Abstract
Diffuse large B cell lymphoma (DLBCL) is one of the most common types of malignancy worldwide. The lack of clear symptoms and early detection make it difficult to diagnose at an early stage, leading to poor prognosis of the patients. Long non-coding RNAs (lncRNAs) have come into focus for their important regulatory roles in fundamental biological processes, particularly in cancer initiation, development and progression. The aim of the present study was to investigate the expression of lncRNA Hox transcript antisense intergenic RNA (HOTAIR) in a cohort of patients with DLBCL to assess its clinical value and biological function in DLBCL. The reverse transcription-polymerase chain reaction was used to detect HOTAIR expression levels and cells were transfected with small interfering RNA to compare cell proliferation, cell cycle progression and apoptosis. Western blotting was also conducted to detect possible signaling pathways. It was first found that the expression levels of HOTAIR were upregulated in DLBCL tumor tissues and cell lines, compared with normal tissues and cells. In addition, HOTAIR was significantly correlated with tumor size, clinical stage, B symptoms and International Prognostic Index scores; and higher expression levels of HOTAIR were correlated with improved prognosis. Univariate and multivariate analyses verified that HOTAIR was a key independent predictive factor for DLBCL prognosis. Furthermore, it was revealed that the knockdown of the expression of HOTAIR led to growth inhibition, cell cycle arrest and apoptosis in vitro, possibly through the phosphoinositide 3-kinase/AKT/nuclear factor-κB pathway. These results suggested that HOTAIR may be regarded as a novel indicator of poor prognosis, and may serve as a potential target for gene therapy in the treatment of DLBCL.
Keywords: diffuse large B cell lymphoma, Hox transcript antisense intergenic RNA, long non-coding RNA, prognosis
Introduction
Non-Hodgkin's lymphoma (NHL) is a type of malignancy, which originates from lymphatic cells (1). NHL can be divided into two groups according to the derivation, namely B cell type and T cell type, among which the B cell type accounts for 90% (2). Diffuse large B cell lymphoma (DLBCL) is one of the most life threatening types of malignancy due to its lack of symptoms in the early period and the lack of efficient therapeutic strategies, leading to poor prognoses (3). Therefore, there is an urgent requirement to identify novel biomarkers for early detection and the prediction of prognosis.
Long non-coding RNAs (lncRNAs), with a size >200 nt, have been an increased area of focus since their identification. Although they were previously considered a 'waste', lncRNA has a specific mechanism in several types of disease, particularly in malignant tumors. Hox transcript antisense intergenic RNA (HOTAIR), has an oncogenic role in several types of cancer, including colorectal cancer (4,5), lung cancer (6–8) and pancreatic cancer (9). However, the role of HOTAIR in DLBCL has not been investigated. In the present study, it was found that HOTAIR was upregulated in DLBCL and was correlated with an invasive phenotype and poor prognosis. Furthermore, it was demonstrated that silencing HOTAIR inhibited cell proliferation, promoted cell apoptosis and induced cell cycle arrest, possibly through the phosphoinositide 3-kinase (PI3K/AKT/nuclear factor(NF)-κB pathway.
Materials and methods
Tissue sample collection and cell line preparation
The present study included 50 lymph node samples from patients diagnosed with DLBCL and 20 individuals with reactive lymph nodes as controls. All samples were collected at The Department of Oncology, Dongying People's Hospital (Dongying, China), between July 2006 and December 2011. All samples were snap-frozen and stored in liquid nitrogen following collection. All the patients were diagnosed and confirmed to have DLBCL by histological examination, and any patients who had received preoperative radiotherapy or chemotherapy were excluded. Follow-up data were obtained by reviewing outpatient charts or through correspondence with the patients. The overall survival (OS) was defined as the time interval between the date of diagnosis and the end of the follow-up, or the date at which the patient succumbed to mortality. The present study was approved by the Research Ethics Committee of Dongying People's Hospital, and informed consent was obtained from each of the participants.
Cell lines and culture
The RCK-8, OCL-LY-10, OCL-LY-7, SU-DHL-6 and SU-DHL-4 DLBCL cell lines were purchased from American Type Culture Collection (Manassas, VA, USA). The cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), in a humidified 5% CO2 incubator at 37°C. Analysis results from normal tissues served as the control for comparison (10).
