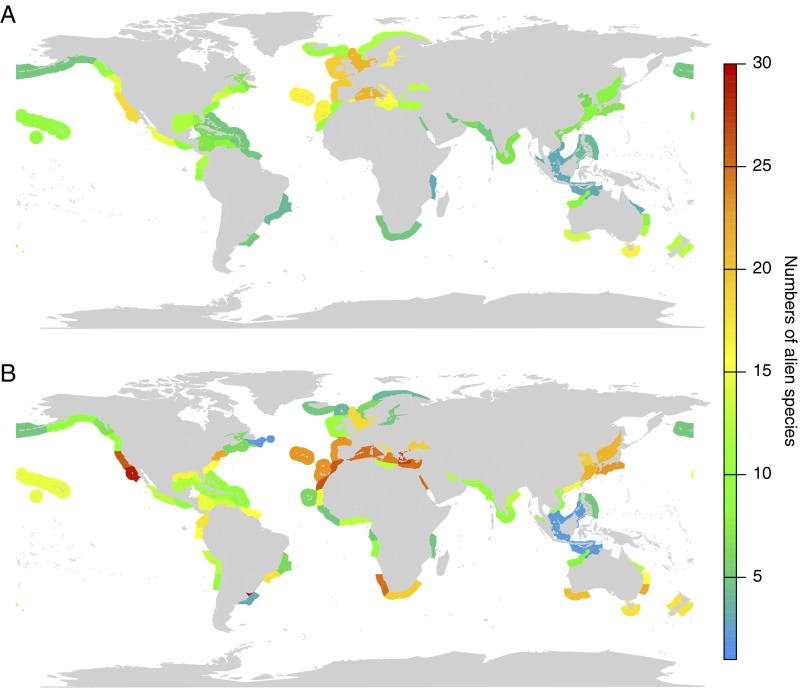
Comparing observed and predicted alien ranges of 40 marine species reveals that the model correctly predicted the presence/absence of the species in an originally alien ecoregion in 77% of all cases (median of all 40 spread simulations; Fig. S1). This value is robust to interspecific variation (e.g., using only 50% of the species reduced the number of correctly predicted ecoregions only to a median of 75%; Fig. S2), model parameterization, model structure, and variation of shipping intensity (SI Text and Table S1). Compared with observational data, the model overpredicts the number of alien species per ecoregion (Fig. S1). Predicted alien species numbers were distinctly higher for Southern Africa and East Asia, which are less studied regions, and thus higher alien species numbers can be expected, but also for the US West Coast.
Fig. S1.
Validation of model results. The figure shows the observed numbers of alien species per marine ecoregions reported in CABI (A) and the numbers of alien species predicted by the model (B). Only those 90 ecoregions are shown that have at least one port covered in this study. Even though the model slightly overpredicts alien species ranges, the presence/absence of an alien species was correctly predicted for 77% of marine ecoregions outside the species native range on average.
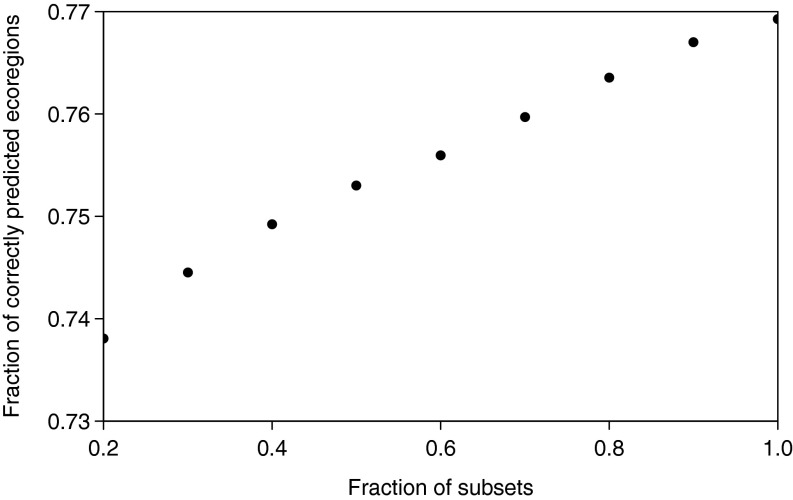
Fig. S2.
Influence of selecting random subsets (20–90%) of the 40 species taken from CABI on model accuracy. For each fraction, the random selection was repeated 1,000 times. Model accuracy was expressed as the median of the fraction of corrected predicted ecoregions of all species and averaged for each fraction (Materials and Methods). The low variation of the model accuracy (from 77% to about 74%) despite a strong reduction of considered species numbers indicates that the model predictions are mostly independent of the selection of species.
Table S1.
Results of the sensitivity analysis
| Model version/parameterization | Description | Fraction of correctly predicted ecoregions | Statistic |
| σT = 2; σS = 6 | Modifying widths of ecological niche | 0.78 | 0.5 |
| Phosphate | Including nutrient concentration to determine environmental niche | 0.77 | 0.51 |
| σT = 2; σS = 10 | Original model parameterization/structure | 0.77 | 0.51 |
| Two ports | Assuming next two ports as being occupied after one simulation step | 0.77 | 0.5 |
| σT = 2; σS = 8 | Modifying widths of ecological niche | 0.77 | 0.5 |
| Five ports | Assuming next five ports as being occupied after one simulation step | 0.77 | 0.49 |
| σT = 1; σS = 10 | Modifying widths of ecological niche | 0.77 | 0.4 |
| σT = 3; σS = 6 | Modifying widths of ecological niche | 0.76 | 0.59 |
| σT = 3; σS = 8 | Modifying widths of ecological niche | 0.76 | 0.59 |
| Nitrate | Including nutrient concentration to determine environmental niche | 0.76 | 0.53 |
| σT = 1; σS = 6 | Modifying widths of ecological niche | 0.76 | 0.39 |
| All ecoregions | Considering all occupied ecoregions to calculate environmental niche | 0.75 | 0.62 |
| σT = 3; σS = 10 | Modifying widths of ecological niche | 0.75 | 0.59 |
| σT = 1; σS = 8 | Modifying widths of ecological niche | 0.75 | 0.38 |
| Silicate | Including nutrient concentration to determine environmental niche | 0.73 | 0.54 |
| Variable σT and σS | Estimating widths of ecological niche from species distribution | 0.68 | 0.52 |
Several model modifications and parameterizations were tested to analyze their influence on model predictions. The model performance was tested using a goodness-of-fit statistic measured as the fraction of correctly predicted ecoregions to all false predictions [i.e., true positives/(false positives + false negatives)]. The model version applied in this study is highlighted in bold. Note that the model version used in this study is not the best-performing one, to be consistent with the parameterization of ref. 10. The differences to the results of the best-performing model were, however, marginal.
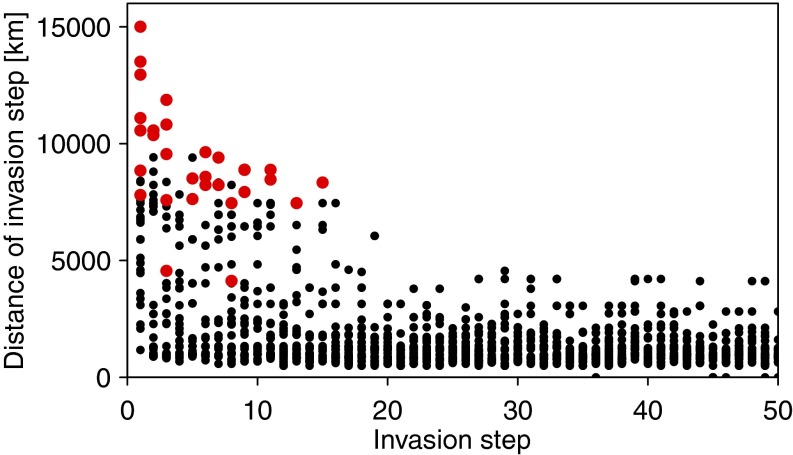
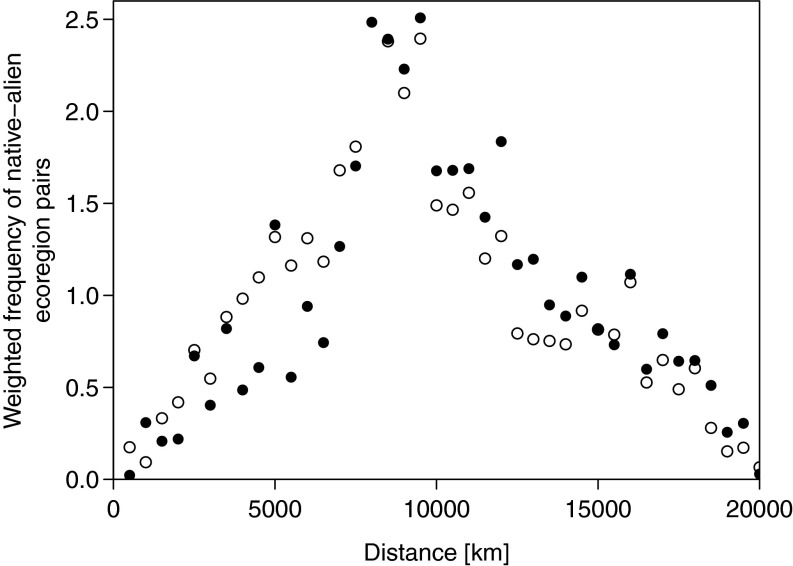
The simulations of the global spreading dynamics of the 40 marine species were used to analyze common patterns of the global spread. For each simulated invasion step of each species, we determined the great circle distance from the centroids of the newly invaded ecoregion to the nearest already occupied ecoregion. This shows that long-distance jumps are very common among the very first invasion steps (Fig. S3). For 44% of all species spreads, the longest jump to a new ecoregion was found among the first three invasion steps with 10,376 km on average. That is, spreading dynamics of these species follow a common pattern: first, the suitable habitats worldwide were invaded irrespective of the distance from where the species originated, and subsequently the species spread to neighboring habitats. This results in a mean geographic distance between observed native and alien ranges of around 10,200 km. Alien species numbers weighted by their number of native and alien regions decreased to shorter and longer distances (Fig. S4). A similar although more complex pattern was also found in ref. 10, which indicates that the models applied in both studies revealed similar results (Materials and Methods).
