Abstract
In yeast, plant, and mammalian somatic cells, short poly(A) tails on mRNAs are subject to uridylation, which mediates mRNA decay. Although mRNA uridylation has never been reported in animal oocytes, maternal mRNAs with short poly(A) tails are believed to be translationally repressed. In this study, we found that 96% of cyclin B mRNAs with short poly(A) tails were uridylated in starfish oocytes. Hormonal stimulation induced poly(A) elongation of cyclin B mRNA, and 62% of long adenine repeats did not contain uridine residues. To determine whether uridylated short poly(A) tails destabilize cyclin B mRNA, we developed a method for producing RNAs with the strict 3′ terminal sequences of cyclin B, with or without oligo(U) tails. When we injected these synthetic RNAs into starfish oocytes prior to hormonal stimulation, we found that uridylated RNAs were as stable as nonuridylated RNAs. Following hormonal stimulation, the 3′ termini of short poly(A) tails of synthesized RNAs containing oligo(U) tails were trimmed, and their poly(A) tails were subsequently elongated. These results indicate that uridylation of short poly(A) tails in cyclin B mRNA of starfish oocytes does not mediate mRNA decay; instead, hormonal stimulation induces partial degradation of uridylated short poly(A) tails in the 3′–5′ direction, followed by poly(A) elongation. Oligo(U) tails may be involved in translational inactivation of mRNAs.
Keywords: mRNA uridylation, poly(A), polyadenylation, oocyte, starfish, oligo(U)
INTRODUCTION
Generally, mRNAs with long poly(A) tails are stable and translationally activated, and shortening of the poly(A) tail decreases mRNA stability (Wilusz and Wilusz 2008; Norbury 2013). Recent work revealed that short poly(A) tails of mRNAs are uridylated in yeast (Rissland et al. 2007; Rissland and Norbury 2009), plant (Sement et al. 2013), and mammalian cells (Chang et al. 2014; Lim et al. 2014). The noncoded uridines decrease the stability of the mRNAs; specifically, the Lsm1–7 complex mediates degradation of 3′ uridylated mRNAs by recruiting decapping factors to 5′ caps (Rissland and Norbury 2009; Norbury 2013). Dis3l2, which interacts with oligo-uridines, is the 3′–5′ exonuclease responsible for the decay of these mRNAs (Malecki et al. 2013). In addition, oligo(U) tails of RNAs such as miRNAs, miRNA-directed cleavage products, and poly(A)− histone mRNA play a role in RNA decay, which is executed by RNA degradation factors including the Lsm1–7 complex, XRN1, the exosome, and Dis3L2 (Song and Kiledjian 2007; Mullen and Marzluff 2008; Chang et al. 2013; Hoefig et al. 2013; Malecki et al. 2013; Ustianenko et al. 2013; Lee et al. 2014; Slevin et al. 2014). Moreover, uridylation of the 3′ end of a polyadenylated luciferase reporter RNA represses translation of the RNA in Xenopus oocytes (Lapointe and Wickens 2013).
Although the poly(A) tails of stored mRNAs in animal oocytes are short, mRNAs such as cyclin B are believed to be stable as maternal mRNAs. In Xenopus oocytes, the poly(A) length of cyclin B1 mRNAs in oocytes is dynamically controlled by uridine-rich cytoplasmic polyadenylation elements (CPE) in their 3′ UTRs and the CPE binding protein (CPEB), which allows binding of other proteins, including poly(A) polymerase (Gld2) and deadenylating enzyme (PARN) (Fox et al. 1989; Fox and Wickens 1990; Hake and Richter 1994; Copeland and Wormington 2001; Kwak et al. 2004). PARN is more active than Gld2, resulting in the shortening of poly(A) tails in immature oocytes (Kim and Richter 2006). After hormonal stimulation, CPEB is phosphorylated, causing the release of PARN from the RNP complex, followed by elongation of poly(A) tails by Gld2 (Paris et al. 1991; Mendez et al. 2000; Kim and Richter 2006; Richter 2007; Radford et al. 2008). Similar elongation of poly(A) tails of mRNAs has been reported in oocytes of mouse, fish, Drosophila, and Spisula (Sheets et al. 1994; Minshall et al. 1999; Tay et al. 2000; Benoit et al. 2008; Yasuda et al. 2010).