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA from tissue samples and cells at ~80–90% confluency were isolated using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and the steps were performed according to the manufacturer's protocol. The acquired RNA was quantified using ultraviolet spectrophotometry (NanoDrop 2000; Thermo Fisher Scientific, Inc.). The quantified RNA was then reverse transcribed into cDNA using the ExScriptRT-PCR kit (Takara Bio, Inc., Otsu, Japan), according to the manufacturer's protocol. qPCR was then performed using an ABI ViiA™ 7 PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) to verify the expression levels of HOTAIR. The expression of GAPDH was also determined and used as an internal control. The thermocycling conditions were as follows: 95°C for 30 sec; followed by 40 cycles of denaturation at 95°C for 3 sec and annealing at 60°C for 30 sec. The primer sequences (obtained from Sangon Biotech Co., Ltd., Shanghai, China) were as follows: HOTAIR, forward 5′-GGT AGA AAA AGC AAC CAC GAAGC-3′ and reverse 5′-ACA TAA ACC TCT GTC TGT GAG TGC C-3′; GAP DH, forward 5′-CCC CGC TAC TCC TCC TCC TAAG-3′ and reverse 5′-TCC ACG ACC AGT TGT CCA TTCC-3′. The relative mRNA expression of each gene was calculated using the comparative quantification cycle (Cq) (2−ΔΔCq) method (11).
Cell transfection assay
RCK-8 cells were selected for the knockdown experiment as they demonstrated the highest basal expression level. The RCK-8 cells (confluency, 30–50%) were transfected with either 50 nM HOTAIR-targeting small interfering (si)RNA or scrambled negative controls (GenePharma, Shanghai, China) using Lipofectamine RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.), according to manufacturer′s protocol. The two HOTAIR RNAi sequences were as follows: 5′-TAA CAA GAC CAG AGA GCT G-3′ (HOTAIR-si1) and 5′-GAA CGG GAG TAC AGA GAG A-3′ (HOTAIR-si2). The scramble sequence was as follows: 5′-UUC UCC GAA CGU GUC ACG UTT -3′. After 24 h of incubation in a humidified atmosphere with 5% CO2 at 37°C, the RNA was isolated, and the efficiency of HOTAIR knockdown was determined using qPCR.
Cell proliferation assay
Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) assays were used to evaluate cell proliferation, according to the manufacturer's protocol. Briefly, the cells (3×103/well) were seeded into 96-well plates in triplicate and incubated in a 5% CO2 humidified atmosphere at 37°C. At specific time points (24, 48 and 72 h), the cells were incubated with 10 µl CCK-8 solution for 2 h at 37°C. The absorbance was measured at a wavelength of 450 nm on a Gen5 microplate reader (BioTek, Winooski, VT, USA). The experiments were repeated in triplicate three times.
Analysis of cell cycle and apoptosis
The HOTAIR RNAi-transfected cells were harvested at a confluency of ~80–90% and washed twice with phosphate-buffered saline (PBS), following which they were fixed with pre-cooled 70% ethanol at 4°C overnight, and washed twice with PBS. The cells were then resuspended in 500 µl PBS, treated with RNase A (50 µg/ml; Beyotime Institute of Biotechnology, Haimen, China) and stained with propidium iodide (25 µg/ml; Beyotime Institute of Biotechnology) for 30 min at 37°C. The distribution of cell-cycle phases were determined using ModFit software (version 4.1; BD Biosciences, Franklin Lakes, NJ, USA). For the analysis of apoptosis, the cells were harvested at 70–80% confluence and incubated with reagent containing Annexin V-fluorescein isothiocyanate and propidium iodide (BD Biosciences) for 15 min in the dark at room temperature. The apoptotic cells were analyzed using a FACS Calibur flow cytometer (BD Biosciences).