Fig. S3.
Geographical distance of invasion steps as a function of the temporal development of the spread for 40 marine species obtained from CABI (dots). The distance of an invasion step was calculated as the great circle distance between the centroids of a newly invaded ecoregion and the nearest already occupied ecoregion. The maximum distance of an invasion step for each species (red dots) was often found among the first invasion steps.
Fig. S4.
Weighted frequency of pairs of observed native ecoregions with observed (circles) and predicted (dots) alien ecoregions for 40 marine alien species as a function of the geographical distance between native and alien ecoregion. The weighted frequencies were calculated by determining the distance between the centroids of all native and alien ecoregions weighted by the number of native–alien ecoregions pairs for each species. The weights were summed for each 500-km bin from 0 to 20,000 km to obtain the frequency of links for each bin. The weighting was done to ensure that all species contributed equally.
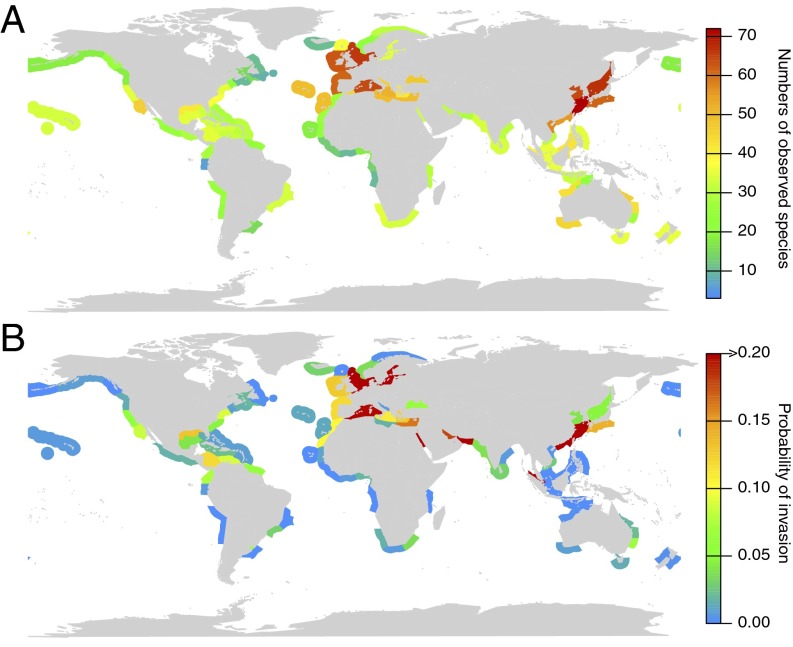
After validation of model predictions, we used the distribution of 97 algal species with a known invasion history obtained from AlgaeBase as initial conditions to determine the species-specific invasion probabilities to unoccupied ecoregions. These species mostly live in temperate to subtropical marine ecoregions in Europe and East Asia (Fig. 1A), corresponding to the known global hot spots of algal diversity (16, 17). The predicted invasion probabilities combined for all of these species reflect the global shipping intensity as marine ecoregions with intense ship traffic such as East Asian Seas or Northern European Seas have a high invasion probability (Fig. 1B). The East Asian Seas and the Northern European Seas are characterized by similar environmental conditions and are well connected by ships, which increases the likelihood of exchanges between both regions. Indeed, the reported native and alien ranges show that species have been mutually exchanged between both regions as many alien species in the Northern European Seas originate from East Asia (n = 11 of 19) and vice versa (4 of 16). Our model calculations revealed rankings of high-risk algal species for each ecoregion, which is exemplarily shown for the North Sea (Table S2). Two of the algal species of the top 10 high-risk species for the North Sea are known from the literature to have naturalized in that ecoregion in recent years [Prorocentrum minimum (18) and Polysiphonia harveyi (19)]. These species are not listed for the North Sea in AlgaeBase probably due to delays in data acquisition. That is, based on data provided by AlgaeBase, the model predicts a high probability that these species will enter the North Sea, which could indeed be confirmed by recent studies.
Fig. 1.
Observed cumulative distribution (native plus alien) of 97 marine algal species with an invasion history (A) and predicted invasion probabilities for further spread of these species to formerly unoccupied marine ecoregions. Colors indicate the number of reported algal species in A and the predicted probability of invasion in B, respectively.
Table S2.
The predicted top 10 high-risk species for the North Sea and their associated probability of invasion, P(Inv), in the North Sea
| Species name | P(Inv) |
| Symphyocladia marchantioides (Harvey), Falkenberg, 1897 | 0.0061 |
| Hypnea spinella (C. Agardh), Kützing, 1847 | 0.0043 |
| Gracilaria tikvahiae, McLachlan, 1979 | 0.0027 |
| Hypnea cornuta (Kützing), J. Agardh, 1851 | 0.0024 |
| Ganonema farinosum (J. V. Lamouroux), K. C. Fan and Yung C. Wang, 1974 | 0.0024 |
| Sargassum fluitans (Børgesen), Børgesen, 1914 | 0.0019 |
| Prorocentrum minimum (Pavillard), J. Schiller, 1933 | 0.0019 |
| Polysiphonia harveyi, Bailey, 1848 | 0.0018 |
| Macrocystis pyrifera (Linnaeus), C. Agardh, 1820 | 0.0017 |
| Petalonia binghamiae (J. Agardh), K. L. Vinogradova, 1973 | 0.0010 |
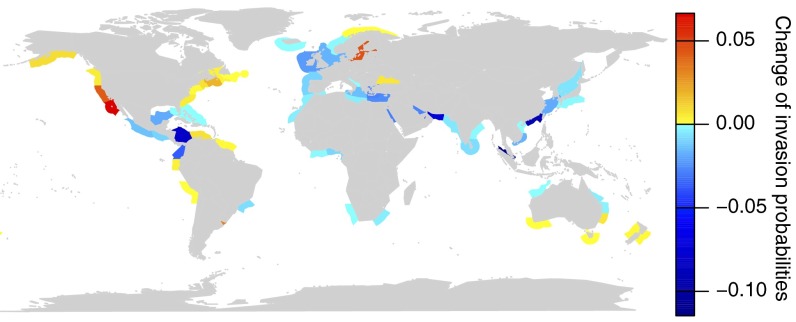
Predicted climate warming will likely modify the similarity of environmental conditions of marine ecoregions and thus will affect the probabilities of invasion. Using predicted mean sea surface temperatures during 2040–2060 revealed a latitudinal gradient in the change of invasion probabilities (Fig. 2): in general, the model predicts a decrease in invasion probabilities in the tropics, and an increase in temperate regions due to climate change. Highest increases in invasion probabilities can be expected for the Northeast Pacific and the Baltic Sea, whereas highest declines arise for ecoregions with high shipping intensity in the tropics such as ecoregions with access to the Panama Canal, the Persian Gulf, the Strait of Malacca, and South China.
Fig. 2.
Predicted changes in invasion probabilities due to climate warming. To simulate climate warming, invasion probabilities for 97 algal species with a known invasion history were calculated using water temperatures during 2040–2060 for the recipient ecoregions. The resulting invasion probabilities were compared with those calculated using current water temperatures, and the deviations are indicated by colors.