In starfish oocytes, the short poly(A) tails of cyclin B mRNA are elongated upon meiotic reinitiation (Hara et al. 2009) induced by a hormonal stimulation of 1-methyladenine (1-MA) (Kanatani et al. 1969). The 1-MA receptor is coupled to a heterotrimeric G protein (Tadenuma et al. 1991, 1992; Chiba et al. 1992), and Gβγ dissociated from Gα activates PI3-kinase (Chiba et al. 1993; Jaffe et al. 1993), PDK1 (Hiraoka et al. 2004), Akt (Okumura et al. 2002), cdc25 phosphatase (Okumura et al. 1996), and cdc2/cyclin B (Kishimoto 2015). Translational activation of cyclin B mRNA is required for meiotic division.
Poly(A) tail length in oocytes is usually experimentally determined by poly(A) test assay (PAT assay), which takes advantage of oligo(dT) annealing to poly(A) tail (Sallés and Strickland 1995). Likewise, the most common method for cDNA synthesis is also based on oligo(dT) annealing. However, if the 3′ terminus of an mRNA contains oligo(U, G, or C) tails, oligo(dT) cannot anneal, resulting in synthesis of a cDNA that does not contain an exact copy of the original mRNA sequence at the 3′ terminus. Therefore, the actual 3′ terminal sequences of cyclin B mRNAs of oocytes remain unknown in many animals. In this study, we applied adaptor ligation to 3′ termini of mRNAs (Wada et al. 2012) in starfish oocytes to determine whether short poly(A) tails of cyclin B mRNAs are modified.
RESULTS AND DISCUSSION
Uridylated cyclin B mRNA in starfish oocytes
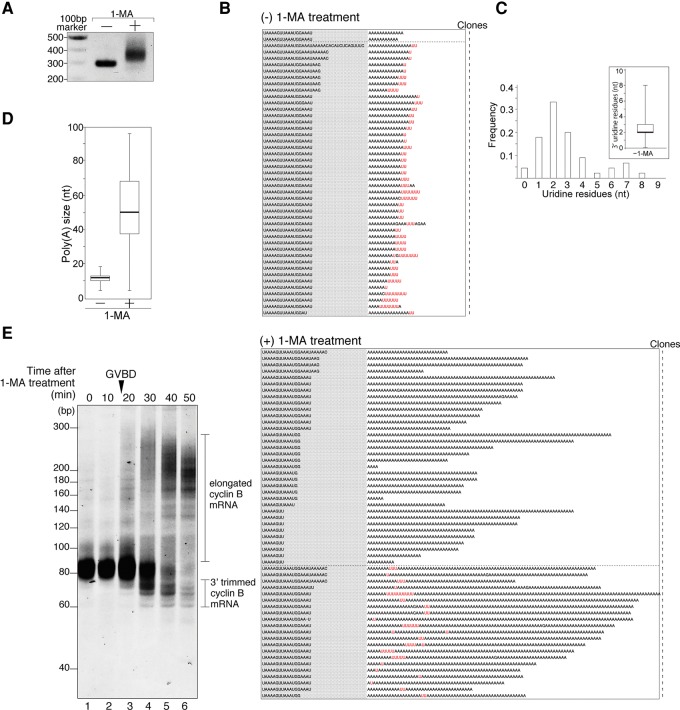
We ligated 23-nucleotide (nt) adaptors to the 3′ ends of total RNA from starfish oocytes, and performed RT-PCR using a 3′ adaptor primer and a cyclin B-specific primer (sfcycB Fwd primer1; see Materials and Methods) designed to hybridize ∼230 nt upstream of the polyadenylation site. Agarose gel electrophoresis of RT-PCR products from oocytes not subjected to hormonal stimulation revealed a single ∼290-bp band (Fig. 1A, −), suggesting that the tail length of cyclin B mRNA was 10–20 nt. The 320–380-bp mobility-shifted band observed in stimulated oocytes with hormone 1-MA (Fig. 1A, +) was consistent with the elongated poly(A) tail length measured by Hara et al. (2009) using the PAT assay.
FIGURE 1.