Western blot analysis
The transfected cells were lysed in radio-immunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology) and, after 48 h, phenylmethanesulfonyl fluoride (Beyotime Institute of Biotechnology) was added and protein concentrations were determined using a standard Bradford assay (Beyotime Institute of Biotechnology). Equal quantities of proteins (20 µg) from each cell line were subjected to western blot analysis. The total proteins were fractionated using SDS-PAGE (Beyotime Institute of Biotechnology) and transferred onto a polyvinylidene fluoride membrane (Beyotime Institute of Biotechnology). The membranes were blocked in milk and then incubated with the indicated primary antibodies at 4°C overnight. The membranes were then incubated with secondary antibodies at room temperature for 1 h, and detection was performed using a chemiluminescence detection system (ImageQuant™ LAS 4000; GE Healthcare Life Sciences). The data were adjusted against the loading control using β-actin. The primary antibodies used for western blot analyses were as follows: Mouse monoclonal anti-total PI3K (1:1,000; cat. no. sc-365404), rabbit polyclonal anti-PI3K-p110 (1:1,000; cat. no. sc-130211), mouse monoclonal anti-total AKT (1:1,000; cat. no. sc-81434), rabbit polyclonal anti-phosphorylated (p)-AKT (Thr308; 1:1,000; cat. no. sc-16646-R), rabbit polyclonal anti-NF-κB p65 (1:1,000; cat. no. sc-372) and rabbit polyclonal anti-p-NF-κB p65 (Ser536; 1:1,000; sc-101752). The secondary antibodies used were goat anti-rabbit (1:5,000; cat. no. sc-2004) and goat anti-mouse (1:5,000; cat. no. sc-2060). All of the antibodies were obtained from Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA).
Statistical analysis
All statistical analysis was performed using SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA). Continuous data were analyzed using an independent t-test, whereas categorical data were analyzed using a χ2 test or Fisher's exact method. Kaplan-Meier analysis was used to compare the OS curves in the different groups. A Cox proportional hazards model was used to examine the significance of the effects of different variables on OS in multivariate analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression levels of HOTAIR are significantly upregulated in DLBCL
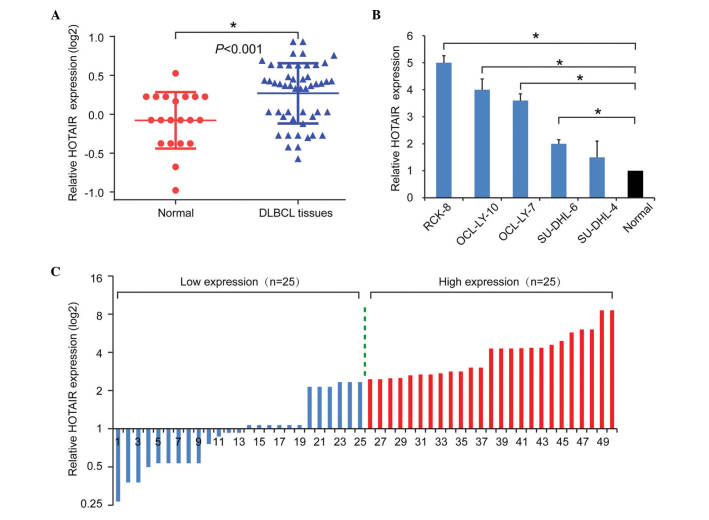
To examine the role of HOTAIR in DLBCL, the present study first examined the expression levels of HOTAIR in tissues using RT-qPCR. As shown in Fig. 1A, HOTAIR was upregulated in the DLBCL tissues, compared with the normal control tissues. To further validate the role of HOTAIR in DLBCL, the expression levels of HOTAIR were also examined in DLBCL cell lines. Consistently, the expression levels of HOTAIR in the DLBCL cell lines were significantly upregulated, compared with the average levels in the normal samples (Fig. 1B). Taken together, these results suggested tat the upregulation of HOTAIR was a frequent event in DLBCL.
Figure 1.
HOTAIR is overexpressed in DLBCL. (A) Expression levels of HOTAIR were significantly upregulated in the cancer tissues, compared with the normal tissues. (B) Expression levels of HOTAIR were significantly upregulated in the DLBCL cell lines, compared with the normal group. (C) Expression of HOTAIR was classified into two groups (low and high). Data are presented as the mean ± standard deviation from three replicate samples. *P<0.05. HOTAIR, Hox transcript antisense intergenic RNA; DLBCL, diffuse large B cell lymphoma.