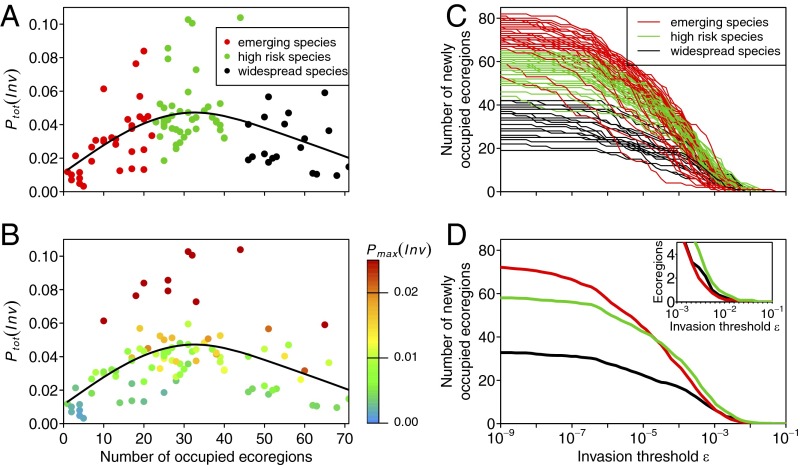
The invasion probabilities distinctly vary among algal species, which—in our model—mainly depends on the species-specific global distribution of algae and their occupancy of ecoregions with high shipping intensity (thereby enhancing the chance of further spread). We found that the total invasion probability Ptot(Inv) (i.e., the probability for a species to invade any unoccupied ecoregion worldwide) is a hump-shaped function of the initial number of occupied marine ecoregions (Fig. 3A). This can be explained by two contrasting mechanisms: increasing the number of occupied ecoregions results (i) in an increase in propagule pressure to unoccupied ecoregions, thereby increasing the likelihood of further spread, and (ii) in a reduction of the number of potential new habitats, which simultaneously decreases the chance of further spread. Combining both relationships results in the described hump-shaped curve. Consequently, species with narrow or wide distributions have a comparatively low total invasion probability, whereas species with an intermediate distribution have the highest invasion probabilities. In addition to the number of occupied ecoregions, the interspecific variation of Ptot(Inv) can be further explained by the maximum invasion probability Pmax(Inv) to any ecoregion (Fig. 3B).
Fig. 3.
Spreading potential of marine alien algae. (A) The total invasion probabilities Ptot(Inv) of algal species follow a hump-shaped curve of the number of initially occupied marine ecoregions, with species of intermediate global distribution having the highest Ptot(Inv). The colors denote the category of spreading potentials classified by cluster analysis (C and D). (B) The same data as in A, but the colors indicate the maximum invasion probability Pmax(Inv) from the occupied range of the species to any unoccupied ecoregion. (C) Invasion curves for each species estimated as the number of ecoregions outside the range, which the species initially occupied, above an invasion threshold ε. Cluster analysis of interspecific variations identified three groups of species with different invasion curves (red, emerging species; green, high-risk species; and black, widespread species). (D) The same as C, but showing the mean invasion curves for each cluster. The Inset indicates a zoom to high invasion threshold values.
To characterize the spreading potential of each species, we introduced an invasion threshold ε and counted the number of ecoregions with an invasion probability above this threshold. Note that a decrease of ε can be interpreted as a general increase in the probability of invasion due to e.g., elevated propagule pressure, interspecific variations in the ability to invade another ecoregion, or increased environmental match due to environmental changes. Decreasing ε (or increasing the invasion probability) leads to a higher number of ecoregions that potentially can be invaded by the species (Fig. 3C). The obtained species-specific invasion curves saturate when the species occupy all initially unoccupied ecoregions worldwide. For example, species with a wide initial distribution can only invade a few remaining unoccupied ecoregions, resulting in a low number of ecoregions at which the invasion curves saturate. Cluster analysis reveals three characteristic groups of invasion curves: the first group, which we call the “emerging species,” has a narrow initial distribution (red dots in Fig. 3A and red lines in Fig. 3 C and D). A decrease in the invasion threshold ε results in a slow increase of the number of unoccupied ecoregions with an invasion threshold above ε. That is, the species have a low chance to invade another ecoregion. Species of the second group, called the “high-risk species,” have an intermediate initial distribution (green dots in Fig. 3A). Their spreading potential increases comparatively steeply even at high invasion thresholds (green lines in Fig. 3 C and D), and thus these species have a high chance to invade other ecoregions. The third group of species, called the “widespread species,” consists of species with a wide initial distribution (black dots in Fig. 3A). Their invasion curves increase with decreasing invasion threshold at a comparatively low rate similar to that of the emerging species (black lines in Fig. 3 C and D), but saturate at a low number of unoccupied ecoregions simply due to the low availability of ecoregion without the species.
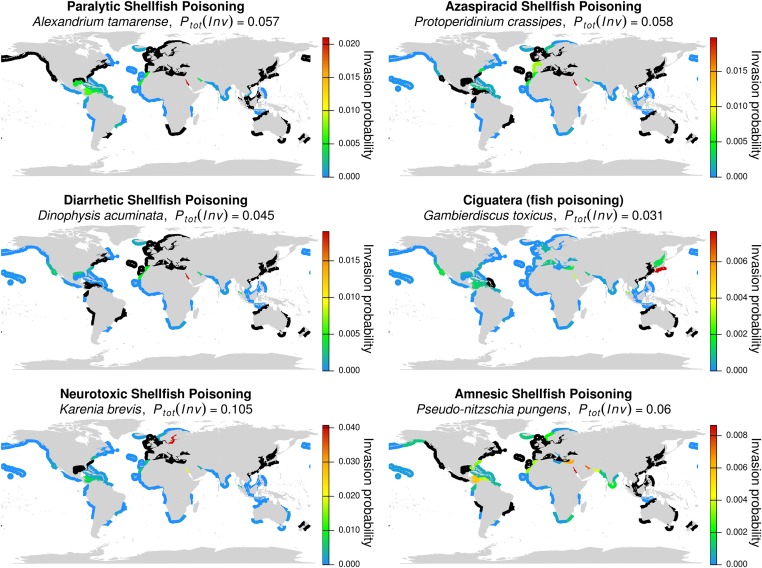
Although the known invasion of a species in an ecoregion is the best predictor for the potential invasiveness of that species for other ecoregions (20, 21), it is interesting to assess the spreading potentials of species, which are not known for their invasiveness but may affect other species and potentially even ecosystems. Some algae produce toxic substances causing fish or shellfish poisoning, which can also be harmful for humans (22). Transportation of toxic algal cells or cysts in ballast water of ships is a likely explanation for harmful algal blooms in previously unaffected regions (22, 23). Although these species may not be known to naturalize outside their native region (i.e., to establish populations viable for years), they may produce harmful algal blooms in these regions at least during one season of favorable conditions. We therefore selected the distributions of six algal species from AlgaeBase responsible for either well-known human diseases such as ciguatera or various shellfish poisonings (24) and calculated invasion probabilities for each species. Most of these species have an intermediate to wide distribution and a comparatively high Ptot(Inv) (Fig. 4). Karenia brevis, for example, causing neurotoxic shellfish poisoning, has a Ptot(Inv) of 0.105, which is in the range of the highest invasion probabilities calculated for species with a known invasion history. The reason is that the K. brevis was found in an intermediate number of ecoregions with intense ship traffic (Fig. 4). According to our classification system for species with known invasion histories (Fig. 3), Gambierdiscus toxicus would fall in the category of emerging species, whereas Protoperidinium crassipes, Dinophysis acuminata, and Karenia brevis could be classified as high-risk species, and the remaining algae would be widespread species.
Fig. 4.
Invasion probabilities for a selection of six algal species causing fish and shellfish poisonings potentially harmful to humans. The selected algae are known to produce toxic compounds causing the respective poisoning of fish or shellfish. Black areas denote the current distribution of species, whereas colors indicate the invasion probability to an unoccupied ecoregion. The total invasion probabilities Ptot(Inv) expressed as the integration of all single invasion probabilities are given in the subheading of each panel.
SI Text
The application of mathematical models always necessitates dealing with a number of assumptions of a simplified world. The basic model applied here was extensively tested in a sensitivity analysis provided in ref. 10, including a test of various model versions, variation of selected parameters, and discussion of major assumptions. We therefore focused our sensitivity analysis here on the application of the model rather than the basic structure and parameterization. To test the robustness of model predictions, we did the following:
-
i)
To determine the ecological niche of species more precisely, additional environmental parameters of nutrient concentrations for phosphate, nitrate, and silicate were added to the model in Eq. 2. The widths of the ecological niches expressed by σ were set to approximately the SD of the respective parameter values observed for all ports reported in our database.
-
ii)
Instead of using constant widths of the ecological niche for all species, σT and σS were taken as the SD of the temperature and salinity means of the native ecoregions.
-
iii)
The widths of the ecological niche σT and σS were modified manually.
-
iv)
Instead of calculating the ecological niche for each species from its respective native ecoregion, we selected temperature and salinity means from all occupied regions provided by CABI, including native and alien ecoregions.
-
v)
Instead of using the port with the highest invasion probability as the next invaded port, we selected the next two, respectively five, ports with the highest invasion probabilities.
-
vi)
To analyze the influence of species selection for model validation, we selected random subsets of species (20–90%) and repeatedly (n = 1,000) calculated the fraction of correctly predicted ecoregions.
None of these modifications improved the agreement between observed and predicted distributions of species distinctly (Table S1). However, for single species, the model results can change, while applying one of the modified calculations of the ecological niche. The overall robustness to model modifications and the selection of species indicates that the number of species was high enough to allow a reliable calculation of the average goodness-of-fit of the model (Fig. S2). Hence, the results presented here are robust to variations in model structure and model application.