Uridylated short poly(A) tail of cyclin B mRNA in starfish oocytes and nonuridylated long poly(A) tail of cyclin B mRNA in oocytes stimulated with the hormone 1-MA. (A) Elongation of the 3′ region of mRNA after hormonal stimulation. Adaptor-ligated cyclin B mRNAs from oocytes treated with (+) or without (−) 1-MA were RT-PCR amplified. The products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. (B) Sequencing results of the 3′ terminal region of cyclin B mRNA from oocytes treated with [(+) 1-MA treatment] or without 1-MA [(−) 1-MA treatment]. Gray shaded sequences show the 3′ terminal portion of the 3′ UTR, and the following sequences are the tail regions of cyclin B mRNA. The number of clones obtained is indicated. (C) Histogram of the number of uridine residues at the 3′ ends of cyclin B mRNAs in starfish oocytes not subjected to hormonal stimulation. Inset shows box-plot analysis of oligo(U) tail length. The 25th and 75th percentiles are shown by box edges, the median value is indicated by the thick line, and whiskers show the maximum and minimum values. (D) Box plot analysis of poly(A) tail length of cyclin B mRNAs in oocytes. Median of poly(A) size increased from 12 to 50 nt after hormonal stimulation. (E) Trimming activity at the 3′ region of cyclin B mRNA after hormonal stimulation. GVBD occurred 18 min after 1-MA treatment. At the indicated time after hormonal stimulation, total RNA was purified from oocytes, and adaptor-ligated cyclin B mRNAs were RT-PCR amplified using 3′ adaptor Rev primer 1 and sfcycB short Fwd primer. The PCR products were subjected to high-resolution acrylamide gel electrophoresis (15%) and visualized by SYBR-Green I staining.
To determine whether 3′ termini of poly(A) tails of starfish cyclin B mRNAs are modified, we cloned the RT-PCR product of cyclin B (Fig. 1A) and sequenced its 3′ terminal region. To our surprise, 96% (n = 45) of cyclin B mRNAs had a 1–8 nt oligo(U) tail (median, 2 nt) downstream from a short (4–18 nt) poly(A) tail (median, 12 nt) (Fig. 1B, [−] 1-MA treatment; 1C; 1D). The most frequent number of uridines was two (Fig. 1C). When we treated oocytes with 1-MA, we could not detect uridine residues in long poly(A) sequences of cyclin B mRNAs in 62% (n = 55) of all reads (Fig. 1B, [+] 1-MA treatment; 1D). The number of clones with uridine residues in the poly(A) regions was significantly reduced following 1-MA treatment (P < 0.01, χ2 test). In some clones, the last several nucleotides immediately preceding the poly(A) tails (“AAAUGGAAAU,” Fig. 1B, [–] 1-MA treatment) were trimmed into shorter sequences after hormonal stimulation (Fig. 1B, [+] 1-MA treatment), suggesting that exonuclease digestion in the 3′–5′ direction may occur prior to polyadenylation. Some long poly(A) tails contained oligo(U) segments (Fig. 1B, [+] 1-MA treatment: 38% [n = 55] of all reads), suggesting that mRNAs with oligo(U) tails can serve as substrates for poly(A) polymerase, and that U tails are not removed as a prerequisite for polyadenylation. In addition, to detect trimming activity in the 3′ region of cyclin B mRNA, we performed 3′ adaptor (23-mer) ligation of mRNA obtained from oocytes before and after hormonal stimulation. We subjected the resultant ligated RNA to RT-PCR using sfcycB short Fwd primer (22-mer), which hybridizes from 21 to 43 nt upstream of the polyadenylation site and the adaptor primer, followed by high-resolution electrophoresis (Fig. 1E). In oocytes without hormonal stimulation, we detected PCR products in a range of sizes (75–87 bp) corresponding to the variation in the length of poly(A) cyclin B mRNAs with oligo(U) tails (Fig. 1B, [−] 1-MA treatment). Immediately after GVBD, both shorter and longer cyclin B mRNAs appeared, corresponding to trimmed and poly(A)-elongated cyclin B mRNAs, respectively. The population of trimmed cyclin B mRNAs increased at 30–40 min and decreased at 50 min, whereas the population of elongated cyclin B mRNAs increased gradually over time. These results suggest that 3′ trimming and polyadenylation occurred simultaneously after GVBD, and that 3′ trimmed cyclin B mRNAs were substrates for polyadenylation. Thus, the majority of cyclin B mRNAs are first trimmed at the 3′ end and subsequently polyadenylated (Fig. 1B,E). In addition, some long poly(A) tails contain oligo(U) segments (Fig. 1B [+] 1-MA), indicating that some mRNAs with oligo(U) tails can also serve as substrates for poly(A) polymerase. These results suggest that removal of U tails is not a strict prerequisite for polyadenylation.