Overexpression of HOTAIR is correlated with the clinicopathological features of DLBCL
To further investigate the clinical role of HOTAIR in DLBCL, the present study analyzed the correlation between the expression of HOTAIR and patient clinicopathological features. The median value was used as a cut-off to divide the expression levels of HOTAIR into a high expression group (n=25) and a low expression group (n=25; Fig. 1C). As shown in Table I, elevated expression levels of HOTAIR were positively associated with advanced clinical stage (P=0.004), tumor volume (P=0.045), B symptoms (P=0.011) and International Prognostic Index (IPI) scores (P=0.009).
Table I.
Correlation between the expression of HOTAIR and clinicopathological characteristics in diffuse large B cell lymphoma.
| Characteristic | n | Expression level of HOTAIR
|
P-value | |
|---|---|---|---|---|
| Low (n) | High (n) | |||
| Age (years) | 0.777 | |||
| <60 | 23 | 11 | 12 | |
| ≥60 | 27 | 14 | 13 | |
| Gender | 0.258 | |||
| Male | 24 | 10 | 14 | |
| Female | 26 | 15 | 11 | |
| Ann Arbor stage | 0.004a | |||
| I–II | 20 | 15 | 5 | |
| III–IV | 30 | 10 | 20 | |
| Extra-nodal status | 0.529 | |||
| <2 | 14 | 8 | 6 | |
| ≥2 | 36 | 17 | 19 | |
| B symptoms | 0.011a | |||
| Absent | 27 | 18 | 9 | |
| Present | 23 | 7 | 16 | |
| Bulk (cm) | 0.045a | |||
| <5 | 21 | 14 | 7 | |
| ≥5 | 29 | 11 | 18 | |
| IPI score | 0.009a | |||
| 0–2 | 19 | 14 | 5 | |
| 3–5 | 31 | 11 | 20 | |
HOTAIR, Hox transcript antisense intergenic RNA; IPI, International Prognostic Index;
P<0.05.
Survival analysis and prognostic significance of HOTAIR
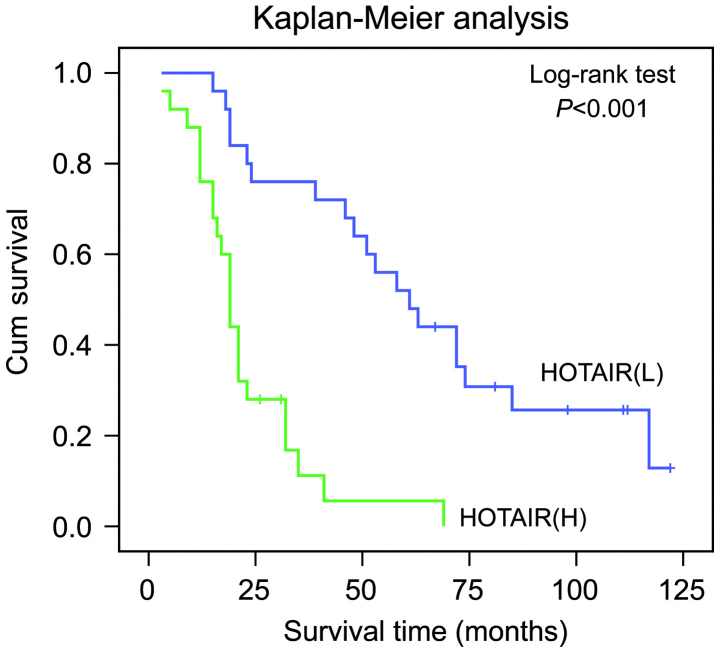
Kaplan-Meier analysis with a log-rank test for OS was performed to determine the possible association between the tumor expression levels of HOTAIR and patient survival rates. The results revealed that patients with higher expression levels of HOTAIR had a poorer prognosis, whereas the low expression group possessed a longer OS (P<0.001; Fig. 2).
Figure 2.