A limitation of our modeling approach originates in the availability of ship movement data. Although the observed distribution of marine species can be the result of decades-long spreading histories, shipping data are only available recently and were assumed to be constant in time. Particularly, spreading processes before 1950 may be less reliably predicted using recent ship movement data. To further analyze whether recent shipping data can be used as a proxy to characterize the historic spread of marine alien species, we need to know when spreading dynamics of those alien species considered here were most intense. For example, if species mostly spread in the 19th century, the ship movement data are far less reliable predictors for alien species spread compared with those that occurred in the late 20th century.
We therefore compiled a list of the years of first records of the alien species in a region from online databases and the literature. These first records were reported for countries and large islands, and we decided to not transform them to marine ecoregions because it was not possible to determine the respective ecoregion if the countries/islands bordered more than one ecoregion. For 38 of the 40 alien species used for model validation, we obtained the year of first record for a total of 312 regions (Table S3), but the first records were not available for the full alien ranges. The years of first records ranged from 1693 to 2014 and were highly skewed toward recent times with a median first record of 1986. This means that 50% of all invasions were recorded in recent decades and 79% after 1945. Thus, the majority of invasions happened within the last decades. Ship movement data are only available during 2007–2008. However, major changes in global shipping dynamics occurred much earlier than 1986 with the Suez Canal opening in 1869, the Panama Canal opening in 1914, and the start of containerized transportation in 1956. Thus, the routes of shipping should not have changed distinctly in recent decades, but the distribution of the intensity of shipping has changed particularly due to the rise of the emerging economies. The ship movements before the rise of these countries are not reflected in our dataset. The quality of our model predictions may therefore be higher if historical ship movement could be considered.
Table S3.
Years of first record of the alien species, which were used for model validation, in a region
| Name | Country | Year | Ref. | Name | Country | Year | Ref. |
| Acanthogobius flavimanus | Littorina littorea | ||||||
| United States | 1963 | www.fishbase.org | Canada | 1840 | www.iucngisd.org | ||
| Australia | 1971 | www.fishbase.org | United States | 1879 | www.cabi.org/isc | ||
| Acentrogobius pflaumii | Marenzelleria neglecta | ||||||
| Australia | 1996 | 39 | Belgium | 2000 | 40 | ||
| Ascidiella aspersa | Denmark | 1990 | 40 | ||||
| Argentina | 1962 | www.iucngisd.org | United Kingdom | 1983 | 40 | ||
| Canada | 2012 | www.iucngisd.org | Russia | 1988 | 40 | ||
| Asterias amurensis | Germany | 1983 | 40 | ||||
| Australia | 1995 | 39 | Latvia | 1988 | 40 | ||
| Tasmania | 1986 | www.cabi.org/isc | Lithuania | 1988 | 40 | ||
| Austrominius modestus | The Netherlands | 1983 | 40 | ||||
| South Africa | 1949 | www.cabi.org/isc | Estonia | 1991 | www.cabi.org/isc | ||
| The Netherlands | 1946 | www.corpi.ku.lt/databases/aquanis | Finland | 1990 | www.cabi.org/isc | ||
| Belgium | 1950 | www.corpi.ku.lt/databases/aquanis | Norway | 1990 | www.cabi.org/isc | ||
| Denmark | 1978 | www.corpi.ku.lt/databases/aquanis | Sweden | 1985 | www.corpi.ku.lt/databases/aquanis | ||
| Germany | 1953 | www.corpi.ku.lt/databases/aquanis | Poland | 1986 | www.corpi.ku.lt/databases/aquanis | ||
| Sweden | 1939 | www.corpi.ku.lt/databases/aquanis | Microcosmus squamiger | ||||
| Ireland | 1957 | www.corpi.ku.lt/databases/aquanis | Egypt | 1919 | www.cabi.org/isc | ||
| United Kingdom | 1941 | www.corpi.ku.lt/databases/aquanis | Morocco | 1982 | www.cabi.org/isc | ||
| France | 1953 | www.corpi.ku.lt/databases/aquanis | Tunisia | 1963 | www.cabi.org/isc | ||
| Caprella mutica | United States | 1986 | www.cabi.org/isc | ||||
| Ireland | 2003 | 41 | France | 1979 | www.cabi.org/isc | ||
| Belgium | 1998 | 40 | Italy | 1971 | www.cabi.org/isc | ||
| United Kingdom | 1991 | 40 | Portugal | 1992 | www.cabi.org/isc | ||
| Germany | 2004 | 40 | Spain | 1978 | www.cabi.org/isc | ||
| The Netherlands | 1993 | 40 | Madeira | 2006 | www.corpi.ku.lt/databases/aquanis | ||
| Canada | 1997 | www.iucngisd.org | Mnemiopsis leidyi | ||||
| Norway | 1996 | www.iucngisd.org | Russia | 1982 | 40 | ||
| United States | 1973 | www.cabi.org/isc | Germany | 2006 | 40 | ||
| New Zealand | 2004 | www.cabi.org/isc | Greece | 1990 | 40 | ||
| Denmark | 2005 | www.corpi.ku.lt/databases/aquanis | Italy | 1980 | 40 | ||
| France | 2004 | www.corpi.ku.lt/databases/aquanis | Romania | 1989 | 40 | ||
| Spain | 2013 | www.corpi.ku.lt/databases/aquanis | Turkey | 1991 | 40 | ||
| Carcinus maenas | Ukraine | 1982 | 40 | ||||
| Argentina | 2003 | www.iucngisd.org | Israel | 2009 | www.iucngisd.org | ||
| Brazil | 1857 | www.iucngisd.org | Norway | 2002 | www.iucngisd.org | ||
| Canada | 1950 | www.iucngisd.org | Denmark | 2005 | www.corpi.ku.lt/databases/aquanis | ||
| Myanmar | 1933 | www.cabi.org/isc | Bulgaria | 1986 | www.corpi.ku.lt/databases/aquanis | ||
| Pakistan | 1971 | www.cabi.org/isc | Georgia | 1997 | www.corpi.ku.lt/databases/aquanis | ||
| Egypt | 1808 | www.cabi.org/isc | Poland | 2007 | www.corpi.ku.lt/databases/aquanis | ||
| Madagascar | 1922 | www.cabi.org/isc | Sweden | 2006 | www.corpi.ku.lt/databases/aquanis | ||
| Panama | 1866 | www.cabi.org/isc | The Netherlands | 1992 | www.corpi.ku.lt/databases/aquanis | ||
| Caulerpa racemosa var. cylindracea | Belgium | 2007 | www.corpi.ku.lt/databases/aquanis | ||||
| Canary Islands | 1989 | 40 | Musculista senhousia | ||||
| Croatia | 2000 | 40 | Israel | 1960 | 40 | ||
| Cyprus | 1973 | 40 | Italy | 1983 | 40 | ||
| Egypt | 1944 | 40 | Romania | 2002 | 40 | ||
| France | 1997 | 40 | Mexico | 1970 | www.iucngisd.org | ||
| Greece | 1956 | 40 | Canada | 1991 | www.iucngisd.org | ||
| Israel | 1955 | 40 | Australia | 1983 | 39 | ||
| Italy | 1993 | 40 | Egypt | 1964 | www.cabi.org/isc | ||
| Lebanon | 1931 | 40 | United States | 1924 | www.cabi.org/isc | ||
| Malta | 1990 | 40 | France | 1978 | www.cabi.org/isc | ||
| Spain | 1998 | 40 | Slovenia | 1997 | www.cabi.org/isc | ||
| Syria | 1954 | 40 | Tasmania | 1995 | www.cabi.org/isc | ||
| Tunisia | 1926 | 40 | New Zealand | 1978 | www.cabi.org/isc | ||
| Turkey | 1980 | 40 | Mytilus galloprovincialis | ||||
| Galapagos | 1934 | 42 | Japan | 1932 | www.iucngisd.org | ||
| Charybdis hellerii | United States | 1993 | 43 | ||||
| Cyprus | 1998 | 40 | Palaemon elegans | ||||
| Egypt | 1933 | 40 | Russia | 1939 | 40 | ||
| Israel | 1924 | 40 | Palaemon macrodactylus | ||||
| Lebanon | 1964 | 40 | The Netherlands | 1999 | 40 | ||
| Syria | 1992 | 40 | Argentina | 2000 | www.iucngisd.org | ||
| Brazil | 1995 | www.iucngisd.org | United States | 1957 | www.cabi.org/isc | ||
| Colombia | 1987 | www.iucngisd.org | Belgium | 1998 | www.cabi.org/isc | ||
| Cuba | 1987 | www.iucngisd.org | Bulgaria | 2009 | www.cabi.org/isc | ||
| Israel | 1924 | www.iucngisd.org | France | 1998 | www.cabi.org/isc | ||
| Turkey | 1981 | 44 | Germany | 2004 | www.cabi.org/isc | ||
| Ciona intestinalis | Portugal | 2004 | www.cabi.org/isc | ||||
| Brazil | 1958 | www.iucngisd.org | Romania | 2002 | www.cabi.org/isc | ||
| Canada | 1900 | www.iucngisd.org | Spain | 1999 | www.cabi.org/isc | ||
