Abstract
MicroRNAs (miRNAs) are a class of highly conserved, single-stranded RNA molecules (length, 18–25 nt) that regulate the expression of their target mRNAs. Previous studies have demonstrated that miRNAs may be novel biomarkers in the diagnosis of certain diseases. In order to evaluate the diagnostic value of miRNAs in childhood tuberculosis (TB), the circulating miRNA profile was determined using microarray analysis. An miRNA-gene network was constructed to identify closely associated miRNAs and these miRNAs were validated using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). A receiver operational curve (ROC) was used to evaluate the diagnostic sensitivity and specificity of confirmed miRNAs. The microarray data demonstrated that 29 miRNAs were altered with 15 upregulated and 14 downregulated. The network showed indicated 14 miRNAs that are critical in childhood TB. RT-qPCR validated that miR-1, miR-155, miR-31, miR-146a, miR-10a, miR-125b and miR-150 were downregulated in while miR-29 was upregulated in children with TB compared with uninfected children. The ROC curve data indicated the diagnostic value of single miRNA was as follows: miR-150>miR-146a>miR-125b>miR-31>miR-10a>miR-1>miR-155>miR-29. Notably, a combination of these miRNAs exhibited increased diagnostic value compared with any single miRNA. To the best of our knowledge, the present study is the first to identify the expression profile of circulating miRNAs in childhood TB and demonstrated that miRNAs may be a novel, non-invasive and effective biomarker for the early diagnosis of childhood TB.
Keywords: childhood tuberculosis, microRNA, microarray, diagnosis, biomarker
Introduction
Tuberculosis (TB) results in high rates of mortality and morbidity globally. According to global TB reports in 2014, an estimated 9.6 million individuals developed TB and 1.5 million mortalities were caused by TB (1). Despite progress being made on TB, childhood TB remains an epidemic in numerous developing countries. There were an estimated 53,000 TB cases among children (<15 years of age) and 74,000 TB-associated mortalities in 2012, which account for the 6 and 8% of the global totals, respectively. It has been reported that there was no marked decrease in the prevalence of childhood TB from 1979 to 2000 in China, and patients with bacteriologically-negative TB remain a large proportion of TB patients (2,3). There remains a long way to go for China to achieve their 2015 targets of eliminating TB. From 2016, the aim is to end the global TB epidemic by implementing the End TB Strategy. Adopted by the World Health Assembly in May 2014, the strategy serves to reduce the number of TB mortalities by 90% by 2030 (compared with 2015 levels), decrease the number of new cases by 80% and ensure that no family is financially burdened due to TB (1). To achieve this goal, novel and effective methods for the early development of childhood TB are required.
MicroRNA (miRNA) is a class of highly conserved, single-stranded RNA molecules (length, 18–25 nt) that regulate expression levels of their target mRNAs (4,5). Emerging research has demonstrated that serum miRNA is stable to repeated freezing and thawing as well as heat, acidic and alkaline conditions and other extremes. It may have potential as a useful biomarker for disease diagnosis, the effects of therapeutics and prognosis (6–10). Previous studies have demonstrated that miRNA is associated with numerous diseases, including cancer and heart, immune and infectious diseases (11–14). Altered levels of miRNAs following Mycobacterium tuberculosis (MTB) infection have been reported, however, these studies focused on adults rather than children (15–18). Therefore, the present study aimed to identify and validate the altered levels of circulating miRNAs in childhood TB. Furthermore, the present study aimed to determine the diagnostic value of single and combined miRNAs in childhood TB.
Materials and methods
Ethics statement
The present study was reviewed and approved by the ethics committee of Chongqing Medical University (Chongqing, China). Written informed consent was obtained from participants' parents at the time of enrollment in the present study.