High levels of HOTAIR are correlated with poor OS in patients with diffuse large B cell lymphoma. Kaplan-Meier analysis with a log-rank test for OS was performed to assess the possible association between tumor expression levels of HOTAIR and patient survival rates. OS, overall survival HOTAIR, Hox transcript antisense intergenic RNA; H, high expression; L, low expression.
Using univariate analysis in the Cox proportional hazards model, decreased OS was associated with the following characteristics: Expression of HOTAIR, IPI score, clinical stage and B symptoms (P<0.05; Table II). Multivariate analysis revealed that the expression of HOTAIR, in addition to IPI scores, was an independent predictor of OS (HR, 3.127; 95% CI, 1.217–8.037; P=0.018). These results suggested that HOTAIR may offer potential as a potent prognostic factor in patients with DLBCL.
Table II.
Univariate and multivariate analyses of prognostic variables of overall survival in patients with diffuse large B cell lymphoma.
| Variable | Univariate analysis
|
Multivariate analysis
|
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Expression of HOTAIR | 0.018a | |||||
| (high, vs. low) | 5.220 | 2.481–10.984 | <0.001a | 3.127 | 1.217–8.037 | |
| Age (≥60, vs. <60 years) | 0.721 | 0.392–1.327 | 0.293 | |||
| Gender (female, vs. male) | 0.778 | 0.424–1.427 | 0.778 | |||
| Ann Arbor stage | ||||||
| (III–IV, vs. I–II) | 2.780 | 1.437–5.377 | 0.002a | |||
| Extra-nodal status | ||||||
| (≥2, vs. <2) | 1.002 | 0.517–1.943 | 0.994 | |||
| B symptoms | ||||||
| (present, vs. absent) | 2.339 | 1.228–4.454 | 0.010a | |||
| Bulk (≥5, vs. <5cm) | 1.491 | 0.793–2.803 | 0.214 | |||
| IPI score (3–5, vs. 0–2) | 2.948 | 1.512–5.750 | 0.002a | 2.806 | 1.392–5.654 | 0.004a |
HOTAIR, Hox transcript antisense intergenic RNA; IPI, International Prognostic Index; HR, hazard ratio; CI, confidence interval.
P<0.05.
HOTAIR knockdown inhibits cell proliferation in vitro
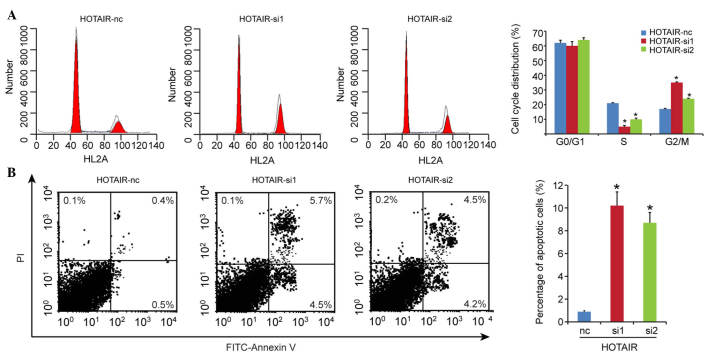
As HOTAIR is a prognostic factor for patients with DLBCL, the present study subsequently investigated the detailed function of HOTAIR in the DLBCL cells. The RCK-8 cells, which exhibited the highest expression level of HOTAIR among the cell lines, was selected as the targeted cell. The expression of HOTAIR was silenced in the RCK-8 cells, and the silencing efficiency was confirmed using qPCR. As shown in Fig. 3A, the si1 and si2 fragments effectively decreased the expression of HOTAIR. A CCK-8 assay was used to detect proliferation in vitro, and the results showed that HOTAIR knockdown inhibited cell proliferation, compared with the scramble group (Fig. 3B).
Figure 3.