| Peru | 1885 | www.iucngisd.org | United Kingdom | 1992 | www.cabi.org/isc | ||
| Australia | 1958 | 39 | Poland | 2014 | www.corpi.ku.lt/databases/aquanis | ||
| United States | 1917 | 43 | Phyllorhiza punctata | ||||
| Hawaiian Islands | 1933 | 45 | Israel | 1965 | 40 | ||
| Crassostrea virginica | Brazil | 1955 | www.iucngisd.org | ||||
| Hawaiian Islands | 1866 | 46 | Philippines | 1921 | www.cabi.org/isc | ||
| Denmark | 1880 | 40 | United States | 1981 | www.cabi.org/isc | ||
| United Kingdom | 1872 | 40 | Hawaiian Islands | 1933 | 45 | ||
| France | 1861 | 40 | Polyandrocarpa zorritensis | ||||
| Germany | 1887 | 40 | Japan | 1991 | www.iucngisd.org | ||
| Ireland | 1870 | www.corpi.ku.lt/databases/aquanis | United States | 1994 | www.cabi.org/isc | ||
| Japan | 1968 | www.cabi.org/isc | Italy | 1974 | www.cabi.org/isc | ||
| United States | 1870 | www.cabi.org/isc | Pseudochattonella verruculosa | ||||
| The Netherlands | 1939 | www.cabi.org/isc | Denmark | 1998 | www.corpi.ku.lt/databases/aquanis | ||
| Romania | 1973 | www.corpi.ku.lt/databases/aquanis | Norway | 1998 | www.corpi.ku.lt/databases/aquanis | ||
| Belgium | 1961 | www.corpi.ku.lt/databases/aquanis | Pterois volitans | ||||
| Crepidula fornicata | Colombia | 2008 | www.iucngisd.org | ||||
| Belgium | 1911 | 40 | Martinique | 2011 | www.iucngisd.org | ||
| Denmark | 1934 | 40 | United States | 1985 | www.fishbase.org | ||
| France | 1949 | 40 | Bermuda | 2000 | www.fishbase.org | ||
| Germany | 1934 | 40 | Anguilla Islands | 2010 | www.cabi.org/isc | ||
| Italy | 1973 | 40 | Bahamas | 1996 | www.cabi.org/isc | ||
| Malta | 1973 | 40 | Barbados | 2011 | www.cabi.org/isc | ||
| The Netherlands | 1929 | 40 | Dominican Rep. | 2008 | www.cabi.org/isc | ||
| Norway | 1958 | www.iucngisd.org | Guadeloupe | 2010 | www.cabi.org/isc | ||
| Spain | 1983 | www.corpi.ku.lt/databases/aquanis | Nicaragua | 2009 | www.cabi.org/isc | ||
| Sweden | 1948 | www.corpi.ku.lt/databases/aquanis | Saint Kitts and Nevis | 2010 | www.cabi.org/isc | ||
| Ireland | 1893 | www.corpi.ku.lt/databases/aquanis | Venezuela | 2009 | www.cabi.org/isc | ||
| United Kingdom | 1872 | www.corpi.ku.lt/databases/aquanis | Puerto Rico | 2002 | www.fishbase.org | ||
| Diplosoma listerianum | Belize | 2001 | www.fishbase.org | ||||
| Canada | 2008 | www.iucngisd.org | Cuba | 2005 | www.fishbase.org | ||
| Hawaiian Islands | 1900 | 45 | Turks and Caicos | 2006 | www.fishbase.org | ||
| Dreissena polymorpha | Haiti | 2007 | www.fishbase.org | ||||
| Austria | 1911 | 40 | Virgin Islands, US | 2008 | www.fishbase.org | ||
| Belgium | 1826 | 40 | Cayman Islands | 2008 | www.fishbase.org | ||
| Croatia | 1995 | 40 | Jamaica | 2008 | www.fishbase.org | ||
| United Kingdom | 1820 | 40 | Costa Rica | 2009 | www.fishbase.org | ||
| Finland | 1995 | 40 | Antigua and Barbuda | 2009 | www.fishbase.org | ||
| France | 1847 | 40 | Panama | 2009 | www.fishbase.org | ||
| Germany | 1824 | 40 | Curacao | 2009 | www.fishbase.org | ||
| The Netherlands | 1826 | 40 | Mexico | 2009 | www.fishbase.org | ||
| Slovenia | 1993 | 40 | Honduras | 2009 | www.fishbase.org | ||
| Spain | 2001 | 40 | Rapana venosa | ||||
| Switzerland | 1850 | 40 | United Kingdom | 1991 | 40 | ||
| Czech Republic | 1890 | www.iucngisd.org | Russia | 1946 | 40 | ||
| Denmark | 1840 | www.cabi.org/isc | France | 1998 | 40 | ||
| Canada | 1986 | www.cabi.org/isc | Greece | 1986 | 40 | ||
| Mexico | 1993 | www.cabi.org/isc | Italy | 1973 | 40 | ||
| United States | 1986 | www.cabi.org/isc | The Netherlands | 2005 | 40 | ||
| Belarus | 1801 | www.cabi.org/isc | Romania | 1961 | 40 | ||
| Estonia | 1801 | www.cabi.org/isc | Slovenia | 1974 | 40 | ||
| Ireland | 1993 | www.cabi.org/isc | Ukraine | 1954 | 40 | ||
| Italy | 1969 | www.cabi.org/isc | Israel | 2002 | www.cabi.org/isc | ||
| Latvia | 1801 | www.cabi.org/isc | Turkey | 1960 | www.cabi.org/isc | ||
| Lithuania | 1801 | www.cabi.org/isc | United States | 1998 | www.cabi.org/isc | ||
| Poland | 1800 | www.cabi.org/isc | Argentina | 1998 | www.cabi.org/isc | ||
| Sweden | 1920 | www.cabi.org/isc | Uruguay | 1998 | www.cabi.org/isc | ||
| Russia | 1824 | www.corpi.ku.lt/databases/aquanis | Spain | 2007 | www.corpi.ku.lt/databases/aquanis | ||
| Ensis directus | Bulgaria | 1956 | www.corpi.ku.lt/databases/aquanis | ||||
| United Kingdom | 1989 | 40 | Georgia | 1947 | www.corpi.ku.lt/databases/aquanis | ||
| Norway | 1982 | www.iucngisd.org | Rhithropanopeus harrisii | ||||
| France | 1991 | www.corpi.ku.lt/databases/aquanis | Belgium | 1985 | 40 | ||
| Belgium | 1986 | www.corpi.ku.lt/databases/aquanis | Denmark | 1953 | 40 | ||
| The Netherlands | 1981 | www.corpi.ku.lt/databases/aquanis | Russia | 1950 | 40 | ||
| Denmark | 1981 | www.corpi.ku.lt/databases/aquanis | France | 1957 | 40 | ||
| Germany | 1979 | www.corpi.ku.lt/databases/aquanis | Germany | 1936 | 40 | ||
| Sweden | 1978 | www.corpi.ku.lt/databases/aquanis | United Kingdom | 1996 | 40 | ||
| Gracilaria salicornia | Italy | 1994 | 40 | ||||
| Hawaiian Islands | 1950 | 45 | Lithuania | 2001 | 40 | ||
| Grateloupia turuturu | The Netherlands | 1874 | 40 | ||||
| France | 1982 | 40 | Poland | 1801 | 40 | ||
| Italy | 1969 | 40 | Romania | 1951 | 40 | ||
| The Netherlands | 1993 | 40 | Spain | 1991 | 40 | ||
| Portugal | 1998 | 40 | Tunisia | 2003 | 40 | ||
| Spain | 1990 | 40 | Ukraine | 1937 | 40 | ||
| United Kingdom | 1973 | 47 | Brazil | 1998 | www.iucngisd.org | ||
| Hemigrapsus sanguineus | Sweden | 2014 | www.corpi.ku.lt/databases/aquanis | ||||
| Tunisia | 2003 | 40 | Estonia | 2011 | www.corpi.ku.lt/databases/aquanis | ||
| United States | 1990 | www.cabi.org/isc | Latvia | 2013 | www.corpi.ku.lt/databases/aquanis | ||
| The Netherlands | 1999 | www.corpi.ku.lt/databases/aquanis | Finland | 2009 | www.corpi.ku.lt/databases/aquanis | ||
| Germany | 2009 | www.corpi.ku.lt/databases/aquanis | Bulgaria | 1934 | www.corpi.ku.lt/databases/aquanis | ||
| Romania | 2008 | www.corpi.ku.lt/databases/aquanis | Schizoporella errata | ||||
| Hemigrapsus takanoi | Canary Islands | 2003 | 40 | ||||
| The Netherlands | 1999 | 40 | Egypt | 1977 | www.cabi.org/isc | ||
| France | 1993 | www.cabi.org/isc | Hawaiian Islands | 1693 | www.cabi.org/isc | ||
| Belgium | 2003 | www.cabi.org/isc | |||||
| Spartina alterniflora | |||||||
| Germany | 2007 | www.cabi.org/isc | France | 1960 | 40 | ||
| Spain | 1997 | www.cabi.org/isc | Spain | 1975 | 40 | ||
| Hemimysis anomala | Japan | 2001 | www.iucngisd.org | ||||
| Austria | 1998 | 40 | United Kingdom | 1816 | 47 | ||
| Belgium | 1999 | 40 | Taiwan | 2008 | 48 | ||
| Russia | 1960 | 40 | China | 1979 | 49 | ||
| France | 2000 | 40 | Styela plicata | ||||
| Germany | 1997 | 40 | Brazil | 1883 | www.iucngisd.org | ||
| The Netherlands | 1997 | 40 | Australia | 1966 | 39 | ||
| Poland | 2002 | 40 | Bermuda | 1882 | www.cabi.org/isc | ||
| Ukraine | 1955 | 40 | United States | 1915 | 43 | ||
| Lithuania | 1960 | www.cabi.org/isc | Ulva pertusa | ||||
| Moldova | 1969 | www.cabi.org/isc | France | 1974 | 40 | ||
| Sweden | 1995 | www.corpi.ku.lt/databases/aquanis | The Netherlands | 1993 | 40 | ||
| Estonia | 2009 | www.corpi.ku.lt/databases/aquanis | Spain | 1990 | www.cabi.org/isc | ||
| Finland | 1992 | www.corpi.ku.lt/databases/aquanis | |||||
| Ireland | 2007 | www.corpi.ku.lt/databases/aquanis | |||||
Regions constitute countries and large islands. Note that only those regions are listed with a known year of first record of the respective species. The full alien ranges of the species are usually larger. For Ciona savignyi and Ulva reticulate, no first records were found.