Diagnostic process
Children with the following clinical manifestations were enrolled as suspected TB cases: i) Cough, fever and weight loss lasting >2 weeks; ii) pneumonia not responding to antibiotics; iii) and other clinical findings, including hydrothorax, tuberculin test (+), interferon-γ release assay (+), anti-tuberculosis antibody (+), PCR (+) and chest radiography suggestive of TB. Preliminary screening was conducted according to previously described guidelines (19). Cases that failed to meet the criteria, had pulmonary infection other than TB or had other preliminary diagnoses were excluded. The patients' sputum and stomach lavage fluid were collected to isolate MTB using Lowenstein-Jensen medium and an acid-fast bacillus test. Cases with successful isolation of MTB were regarded as culture-positive tuberculosis, while cases with no successful isolation of MTB were regarded as culture-negative tuberculosis. Healthy children were selected from a child care center at the Children's Hospital of Chongqing Medical University (Chongqing, China) between February 2012 and July 2013. Written informed consent was obtained from the patient's family. These children did not recently suffer from any infectious diseases and did not have any immunodeficiency diseases.
Sample collection and handing
A total of 129 children were recruited from the Children's Hospital of Chongqing Medical University (Chongqing, China). Following preliminary screening, 28 patients and 24 healthy children were included in the present study. Three active pulmonary TB cases (confirmed by MTB culture) and three healthy samples were used for microarray detection. The remaining 25 patients and 21 healthy children had samples taken for use in reverse transcription-quantitative polymerase chain reaction validation. There was no significant difference in age or gender between the two groups. The demographic and clinical characteristics of patients are summarized in Table I. Fresh blood samples were collected from the participants in 2.0 ml anticoagulant tubes. Within 4 h of collection, red blood cell lysis buffers produced in our laboratory were used to collect peripheral white blood cells for total RNA extraction. A total of 3 ml 1X red blood cell lysis buffer per ml of blood was added and mixed sufficiently. Centrifugation was then performed at 6,000 x g at room temperature for 8–10 min and the supernatant was discarded. Subsequently, 1 ml 1X red blood cell lysis buffer per tube was added, mixed and centrifuged at 6,000 × g at room temperature for 5 min. The supernatant was discarded and the sediment was white blood cells.
Table I.
Clinical characteristics of childhood TB.
| Characteristic | Culture-positive TB | Culture-negative TB |
|---|---|---|
| Total | 14 | 14 |
| Age (years, mean ± SD) | 8.4±5.7 | 6.7±4.9 |
| Gender (male/female) | 11/3 | 5/9 |
| History of close TB contact | 7 | 4 |
| TST positive | 4 | 10 |
| Cough for >2 weeks | 8 | 5 |
| Fever for >2 weeks | 8 | 10 |
| Night sweats for >2 weeks | 0 | 0 |
| Weight loss/failure to thrive | 0 | 2 |
| Radiographic features of TB | 14 | 14 |
| Culture-positive TB | 14 | 0 |
| HIV-positive | 0 | 0 |
| HBV-positive | 0 | 0 |
TB, tuberculosis; TST, tuberculin skin test; SD, standard deviation; HIV, human immunodeficiency virus; HBV, hepatitis B virus.
RNA extraction
Total RNA was isolated from peripheral white blood cells using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocols. Each sample was dissolved in 50 µl RNAase-free water. RNA quantity and purity was measured using a NanoDrop spectrophotometer (NANODROP 1000; Thermo Fisher Scientific, Inc.). In solution, the high quantity RNA had an A260/A280 ratio of 1.8–2.1. The total RNA concentration was 100–400 ng/µl.