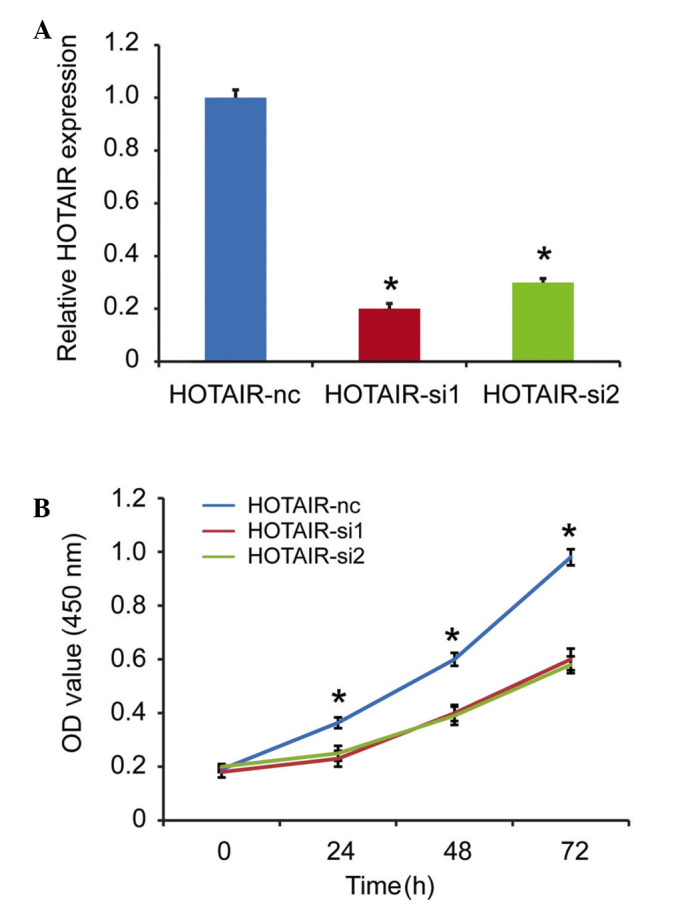
Silencing of HOTAIR in RCK-8 cells inhibits cell proliferation. (A) Relative expression levels of HOTAIR in RCK-8 cells transfected with siRNA control (nc) or siRNA HOTAIR, validated using reverse transcription-quantitative polymerase chain reaction analysis. (B) Cell Counting Kit-8 assays were performed to determine the proliferation of the RCK-8 cells. Data are presented as the mean ± standard deviation from three replicate samples. *P<0.05. HOTAIR, Hox transcript antisense intergenic RNA; DLBCL, diffuse large B cell lymphoma; siRNA, small interfering RNA; OD, optical density.
HOTAIR knockdown induces G2/M cell cycle arrest and promotes cell apoptosis
As the induction of cell cycle arrest is an important anti-proliferative mechanism, the present study investigated whether the inhibitory activity of HOTAIR knockdown on growth involved the control of cell cycle progression. The results of the cell cycle analysis showed that the G2/M-phase fraction increased between 17.0±1.5% in the HOTAIR-nc group, and 35.0±0.8% and 24.0±0.3% in the HOTAIR-si1 and HOTAIR-si2 groups, respectively (Fig. 4A), which suggested the induction of G2/M cell cycle arrest following HOTAIR knockdown.
Figure 4.
HOTAIR knockdown induces G2/M cell cycle arrest and promotes cell apoptosis. (A) Cell cycle and (B) apoptosis analyses were performed using flow cytometry in RCK-8 cells transfected with control (nc) or HOTAIR siRNA. Data are presented as the mean ± standard deviation from three replicate samples. *P<0.05. HOTAIR, Hox transcript antisense intergenic RNA; DLBCL, diffuse large B cell lymphoma; siRNA, small interfering RNA; FITC, fluorescein isothiocyanate; PI, propidium iodide.
To determine whether apoptosis contributed to the cell growth inhibition by HOTAIR knockdown, the present study evaluated the effect of the expression of HOTAIR on cell apoptosis. Compared with the HOTAIR-nc group (0.9±0.1%), cell apoptosis was significantly enhanced in the HOTAIR knockdown groups (HOTAIR-si1, 10.2±1.2%; HOTAIR-si2, 8.7±0.9%), as shown in Fig. 4B. These results indicated that cell cycle arrest and increased apoptosis contributed to the inhibition of proliferation in DLBCL on silencing HOTAIR.