Discussion
The mitigation of further introductions of alien species is a challenging task mostly due to the complexity of the invasion process and the lack of data. Models can help to improve mitigation strategies by the determination of hot spots of biological invasions, the determination of high-risk pathways, and the identification of species likely to arrive next. Several studies have tried to predict the likelihood of new invasions (10, 13, 25–28); however, the common paucity of model validations hampers the assessment of the quality of model predictions and their comparison between different approaches. Here, we provide a robust approach for the validation of colonization models, simulating the spread of alien species, which may serve as an example for other models and taxonomic groups. We show that our model correctly predicts the invasion status of species in ecoregions outside their native range in good agreement with observations (77% of the presence/absence of an alien species correctly predicted). This is particularly striking because of the simplicity of the model, considering only habitat matches and maritime traffic intensity and lacking seemingly important predictors like species-specific characteristics, biotic interactions, additional environmental variables, or historic shipping data. Indeed, the consideration of additional parameters in the model such as further environmental variables did not improve the model fit (Table S1). The quality of the fit ranges between those found in other modeling studies [e.g., R2 = 0.64 for the global spread of terrestrial vascular plants (29) or an 87–94% accuracy for the introduction of fishes into the Great Lakes (27)], but other models were either more complex or restricted to much smaller geographic areas or a single taxonomic group. We are not aware of any study, simulating the global spread of marine species from various taxonomic groups simultaneously as done here, which renders a direct comparison difficult. Probably the best indication of the model’s quality is the fact that two of the algal species predicted to be among the top 10 high-risk species for the North Sea are known to have naturalized in that ecoregion in recent years. That is, in two cases our species-specific predictions could be confirmed by recent samplings, which are not yet included in AlgaeBase. This also shows that it is possible to explain the spread of alien species to a large degree by simple vector-based invasion models. The sensitivity analysis (SI Text) highlights that our model predictions are robust to the choice of species for model validation, and to the variation in model structure, parameterization, and input datasets.
For the 97 marine algal species considered in this study, the highest probability of invasion arises in Asian and European ecoregions (Fig. 1B), mostly along the major shipping routes. The Asian Seas were also identified as invasion hot spots in a previous analysis of marine invasion, thereby ignoring observed species distributions (10). In contrast to our study, the invasion probabilities of Northern European Seas were very low in ref. 10, which was explained by the, on average, low environmental similarity to most other ecoregions worldwide. The reason for the elevated invasion probabilities found in this study is that many algal species predicted to be at high risk in Northern European Seas are native to East Asian waters, which provide similar environmental conditions to Northern European Seas. As both regions are highly interconnected due to intense ship traffic, the probability to invade from one region to the other is also high, resulting in the described mutual exchange of species between these regions. This shows that, although the overall patterns of both studies were similar, observational data are important to further improve the predictions of invasion hot spots.
Our model predicts that climate warming will lead to reduced invasion probabilities in the tropics and elevated ones in temperate regions, particularly in North America (Fig. 2). The reason is that many species used in this study live in temperate to subtropical ecoregions and were assumed to be adapted to water temperatures of those regions. If water temperatures increase, these species will find appropriate environmental conditions at higher latitudes than inhabited now, whereas environmental conditions will get less suitable in tropical regions. The highest increase in invasion probability arises for the Northeast Pacific. For this ecoregion, the by-far most important donor area constitutes the Northwest Pacific (10), and an elevation of water temperatures in the Northeast Pacific will increase the environmental match of both regions, resulting in increased invasion probabilities. Indeed, a recent finding of the Asian macroalga Sargassum horneri at the West Coast of Mexico and the United States is likely is a consequence of this increased environmental match (30), which further supports our model predictions.
Our model predicts the total invasion probability of a species to be a hump-shaped function of the initially occupied number of ecoregions (Fig. 3 A and B), the reason being that species with an intermediate global distribution pose a large propagule pressure to a comparatively large number of unoccupied ecoregions. This pattern corresponds to the relationship of the colonization rate on the number of occupied patches of classical metacommunities models (31). In these models, the colonization rate of new patches has to be zero if either none or all patches are occupied by a species and positive in between, which indicates the close relationship between both types of models.
The reliable prediction of invasion dynamics remains one of the biggest challenges in invasion ecology. We here demonstrate an approach for a robust validation of global invasion models, which allows the assessment of the quality of model predictions. This study highlights that the combination of invasion models with observational data can essentially improve the predictions of invasion probabilities. The model applied here can be easily adopted to simulate the spread of other taxonomic groups (see ref. 29 for an example of terrestrial vascular plants). Many online databases emerged in recent years. Notable examples are the Delivering Alien Invasive Species Inventories for Europe (DAISIE) (www.europe-aliens.org), the Invasive Species Compendium by the Centre for Agriculture and Bioscience International (CABI), or the upcoming Global Register of Invasive Species (GRIS) by the Invasive Species Specialists Group (www.issg.org), all of which provide native and alien ranges of numerous species. It is therefore the logical consequence to combine invasion models with available distributional data to improve the predictability of invasion dynamics as shown here. The prediction of future invasions is a prerequisite of efficient mitigation strategies, and the determination of the species identity enables a targeted monitoring of potential high-risk species.
Materials and Methods
Data.
As input variables, the model requires data about the arrival and departure dates of single ships moving between ports, ship type, ship size, ballast tank volumes, and environmental data of ports. The ship and route-specific data (i.e., ship size, ship type, and arrival and departure dates) were obtained from a large dataset of ship movements and ship characteristics provided by Lloyd’s Register Fairplay (www.ihs.com), consisting of nearly 3 million ship voyages of 32,511 ships between 1,469 ports during 2007–2008 (10), and ballast tank volume data taken from the American Bureau of Shipping (32). The shipping intensity was assumed to be constant in time (but see the discussion about the reliability of using recent ship movement data to predict historic spreading dynamics in SI Text). Sea surface temperatures and nutrient concentrations (nitrate, phosphate, and silicate) were obtained from the World Ocean Atlas (WOA) (www.nodc.noaa.gov), providing 50-y averages at 1° spatial resolution. Surface salinity data of ports were calculated from port-specific data of water densities provided by Lloyd’s Register Fairplay (www.sea-web.com/portguide.html) for most of the ports (69%). Water densities were recalculated to salinities using temperatures taken from the WOA. For the remaining ports, salinities were taken from the WOA. Future sea surface temperatures were obtained from the Coupled Model Intercomparison Project (CMIP5) (cmip-pcmdi.llnl.gov/cmip5/) providing predictions of sea surface temperatures using an Intergovernmental Panel on Climate Change scenario of intermediate greenhouse gas concentration (RCP6) (33). The average sea surface temperatures were calculated for each marine ecoregion for 2040–2060.
Model.
We applied a vector-based model of marine invasion to calculate the probability of invasion by global shipping between 1,469 major ports worldwide (10). In the original form, the model consisted of three independent probabilities: the probability to be alien, P(Alien); the probability of introduction, P(Intro); and the probability of establishment, P(Estab). Here, however, when we combine the model with observational data of species distributions, the term P(Alien), constituting a theoretical estimator for biogeographical dissimilarity is not required anymore. We therefore removed P(Alien) from the model.
According to ref. 10, the probability of introduction,
| [1] |
is a function of exchanged ballast water volume Br on route r between donor port i and recipient port j and travel time Δt. Pr(Intro) is obtained for each single ship on a certain route r taken from the ship movement dataset. Br is calculated as , with Wr being the ship type- and ship size-specific amount of released ballast water in cubic meters obtained from regression fits shown in figure S1 in ref. 10, Vr denoting the ship size-specific mean volume of ballast tanks of a ship, δr representing the number of stop-over ports on route r, and z being the fraction of nonzero releases depending on the type and the size of a ship. Note that in ref. 10, z was falsely described as the fraction of zero releases, although it has to be the fraction of nonzero releases as stated here.