Analysis of miRNA microarray data
miRNA microarray assays were performed using the Agilent Human miRNA microarray platform (Agilent Technologies, Inc., Santa Clara, CA, USA). Labeling and hybridization were performed according to the protocols in the Agilent miRNA microarray system. The microarray was scanned by Agilent miRNA Scanner and the microarray data was normalized using R-language programming (http://www.r-project.org). The normalized data was analyzed according to log 2-transformed expression level and the expression levels were tested by Student's t-test and the Wilcoxon signed rank sum test. The miRNAs that indicated significantly different expression in the two tests and had at least a 2-fold changes were filtered. An miRNA may target different genes and a gene may be targeted by different miRNAs. In order to understand the complicated associations between miRNAs and target genes, Targetscan (http://genes.mit.edu/targetscan/index.html) and Pictar (http//pictar.bio.nyu.edu) databases were used to predict the target genes. The miRNA-gene network was built to observe closely associated miRNAs following TB infection using Cytoscape (version 3.0.1; http://www.cytoscape.org/).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA (500 ng) was isolated and miRNA was obtained by poly (A) tail method. miRNAs in the center of the network were selected for validation using a SYBR green RT-qPCR assay (All-In-One miRNA qRT-PCR Detection kit, GeneCopoeia, Inc., Rockville, MD, USA). The miRNA primers were synthesized by GeneCopoeia, Inc. and the expression level was normalized to U6. The miRNA was polyadenylated by poly(A) polymerase and reverse transcribed to cDNA under conditions of 37°C for 60 min followed by 85°C for 5 min. The RT reaction was conducted in a reaction volume of 10 µl, which contained 2.5 U/µl PolyA polymerase (0.4 µl), 0.4 µl RTase mix, 2 µl 5X Reaction Buffer, 500 ng total RNA and variable volume of RNase Free dH2O. Synthesized cDNA was diluted 5 times for Real-Time PCR reaction. The total volume of the reaction mixture was 20 µl, including 10 µl of 2X All-In-One qPCR mix, 2 µl All-In-One qPCR Primer (2 µM), 2 µl Universal Adaptor PCR Primer (2 µM), 2 µl cDNA and 4 µl dH2O. The PCR was conducted with incubation at 95°C for 10 min, and 40 cycles of 95°C for 10 sec, 55°C for 20 sec and 72°C for 10 sec using a CFX96 Real-Time system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Each sample was run in triplicate. For the RT-qPCR data, the relative expression level of each miRNA was calculated according to the different cycle quantification (Cq) values between the target miRNAs and miR-U6 by using the 2−ΔΔCq method (20).
Statistical analysis
Statistical analysis was performed with SPSS software (version 21; IBM SPSS, Armonk, NY, USA). The data are presented as the mean ± standard deviation. An unpaired t-test was used for statistical analysis and P<0.05 was considered to indicate a statistically significant difference. Receiver operating characteristic (ROC) curve was generated to evaluate the diagnostic value of miRNAs. The area under the curve (AUC) and 95% confidence intervals (CI) were calculated to determine the specificity and sensitivity. To increase the diagnostic accuracy, a combination of circulating miRNAs was analyzed. The ROC curve and multivariate logistic regression in the ROC curves were calculated with MedCalc 11.4.3.0 statistical software (MedCalc Software bvba, Ostend, Belgium).
Results
Identification of circulating miRNAs from children with TB
Following preliminary screening, a total of 29 miRNAs were altered between the TB group and the control. Of these, 15 miRNAs were upregulated and 14 were downregulated when compared with the healthy control group (Table II). Cluster analysis based on differentially expressed miRNAs indicated marked distinctions between the two groups (Fig. 1).
Table II.
FC of miRs between children with tuberculosis and healthy controls.
| ID | logFC |
|---|---|
| miR-142-5p | 1.165432929 |
| miR-29b | 1.289097829 |
| miR-21* | 1.67673294 |
| miR-542-5p | 1.812354699 |
| miR-32 | 1.86960397 |
| miR-142-3p | 1.887657013 |
| miR-95 | 2.410210413 |
| miR-144 | 2.099441438 |
| miR-17* | 2.128855333 |
| miR-141 | 2.278684837 |
| miR-33a | 2.372570115 |
| miR-136 | 2.571642478 |
| miR-324-5p | 2.845130237 |
| miR-193a-3p | 3.94172976 |
| miR-503 | 4.518278633 |
| miR-31 | −5.138973159 |
| miR-564 | −3.577222134 |
| miR-1 | −2.229848349 |
| miR-181a-2* | −2.048150209 |
| miR-874 | −1.99302363 |
| miR-1305 | −1.894597861 |
| miR-10a | −1.824343578 |
| miR-1275 | −1.756667644 |
| miR-125b | −1.675341028 |
| miR-342-5p | −1.637504768 |
| miR-150 | −1.370167992 |
| miR-155 | −1.282465779 |
| miR-342-3p | −1.131121391 |
| miR-146a | −1.00258041 |
FC, fold change; miR, microRNA.