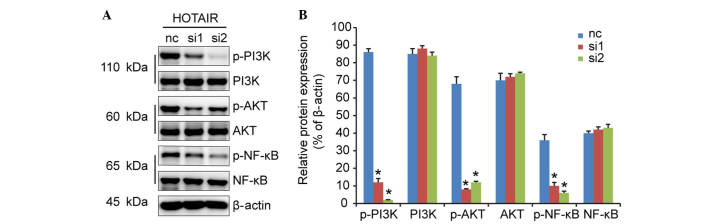
PI3K/AKT/NF-κB signaling pathway contributes to cell proliferation mediated by HOTAIR
The serine/threonine kinase, AKT, a downstream effector of PI3K, is involved in cell survival and anti-apoptotic signaling. A previous study demonstrated that cell signaling is mediated by PI3K/AKT via the induction of the NF-κB transcription factor, which is associated with cell proliferation (12). In the present study, no changes were found in the protein levels of total PI3K, AKT or NF-κB in response to HOTAIR knockdown (Fig. 5). However, the levels of PI3K, AKT and NF-κB phosphorylation were markedly decreased. These results indicated that the suppression of HOTAIR inhibited the activation of p-Akt and p-NF-κB, leading to decreased cell proliferation.
Figure 5.
HOTAIR silencing inhibits DLBCL cell proliferation, possibly through the PI3K/AKT/NF-κB signaling pathways. (A) Western blot analysis was performed to detect the expression levels of PI3K, AKT, NF-κB, p-PI3K, p-AKT and p-NF-κB, and (B) relative protein expression levels were calculated. *P<0.05 vs. the nc group. HOTAIR, Hox transcript antisense intergenic RNA; DLBCL, diffuse large B cell lymphoma; nc, negative control; PI3K, phos-phoinositide 3-kinase; NF-κB, nuclear factor-κB; p-, phosphorylated; nc, negative control.
Discussion
DLBCL is the most common subtype of lymphoid neoplasm, accounting for 30–40% of cases (13), and threatens the health of individuals worldwide. The pathogenesis and carcinogenesis of DLBCL are multi-step and heterogeneous processes, and involve different genetic and epigenetic changes. Despite increasing investigations focused on elucidating the complex progress, the majority focus on protein-coding genes rather than another crucial factor, lncRNA, which have been previously regarded as 'noise' in the process of the disease. HOTAIR, was first described by Rinn et al (14) as 2,158 nucleotides and 6 exons in length, and reported to repress transcription in trans across 40 kilobases of the HOXD locus. A substantial quantity of data have demonstrated that HOTAIR is involved in several processes in carcinogenesis and the promotion of malignancy (15), including those affecting mobility, invasion, proliferation, apoptosis, aggression and metastasis (16–18). In response to these findings, the present study investigated whether HOTAIR is involved in the malignant processes of DLBCL.
In the present study, it was found that HOTAIR was upregulated in DLBCL tumor samples, compared with normal tissues. Consistently, the same trend was observed in the DLBCL cell lines. The upregulation of HOTAIR was correlated with certain critical clinicopathological features, including clinical stage, B symptoms, IPI scores and tumor volumes. Additionally, it was demonstrated that higher expression levels of HOTAIR in tumor tissues predicted a poor prognosis in the patients with DLBCL. Univariate and multivariate analyses confirmed that HOTAIR may be used as a promising biomarker for patients with DLBCL.
The present study also investigated the biological function of HOTAIR in DLBCL cell lines. RCK-8 was selected due to its higher expression level of HOTAIR. HOTAIR knockdown significantly inhibited cell proliferation. A previously study showed that inducing cell cycle arrest and promoting cell apoptosis may be the possible anti-proliferative mechanisms (19). Therefore, the present study performed cell cycle and apoptosis analyses, and the results revealed that HOTAIR knockdown arrested the cell cycle in the G2/M period and induced cell apoptosis. These may have contributed to the inhibition of proliferation. The present study also examined the possible pathways involved in this process, and it was found that the silencing of HOTAIR inhibited the phosphorylation of PI3K, AKT and NF-κB. Taken together, HOTAIR inhibited cell proliferation, partly through the PI3K/AKT/NF-κB pathways, which were downstream of the lncRNA-specific gene.