The probability of establishment,
| [2] |
was modeled as a Gaussian function of differences of water temperature ΔTij and salinity ΔSij of donor port i and recipient port j standardized by the width of the respective ecological niche σT and σS. α represents the basic probability of establishment. To analyze the influence of climate warming on model predictions, Pij(Estab) was calculated using future sea surface temperature (2040–2060; Data) for the recipient regions.
The probability of invasion from port i to port j is given by the complement of species failing to invade on all ship routes rij connecting both ports,
| [3] |
The invasion probabilities between ports were aggregated accordingly to obtain the invasion probabilities Pj(Inv) from the ecoregions occupied by the respective species to each unoccupied ecoregion j. The total invasion probability was calculated for each species as Ptot(Inv) = 1 − ∏j[1 − Pj(Inv)] and the maximum probability of invasion as Pmax(Inv) = max(Pj(Inv)).
Although some modifications of the model structure and parameterizations slightly improved the accuracy of model predictions (SI Text and Table S1), we adopted the basic model version and the parameterization provided by ref. 10 for consistency. A more detailed description of the model, its parameterization, the input variables, and tests of the robustness of this approach are provided in ref. 10.
Model Validation.
For model validation (i.e., the quantitative assessment of the quality of model predictions using observed data), we compiled data of native and alien distributions of marine alien species from the CABI Invasive Species Compendium (www.cabi.org/isc/). In a first step, we used the implemented search engine of CABI with the search term “(HAB marine) AND ballast water” to filter for species in marine habitats likely to be transported by ballast water. We manually removed species not fitting to this category as, e.g., freshwater species with a high salinity tolerance were included as well, with the exception of Dreissena polymorpha as this species is known to be transported by open-sea vessels (34). We also removed regions with uncertain invasion status of a species, which resulted in a total of 40 species with known native and alien distributions from various taxonomic groups ranging from algae to fish. The following species were used for model validation: Acanthogobius flavimanus, Acentrogobius pflaumii, Ascidiella aspersa, Asterias amurensis, Austrominius modestus, Caprella mutica, Carcinus maenas, Caulerpa racemosa var. cylindracea, Charybdis hellerii, Ciona intestinalis, Ciona savignyi, Crassostrea virginica, Crepidula fornicata, Diplosoma listerianum, Dreissena polymorpha, Ensis directus, Gracilaria salicornia, Grateloupia turuturu, Hemigrapsus sanguineus, Hemigrapsus takanoi, Hemimysis anomala, Littorina littorea, Marenzelleria neglecta, Microcosmus squamiger, Mnemiopsis leidyi, Musculista senhousia, Mytilus galloprovincialis, Palaemon elegans, Palaemon macrodactylus, Phyllorhiza punctata, Polyandrocarpa zorritensis, Pseudochattonella verruculosa, Pterois volitans, Rapana venosa, Rhithropanopeus harrisii, Schizoporella errata, Spartina alterniflora, Styela plicata, Ulva pertusa, and Ulva reticulate. CABI provides distributions usually on a country basis, but in some cases also subnational units such as states, provinces, or islands are provided. We translated the distributional data provided by CABI into marine ecoregions, thereby assuming that if a species was listed for a geographic unit bordering or located in a certain ecoregion, the species can be found in the whole ecoregion including the ports. This may result in an overprediction of the actual distribution of species, but given the highly patchy sampling coverage of the world’s coastlines, simulations can only be done on a coarse geographic resolution, which necessitates such simplifications. A marine ecoregion represents an area of similar environmental conditions and species assemblages, and thus it seems likely that a species can also be found in other parts of that ecoregion. We adopted the classification of “marine ecoregions of the world” by Spalding et al. (35) but considered only those ecoregions that have at least one port listed in our shipping database, resulting in a total of 90 marine ecoregions. To assess the reliability of using recent ship movement data for the prediction of historic spreading dynamics of alien species, we compiled a list of the years of first record of the alien species in a region (Table S3). Inevitably, the year of first record is not provided for all species and all regions due to the lack of data.
For each species, we calculated the mean temperature and salinity requirements from their known native distribution by taking the average of all native ecoregion means of temperature and salinity. The obtained species-specific ecological niche was used as input variables Ti and Si in Eq. 2. Note that i now represents the geographic area used to calculate the ecological niche of a species. As initial condition, we set all ports where the species is considered to be native as “occupied” and all other ports to “unoccupied.” We then applied the model to calculate the invasion probability to all unoccupied ports. The port with the highest invasion probability was identified and set to occupied. This was repeated until a threshold of a very low invasion probability was reached, which corresponds to a hypothetical nearly worldwide coverage of the species. The sequence of invaded ports was transformed to the sequence of invaded ecoregions, thereby removing duplicated ecoregions. For each time step of the modeled spread, we compared the predicted alien distribution of the species with the observed alien distribution obtained from CABI and calculated the goodness-of-fit of predicted and observed alien distributions. The goodness-of-fit was estimated by the number of correctly predicted invaded ecoregions (true positives) divided by the sum of false predictions (false positives plus false negatives). At the time step of the largest goodness-of-fit value, the deviation between predicted and observed distribution was lowest, and the respective distribution was selected as the best fit. This procedure was repeated for each of the 40 species and the median of the resulting 40 goodness-of-fit values was taken as the overall goodness-of-fit of the model.
To test the robustness of our model predictions, we performed an extensive sensitivity analysis, thereby varying the simulation process, the model parameterization, and the distributional data used for model validation, and discuss the reliability of the ship movement data for our modeling purpose (SI Text).
Model Application.
After model validation, we calculated invasion probabilities using the global distribution of marine algae taken from AlgaeBase (www.algaebase.org) to identify future invasion hot spots and high-risk species of marine algae. We used the species names listed as “accepted names” in the database. AlgaeBase provides detailed information for numerous algal species, including their current global distribution, but AlgaeBase does not provide the information whether a species is native or alien. We therefore adopted the finding of previous studies that the best forecasting tool to predict the invasion risk is whether the species has naturalized elsewhere (20), which is sometimes even taken as the only permanent predictor of invasion success (21). We screened the literature and other databases to compile a list of algal species with a known invasion history. We started with a list of algal species provided by Molnar et al. (36) and added other species from the literature and online databases and finally ended up with 97 algal species with a known invasion history. For these species, we selected their current distribution from AlgaeBase. For each species, the ecological niche represented by Ti and Si was calculated as the mean temperatures and salinities of all occupied ecoregions. We standardized the geographic units to the level of a country, island, or marine ecoregion depending on the level of detail provided by AlgaeBase. For large countries (United States, Canada, Russia, and Australia), we distinguish subnational units such as provinces or states if this information was provided. For example, if a species was listed for Australia, we assigned all ecoregions surrounding Australia to this species, whereas if the species was found only at the island of Tasmania or at the coast of Queensland, we only selected the respective ecoregions. Assuming that the species is homogeneously distributed in the respective geographic unit (along a country coast or in a marine ecoregion), all ports in that area were set to occupied. For instance, if the species was found in the North Sea, we set all North Sea ports to occupied, whereas if the species was listed for Germany, we selected the North Sea ports and Baltic Sea ports located in Germany as occupied ports. For each species, we then calculated the invasion probabilities to any unoccupied port.
Spreading Potentials of Species.
The invasion probabilities obtained from the model applied to the AlgaeBase data were used to characterize the spreading potential of each species. This was done by introducing a threshold ε of invasion probability ranging from 10−9 to 10−1. For each species, the ecoregions outside the species’ native range with an invasion probability above ε were counted. This can be interpreted as those ecoregions that can be invaded by the species given a certain threshold value. While successively reducing ε, the number of these ecoregions with an invasion probability above ε increased, but at different rates depending on the species-specific spreading potentials resulting in species-specific invasion curves. To characterize the interspecific differences, we calculated the area bounded between all pairs of invasion curves and applied a hierarchical clustering algorithm using the open-source software package R (37). The distances between clusters were calculated with “Ward's minimum variance method” (38), which aims at finding compact spherical clusters. We tested other clustering algorithms as well, which, however, did not change the results significantly.
Acknowledgments
H.S. and B.B. acknowledge financial support from the Volkswagen Foundation. H.S. was further supported by the Austrian Research Foundation (FWF) Grant I2096-B16 and by the German Research Foundation (DFG) Grant SE 1891/2-1, and N.S. by the Research-Oriented Teaching Project (Forschungsorientierte Lehre) of the University of Oldenburg.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 5470.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1524427113/-/DCSupplemental.