Figure 1.
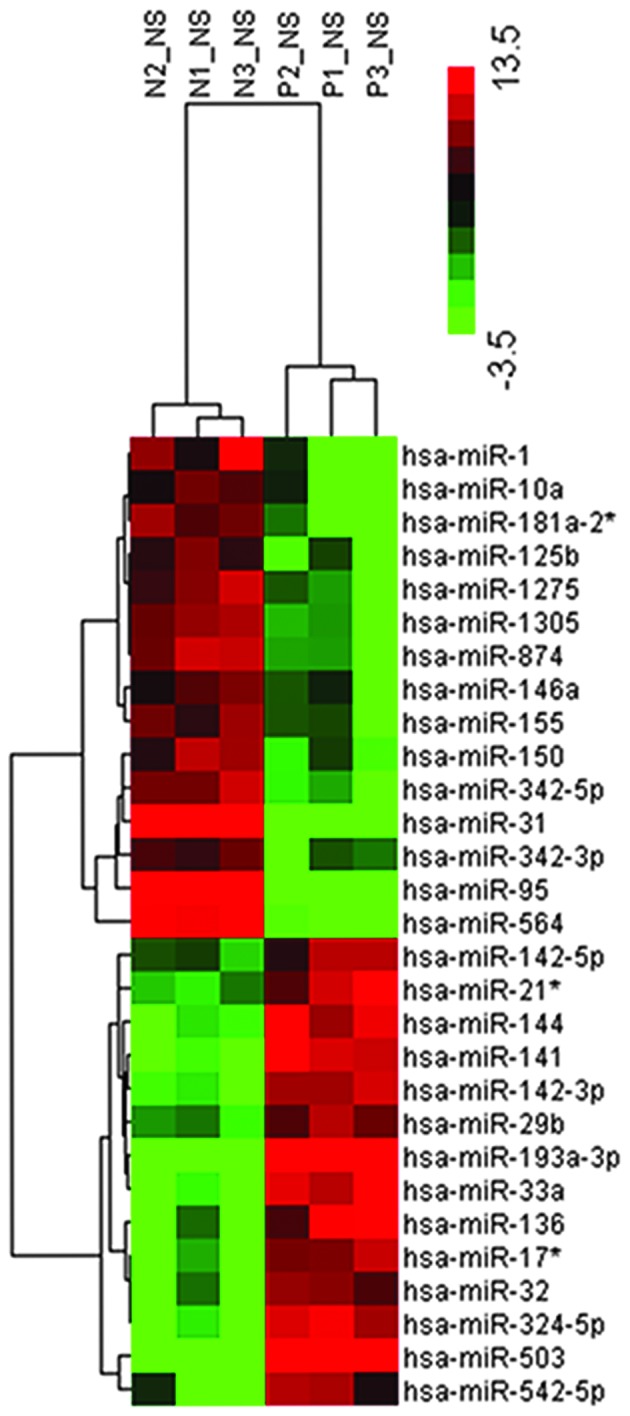
Hierarchical clustering of circulating miRs from childhood TB and control groups. Peripheral white blood cells were clustered according to the expression profiles of 29 differentially expressed miRs between children with TB and healthy controls. The analyzed samples and the miRs were presented in rows. The miRNA clustering tree is shown on the right, and the sample clustering tree is presented at the top. The color scale in the right top corner indicates the relative expression level of the miRs, with red representing a high expression level and green representing a low expression level. miR, microRNA; TB, tuberculosis; N, healthy children; P, TB patients.