In conclusion, the present study provided novel evidence that the overexpression of HOTAIR was a common event underlying DLBCL, representing a key pro-oncogenic role. It also functioned as an indicator of poor survival rates in the patients with DLBCL. In addition, the present study demonstrated that HOTAIR was imperative for DLBCL carcinogenesis by affecting cell proliferation, cell cycle and apoptosis. Taken together, the data obtained in the present study demonstrated the critical role of HOTAIR in the molecular etiology of DLBCL, and may provide a therapeutic regimen, dependent on lncRNA, directed against this life-threatening disease.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Shankland KR, Armitage JO, Hancock BW. Non-Hodgkin lymphoma. Lancet. 2012;380:848–857. doi: 10.1016/S0140-6736(12)60605-9. [DOI] [PubMed] [Google Scholar]
- 3.Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC, Gaasenbeek M, Angelo M, Reich M, Pinkus GS, et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- 4.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 5.Wu ZH, Wang XL, Tang HM, Jiang T, Chen J, Lu S, Qiu GQ, Peng ZH, Yan DW. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep. 2014;32:395–402. doi: 10.3892/or.2014.3186. [DOI] [PubMed] [Google Scholar]
- 6.Zhao W, An Y, Liang Y, Xie XW. Role of HOTAIR long noncoding RNA in metastatic progression of lung cancer. Eur Rev Med Pharmacol Sci. 2014;18:1930–1936. [PubMed] [Google Scholar]
- 7.Nakagawa T, Endo H, Yokoyama M, Abe J, Tamai K, Tanaka N, Sato I, Takahashi S, Kondo T, Satoh K. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun. 2013;436:319–324. doi: 10.1016/j.bbrc.2013.05.101. [DOI] [PubMed] [Google Scholar]
- 8.Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng W, Wu K, Feng J. Long noncoding RNA HULC predicts poor clinical outcome and represents pro-oncogenic activity in diffuse large B-cell lymphoma. Biomed Pharmacother. 2016;79:188–193. doi: 10.1016/j.biopha.2016.02.032. [DOI] [PubMed] [Google Scholar]
- 11.Zhou W, Wang G, Zhao X, Xiong F, Zhou S, Peng J, Cheng Y, Xu S, Xu X. A multiplex qPCR gene dosage assay for rapid genotyping and large-scale population screening for deletional α-thalassemia. J Mol Diagn. 2013;15:642–651. doi: 10.1016/j.jmoldx.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Gilmore TD. Introduction to NF-kappaB: Players, pathways, perspectives. Oncogene. 2006;25:6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- 13.A clinical evaluation of the international lymphoma study group classification of non-Hodgkin's lymphoma. The Non-Hodgkin's lymphoma classification project. Blood. 1997;89:3909–3918. [PubMed] [Google Scholar]
- 14.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiyomaru T, Fukuhara S, Saini S, Majid S, Deng G, Shahryari V, Chang I, Tanaka Y, Enokida H, Nakagawa M, et al. Long non-coding RNA HOTAIR is targeted and regulated by miR-141 in human cancer cells. J Biol Chem. 2014;289:12550–12565. doi: 10.1074/jbc.M113.488593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hajjari M, Salavaty A. HOTAIR: An oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12:1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou X, Chen J, Tang W. The molecular mechanism of HOTAIR in tumorigenesis, metastasis, and drug resistance. Acta Biochim Biophys Sin (Shanghai) 2014;46:1011–1015. doi: 10.1093/abbs/gmu104. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y, Zhang L, Wang Y, Li H, Ren X, Wei F, Yu W, Wang X, Zhang L, Yu J, Hao X. Long noncoding RNA HOTAIR involvement in cancer. Tumour Biol. 2014;35:9531–9538. doi: 10.1007/s13277-014-2523-7. [DOI] [PubMed] [Google Scholar]
- 19.Jiao F, Hu H, Yuan C, Wang L, Jiang W, Jin Z, Guo Z, Wang L. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol Rep. 2014;32:2485–2492. doi: 10.3892/or.2014.3518. [DOI] [PubMed] [Google Scholar]