References
- 1.Simberloff D, et al. Impacts of biological invasions: What’s what and the way forward. Trends Ecol Evol. 2013;28(1):58–66. doi: 10.1016/j.tree.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 2.Pyšek P, et al. A global assessment of invasive plant impacts on resident species, communities and ecosystems: The interaction of impact measures, invading species’ traits and environment. Glob Change Biol. 2012;18(5):1725–1737. [Google Scholar]
- 3.McGeoch MA, et al. Global indicators of biological invasion: Species numbers, biodiversity impact and policy responses. Divers Distrib. 2010;16(1):95–108. [Google Scholar]
- 4.Butchart SHM, et al. Global biodiversity: Indicators of recent declines. Science. 2010;328(5982):1164–1168. doi: 10.1126/science.1187512. [DOI] [PubMed] [Google Scholar]
- 5.Millennium Ecosystem Assessment . Ecosystems and Human Well-Being: Biodiversity Synthesis. Island Press; Washington, DC: 2005. [Google Scholar]
- 6.Pyšek P, Richardson DM. Invasive species, environmental change and management, and health. Annu Rev Environ Resour. 2010;35(1):25–55. [Google Scholar]
- 7.Puth LM, Post DM. Studying invasion: Have we missed the boat? Ecol Lett. 2005;8(7):715–721. [Google Scholar]
- 8.Levine JM, D’Antonio CM. Forecasting biological invasions with increasing international trade. Conserv Biol. 2003;17(1):322–326. [Google Scholar]
- 9.Hulme PE, et al. Grasping at the routes of biological invasions: A framework for integrating pathways into policy. J Appl Ecol. 2008;45(2):403–414. [Google Scholar]
- 10.Seebens H, Gastner MT, Blasius B. The risk of marine bioinvasion caused by global shipping. Ecol Lett. 2013;16(6):782–790. doi: 10.1111/ele.12111. [DOI] [PubMed] [Google Scholar]
- 11.Keller RP, Drake JM, Drew MB, Lodge DM. Linking environmental conditions and ship movements to estimate invasive species transport across the global shipping network. Divers Distrib. 2010;17(1):93–102. [Google Scholar]
- 12.Drake JM, Lodge DM. Global hot spots of biological invasions: Evaluating options for ballast-water management. Proc Biol Sci. 2004;271(1539):575–580. doi: 10.1098/rspb.2003.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jerde CL, Lewis MA. Waiting for invasions: A framework for the arrival of nonindigenous species. Am Nat. 2007;170(1):1–9. doi: 10.1086/518179. [DOI] [PubMed] [Google Scholar]
- 14.Williams SL, Smith JE. A global review of the distribution, taxonomy, and impacts of introduced seaweeds. Annu Rev Ecol Evol Syst. 2007;38:327–359. [Google Scholar]
- 15.Wallentinus I, Nyberg CD. Introduced marine organisms as habitat modifiers. Mar Pollut Bull. 2007;55(7-9):323–332. doi: 10.1016/j.marpolbul.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 16.Bolton JJ. Global seaweed diversity: Patterns and anomalies. Bot Mar. 1994;37(3):241–246. [Google Scholar]
- 17.Kerswell AP. Global biodiversity patterns of benthic marine algae. Ecology. 2006;87(10):2479–2488. doi: 10.1890/0012-9658(2006)87[2479:gbpobm]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 18.Hoppenrath M. A revised checklist of planktonic diatoms and dinoflagellates from Helgoland (North Sea, German Bight) Helgol Mar Res. 2004;58(4):243–251. [Google Scholar]
- 19.Maggs CA, Stegenga H. Red algal exotics on North Sea coasts. Helgol Meersunters. 1998;52(3-4):243–258. [Google Scholar]
- 20.Kulhanek SA, Ricciardi A, Leung B. Is invasion history a useful tool for predicting the impacts of the world’s worst aquatic invasive species? Ecol Appl. 2011;21(1):189–202. doi: 10.1890/09-1452.1. [DOI] [PubMed] [Google Scholar]
- 21.Williamson M. Invasions. Ecography (Cop) 1999;22(1):5–12. [Google Scholar]
- 22.Hallegraeff GM. A review of harmful algal blooms and their apparent global increase. Phycologia. 1993;32(2):79–99. [Google Scholar]
- 23.Van Dolah FM. Marine algal toxins: Origins, health effects, and their increased occurrence. Environ Health Perspect. 2000;108(Suppl 1):133–141. doi: 10.1289/ehp.00108s1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D-Z. Neurotoxins from marine dinoflagellates: A brief review. Mar Drugs. 2008;6(2):349–371. doi: 10.3390/md20080016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herborg LM, Jerde CL, Lodge DM, Ruiz GM, MacIsaac HJ. Predicting invasion risk using measures of introduction effort and environmental niche models. Ecol Appl. 2007;17(3):663–674. doi: 10.1890/06-0239. [DOI] [PubMed] [Google Scholar]
- 26.Paini DR, Yemshanov D. Modelling the arrival of invasive organisms via the international marine shipping network: A Khapra beetle study. PLoS One. 2012;7(9):e44589. doi: 10.1371/journal.pone.0044589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolar CS, Lodge DM. Ecological predictions and risk assessment for alien fishes in North America. Science. 2002;298(5596):1233–1236. doi: 10.1126/science.1075753. [DOI] [PubMed] [Google Scholar]
- 28.Gallien L, Munkemuller T, Albert CH, Boulangeat I, Thuiller W. Predicting potential distributions of invasive species: Where to go from here? Divers Distrib. 2010;16(3):331–342. [Google Scholar]
- 29.Seebens H, et al. Global trade will accelerate plant invasions in emerging economies under climate change. Glob Change Biol. 2015;21(11):4128–4140. doi: 10.1111/gcb.13021. [DOI] [PubMed] [Google Scholar]
- 30.Marks LM, et al. Range expansion of a non-native, invasive macroalga Sargassum horneri (Turner) C. Agardh, 1820 in the eastern Pacific. BioInvasions Rec. 2015;4(4):243–248. [Google Scholar]
- 31.Levins R. Some demographic and genetic consequences of environmental heterogeneity for biological control. Bull Entomol Soc Am. 1969;15:237–240. [Google Scholar]
- 32.American Bureau of Shipping 2011 Ballast Water Treatment Advisory (American Bureau of Shipping, Houston). Available at ww2.eagle.org/content/eagle/en.html. Accessed October 23, 2012.
- 33.Moss R, et al. Towards New Scenarios for Analysis of Emissions, Climate Change, Impacts, and Response Strategies. Intergovernmental Panel on Climate Change; Geneva: 2008. [Google Scholar]
- 34.Ricciardi I, MacIsaac HJ. Recent mass invasion of the North American Great Lakes by Ponto-Caspian species. Trends Ecol Evol. 2000;15(2):62–65. doi: 10.1016/s0169-5347(99)01745-0. [DOI] [PubMed] [Google Scholar]
- 35.Spalding MD, et al. Marine ecoregions of the world: A bioregionalization of coastal and shelf areas. Bioscience. 2007;57(7):573–583. [Google Scholar]
- 36.Molnar JL, Gamboa RL, Revenga C, Spalding MD. Assessing the global threat of invasive species to marine biodiversity. Front Ecol Environ. 2008;6(9):485–492. [Google Scholar]
- 37.R Core Team 2014 R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna), Version 3.2.3. Available at www.r-project.org.
- 38.Ward JH. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963;58(301):236–244. [Google Scholar]
- 39.Hewitt CL, et al. Introduced and cryptogenic species in Port Phillip Bay, Victoria, Australia. Mar Biol. 2004;144(1):183–202. [Google Scholar]
- 40.DAISIE . Handbook of Alien Species in Europe. Springer; Dordrecht, The Netherlands: 2009. [Google Scholar]
- 41.O’Flynn C, Kelly J, Lysaght L. Ireland’s invasive and non-native species—trends in introductions. Natl Biodivers Data Cent Ser. 2014;2:1–50. [Google Scholar]
- 42. Bungartz F, et al., eds (2009) Charles Darwin Foundation Galapagos Species Checklist—Lista de Especies de Galápagos de la Fundación Charles Darwin. Available at www.darwinfoundation.org/datazone/checklists/. Accessed November 9, 2015.
- 43.Williams SL. Introduced species in seagrass ecosystems: Status and concerns. J Exp Mar Biol Ecol. 2007;350(1-2):89–110. [Google Scholar]
- 44.Çinar ME, Bilecenoğlu M, Öztürk B, Katagan T, Aysel V. Alien species on the coasts of Turkey. Mediterr Mar Sci. 2005;6(2):119–146. [Google Scholar]
- 45.Carlton JT, Eldredge LG. Marine bioinvasions of Hawai’i: The introduced and cryptogenic marine and estuarine animals and plants of the Hawaiian Archipelago. Bish Museum Bull Cult Environ Stud. 2009;4:1–203. [Google Scholar]
- 46.Coles SL, DeFelice RC, Eldredge LG, Carlton JT. Historical and recent introductions of non-indigenous marine species into Pearl Harbor, Oahu, Hawaiian Islands. Mar Biol. 1999;135(1):147–158. [Google Scholar]
- 47.Roy HE, et al. Non-Native Species in Great Britain: Establishment, Detection and Reporting to Inform Effective Decision Making. Non-Native Species Secretariat; York, UK: 2012. [Google Scholar]
- 48.Wu S, et al. Insights of the latest naturalized flora of Taiwan: Change in the past eight years. Taiwania. 2010;55(2):139–159. [Google Scholar]
- 49.Xu H, et al. An inventory of invasive alien species in China. NeoBiota. 2012;15:1–26. [Google Scholar]