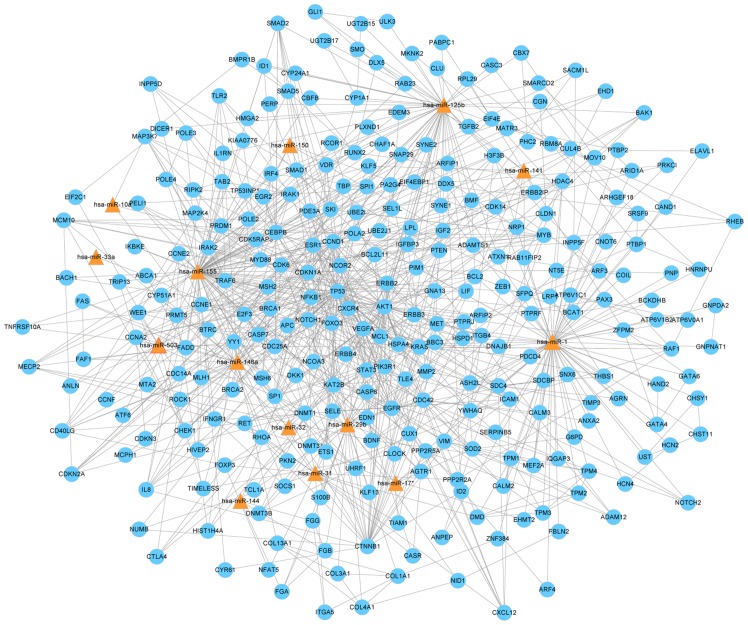
Construction of the miRNA-gene network
To investigate whether the unique miRNA profile and their target genes are associated with the progression of TB infection, and to identify miRNAs that are closely associated with childhood TB, an miRNA-gene network was constructed. miRNAs and their target genes have a complex association as miRNAs may target genes directly or indirectly. The miRNAs and genes closest to the center are more likely to have altered expression levels. The network demonstrated miR-1, miR-155, miR-31, miR-146a, miR-10a, miR-125b, miR-150, miR-29b, miR-141, miR-17*, miR-32, miR-33a, miR-503 and miR-144 were in the center of the network (Fig. 2).
Figure 2.
miR-gene network of childhood tuberculosis. The different target genes are shown in blue and the different miRs are shown by the triangles. The associations between miRs and genes are presented using straight lines. All miRs with differential expression and their target genes were analyzed, the remaining 15 miRs were distributed sporadically and, thus, excluded from the network. miR, microRNA.
Validation of the altered miRNAs in independent samples
Fourteen candidate miRNAs were selected for further verification in 25 children with TB and 21 healthy controls. When compared with healthy controls, miR-1, miR-155, miR-31, miR-146a, miR-10a, miR-125b and miR-150 were downregulated while miR-29 was upregulated in the TB cases (Fig. 3).
Figure 3.
Validation of miRs with different expression levels in children with tuberculosis and healthy controls. miRs in the center of the network were selected for validation. The 2−ΔΔCq method was used to normalize the relative gene expression data in the reverse transcription-quantitative polymerase chain reaction assay and U6 served as the reference gene. **P<0.01; *P<0.05. miR, microRNA.
Circulating miRNA to diagnose childhood TB
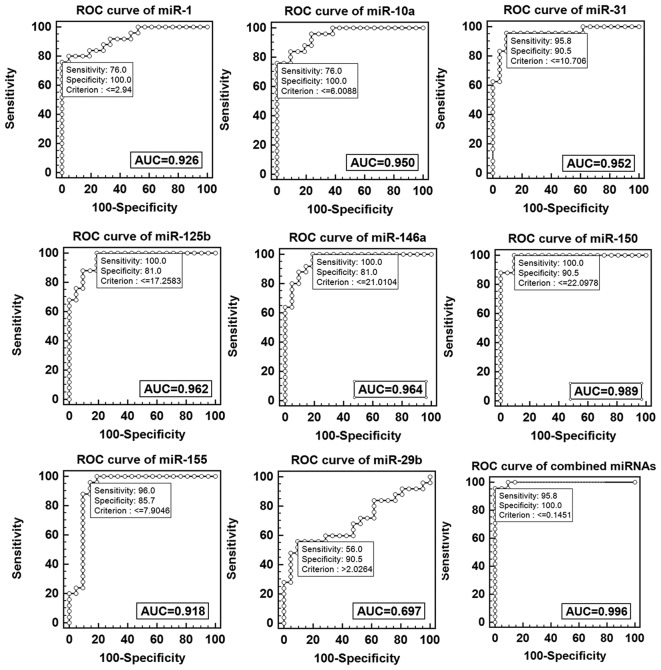
ROC was performed to evaluate the potential of microRNA as a biomarker to diagnose childhood TB. The data demonstrated the diagnostic value of single miRNA was as follows: miR-1 50>miR-146a>miR-125b>miR-31>miR-10a>miR-1>miR-155>miR-29. The data demonstrated miR-1, miR-10a, miR-125b miR-146a, miR-150, miR-155 and miR-31 exhibited a reliable discrimination yielding AUC values of 0.926 (95% CI, 0.809–0.982), 0.950 (95% CI, 0.843–0.993), 0.962 (95% CI, 0.860–0.996), 0.964 (95% CI, 0.862–0.997), 0.989 (95% CI, 0.902–1.0), 0.918 (95% CI, 0.799–0.978) and 0.952 (95% CI, 0.844–0.994) respectively. Using an optical cutoff (maximum Youden index; sensitivity +, specificity −1), the sensitivity of the seven miRNAs was 76, 76, 100, 100, 100, 96 and 95.8% respectively and the specificity was 100, 100, 81, 81, 90.5, 85.7 and 90.5% respectively. The miR-29b had a lower discrimination yielding an AUC value of 0.697 (95% CI, 0.544–9.824), with optical cutoff, the sensitivity and specificity were 56 and 90.5% respectively. Logistic regression analysis with the ROC curves was used to identify combined miRNAs, which may aid in the diagnosis of childhood TB, the result indicated marked separation between TB and healthy groups with an AUC value of 0.996 (95% CI, 0.914–1.0), the sensitivity was 95.8% and the specificity was 100% (Fig. 4).
Figure 4.
ROC curve analysis of single and combined miRs. AUC, area under curve; ROC, receiver operational curve; miR, microRNA.
Discussion
Childhood TB cases are currently diagnosed by history of close contact with a patient with TB, clinical manifestations, chest radiography, tuberculin test, interferon-γ release assay, anti-tuberculosis antibody or PCR. However, children typically lack clinical symptoms, with >50% children asymptomatic at early stages of the disease (21,22). The gold standard to diagnose TB is microbiological isolation of MTB; however, the load of MTB is often low in tested fluids, including sputum, stomach lavage fluid and bronchoalveolar lavage fluid. In addition, the culture of clinical specimens requires long incubation times (4–8 weeks), by which point it is too late to influence clinical strategies of treatment and the success rate of the treatment is low, <30–40% (17,23,24). Identification of a novel biomarker to allow early diagnosis of childhood tuberculosis is required.
There are numerous studies that describe the miRNA profile following TB infection and miR-155, miR-155, miR-200C, miR-193a-3p, miR-595, miR-432, miR-9, miR-582-5p, miR-144 and miR-29b have been validated in other studies (15,18,25–28). The microarray data from the present study is largely inconsistent with previous studies (17,27,29,30). For example, data from the present study indicated that miR-155 was downregulated in the TB group, however, Wu et al (30) demonstrated that miR-155 was upregulated in the purified protein derivative challenged peripheral blood mononuclear cells of patients with active TB. Consistent with previous studies, miR-141, miR-32 and miR-29b were overexpressed in the TB group of the present study. The expression level of miR-144 has varied in previous studies, Wang et al (27) suggested miR-144 is upregulated in TB patients, while Wu et al (30) observed downregulation. In the present study, no significant difference was observed in the miRNA between TB children and healthy children. Furthermore, microarray data from the present study has suggested a difference in expression in a number of miRNAs that have not yet been reported, including miR-31, miR-342-5p, miR-193a-3p, miR-10 and miR-33a. This may be a result of different experimental protocols, experimental conditions and samples.
The miRNA-gene network was constructed to observe key miRNAs. The further validation data demonstrated eight of the miRNAs in the center of the network had significantly different expression levels. Of these miRNAs, miR-1, miR-155, miR-31, miR-146a, miR-10a, miR-125b, miR-150, miR-141, miR-144, miR-17*, miR-32 and miR-503 were downregulated while miR-29 was upregulated in the TB group. The diagnostic values of these miRNAs were analyzed using ROC curves. The data demonstrated each of the miRNAs, excluding miR-29b, had a reliable diagnostic value and they all exhibited moderate specificity and sensitivity. Furthermore, the ROC curve data indicated the diagnostic value of single miRNA was as follows: miR-150>miR-146a>miR-125b>miR-31> miR-10a>miR-1>miR-155>miR-29.
However, a combination of the eight miRNAs demonstrated increased diagnostic value with an AUC of 0.996 (95% CI, 0.914–1.0), the sensitivity and specificity were 95.8 and 100%, respectively. It indicated combined identification of miR-1, miR-155, miR-31, miR-146a, miR-10a, miR-125b, miR-150 and miR-29 may be a novel early diagnostic biomarker.
In conclusion, the present study is the first, to the best of our knowledge, to analyze the circulating miRNA profile in childhood TB. Results from the current study suggest multiple miRNAs may be suitable to serve as potential biomarkers for the diagnosis of childhood TB.
Acknowledgments
The authors would like to thank all the patients and healthy children who participated in the present study at the Children's Hospital of Chongqing. The present study was supported by NSFC (grant no. 81071406) and the Medical Scientific Research of Chongqing (grant no. 20142046).
Abbreviations
- miRNA
microRNA
- TB
tuberculosis
- MTB
Mycobacterium tuberculosis
- ROC
receiver operational curve
- AUC
area under the curve
- CI
confidence interval
References
- 1.World Health Organization . Global Tuberculosis Report 2015. World Health Organization; 2015. 20th edition. [Google Scholar]
- 2.Li L. The national epidemiological sampling survey analysis of childhood tuberculosis from 1979 to 2000. Chinese Medical Journal. 2004;80:1678. In Chinese. [Google Scholar]
- 3.World Health Organisation . Global tuberculosis report 2013. 2013. [Google Scholar]
- 4.Krishnarao A, editor. MicroRNAs – From Basic Science to Disease Biology. Cambridge University Press; Cambridge, UK: 2010. [Google Scholar]
- 5.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 6.Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, Lo YM. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54:482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 7.Alevizos I, Illei GG. MicroRNAs as biomarkers in rheumatic diseases. Nat Rev Rheumatol. 2010;6:391–398. doi: 10.1038/nrrheum.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, et al. Serum microRNAs are promising novel biomarkers. PloS One. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Hu Z, Wang W, Ba Y, Ma L, Zhang C, Wang C, Ren Z, Zhao Y, Wu S, et al. Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int J Cancer. 2012;130:1620–1628. doi: 10.1002/ijc.26177. [DOI] [PubMed] [Google Scholar]
- 10.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, Sunakozaka H, Sakai Y, Horimoto K, Kaneko S. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098–1112. doi: 10.1002/hep.22749. [DOI] [PubMed] [Google Scholar]
- 13.Gidlöf O, Erlinge D. MicroRNAs in the failing heart-novel therapeutic targets? Scand Cardiovasc J. 2014;48:328–334. doi: 10.3109/14017431.2014.983965. [DOI] [PubMed] [Google Scholar]
- 14.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, et al. MicroRNA expression, survival and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Y, Yi Z, Wu X, Li J, Xu F. Circulating microRNAs in patients with active pulmonary tuberculosis. J Clin Microbiol. 2011;49:4246–4251. doi: 10.1128/JCM.05459-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spinelli SV, Diaz A, D'Attilio L, Marchesini MM, Bogue C, Bay ML, Bottasso OA. Altered microRNA expression levels in mononuclear cells of patients with pulmonary and pleural tuberculosis and their relation with components of the immune response. Mol Immunol. 2013;53:265–269. doi: 10.1016/j.molimm.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, Guo J, Fan S, Li Y, Wei L, Yang X, Jiang T, Chen Z, Wang C, Liu J, et al. Screening and identification of six serum microRNAs as novel potential combination biomarkers for pulmonary tuberculosis diagnosis. PloS One. 2013;8:e81076. doi: 10.1371/journal.pone.0081076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abd-El-Fattah AA, Sadik NA, Shaker OG, Aboulftouh ML. Differential microRNAs expression in serum of patients with lung cancer, pulmonary tuberculosis and pneumonia. Cell Biochem Biophys. 2013;67:875–884. doi: 10.1007/s12013-013-9575-y. [DOI] [PubMed] [Google Scholar]
- 19.Subspecialty Group of Respiratory Diseases. Society of Pediatrics, Chinese Medical Association Editorial Board, Chinese Journal of Pediatrics: Diagnostic standards and therapeutic recommendations for pulmonary tuberculosis in children. Zhonghua Er Ke Za Zhi. 2006;44:249–251. In Chinese. [PubMed] [Google Scholar]
- 20.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 21.Anderson ST, Kaforou M, Brent AJ, Wright VJ, Banwell CM, Chagaluka G, Crampin AC, Dockrell HM, French N, Hamilton MS, et al. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N Engl J Med. 2014;370:1712–1723. doi: 10.1056/NEJMoa1303657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eamranond P, Jaramillo E. Tuberculosis in children: Reassessing the need for improved diagnosis in global control strategies. Int J Tuberc Lung Dis. 2001;5:594–603. [PubMed] [Google Scholar]
- 23.Zar HJ, Connell TG, Nicol M. Diagnosis of pulmonary tuberculosis in children: New advances. Expert Rev Anti Infect Ther. 2010;8:277–288. doi: 10.1586/eri.10.9. [DOI] [PubMed] [Google Scholar]
- 24.Wallis RS, Pai M, Menzies D, Doherty TM, Walzl G, Perkins MD, Zumla A. Biomarkers and diagnostics for tuberculosis: Progress, needs and translation into practice. Lancet. 2010;375:1920–1937. doi: 10.1016/S0140-6736(10)60359-5. [DOI] [PubMed] [Google Scholar]
- 25.Maertzdorf J, Weiner J, III, Mollenkopf HJ, TBornotTB Network. Bauer T, Prasse A, Müller-Quernheim J, Kaufmann SH. Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc Natl Acad Sci USA. 2012;109:7853–7858. doi: 10.1073/pnas.1121072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi Y, Cui L, Ge Y, Shi Z, Zhao K, Guo X, Yang D, Yu H, Cui L, Shan Y, et al. Altered serum microRNAs as biomarkers for the early diagnosis of pulmonary tuberculosis infection. BMC Infect Dis. 2012;12:384–393. doi: 10.1186/1471-2334-12-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C, Yang S, Sun G, Tang X, Lu S, Neyrolles O, Gao Q. Comparative miRNA expression profiles in individuals with latent and active tuberculosis. PloS One. 2011;6:e25832. doi: 10.1371/journal.pone.0025832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yi Z, Fu Y, Ji R, Li R, Guan Z. Altered microRNA signatures in sputum of patients with active pulmonary tuberculosis. PloS One. 2012;7:e43184. doi: 10.1371/journal.pone.0043184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Wang X, Jiang J, Cao Z, Yang B, Cheng X. Modulation of T cell cytokine production by miR-144* with elevated expression in patients with pulmonary tuberculosis. Mol Immunol. 2011;48:1084–1090. doi: 10.1016/j.molimm.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 30.Wu J, Lu C, Diao N, Zhang S, Wang S, Wang F, Gao Y, Chen J, Shao L, Lu J, et al. Analysis of microRNA expression profiling identifies miR-155 and miR-155* as potential diagnostic markers for active tuberculosis: A preliminary study. Hum Immunol. 2012;73:31–37. doi: 10.1016/j.humimm.2011.10.003. [DOI] [PubMed] [Google Scholar]