Poly(A) elongation is controlled by 3′ UTR sequence of cyclin B mRNAs
Because synthesized RNAs containing AAUAAA and U-rich sequences are polyadenylated in Xenopus oocytes (Fox et al. 1989), we investigated whether the 3′ UTR of cyclin B mRNA containing such sequences (Supplemental Fig. S1) was sufficient for poly(A) elongation. Using T7 RNA polymerase and blunt-end DNA templates, we obtained synthesized RNA with unexpected complementary sequences (Supplemental Fig. S2A), whereas polyadenylation of the microinjected RNAs occurred after hormonal stimulation (Supplemental Fig. S2B,C). When we used a DNA template with 5′-protruding termini, we obtained RNAs with extra C and U residues (Supplemental Table S1; Supplemental Fig. S3A-No. 1; Milligan et al. 1987; Krupp 1988). To decrease the number of extra bases, we increased the ATP concentration in the transcription reaction (Supplemental Table S1; Fig. S3A-No. 4) and purified the RNA designated cycB-oligo(A) (Supplemental Fig. S4-[3]). To produce cycB-oligo(A) with an oligo(U) tail (Supplemental Fig. S4-[4]), we used poly(U) polymerase and a fluorescently labeled RNA (42 nt) as a marker substrate to determine the number of uridines by electrophoresis (Supplemental Fig. S3B,C). As expected, we could produce cycB-oligo(A) with one to four uridines (median, 2 nt), which we named cycB-oligo(A) + U (Supplemental Fig. S3D,E).
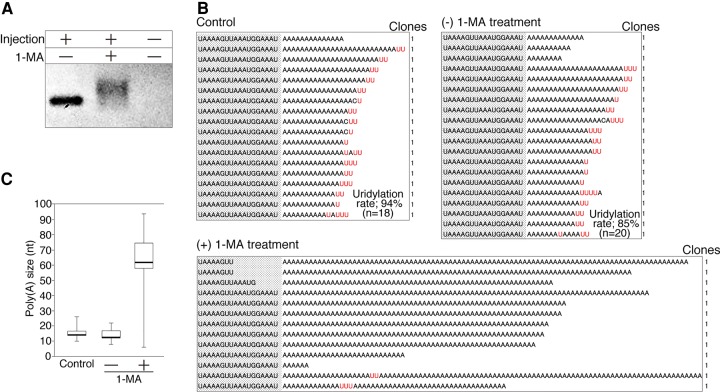
To determine whether the 3′ termini of synthetic cyclin B mRNAs would be polyadenylated, we microinjected cycB-oligo(A) + U into oocytes. After the oocytes were stimulated with or without 1-MA, we recovered total RNA and performed adaptor ligation. To amplify the injected RNA, we performed RT-PCR using primers against the exogenous sequence (Supplemental Fig. S4, exogenous sequence tag) and the adaptor sequence. We detected a single band from oocytes not subjected to hormonal stimulation and a shifted band from stimulated oocytes (Fig. 2A, left and middle lanes), but no band from oocytes that did not receive cycB-oligo(A) + U (Fig. 2A, right lane). Therefore, in vitro synthesized cycB-oligo(A) + U was polyadenylated like endogenous cyclin B mRNA after hormonal stimulation. Next, we cloned and sequenced these RT-PCR products in Figure 2A to check the sequences of the 3′ regions of injected cycB-oligo(A) + U with or without stimulation by 1-MA. In stimulated oocytes, the median length of the poly(A) tail increased to 62 nt, and most reads did not contain uridine residues in the long adenine repeat (Fig. 2B, [+] 1-MA; Fig. 2C, +1-MA). In contrast, uridine residues were stable in unstimulated oocytes, and the median length of oligo(A) was 13 nt (Fig. 2B, [−] 1-MA; Fig. 2C, −1-MA), almost identical to that of the uninjected control (Fig. 2B, Control; Fig. 2C, Control). These results indicate that endogenous polyadenylation of cyclin B mRNAs could be recapitulated using in vitro synthesized cycB-oligo(A) + U. On the basis of these findings, we conclude that the 3′ UTR alone is sufficient for polyadenylation of cyclin B mRNA during oocyte maturation.
FIGURE 2.
The 3′ UTR of cyclin B mRNAs is sufficient to recapitulate endogenous poly(A) elongation. (A) Band shift of cycB-oligo(A) + U in injected oocytes after hormonal stimulation (left and middle lanes), and the control RT-PCR product from oocytes not subjected to RNA injection (right lane). (B) Sequencing results of the 3′ terminal region of cycB-oligo(A) + U before injection (Control), after injection without hormonal stimulation ([−] 1-MA treatment), and after injection with hormonal stimulation ([+] 1-MA treatment). (C) Quantitation of poly(A) tail lengths. Layout and labeling are the same as in B.
mRNA stabilization and decay in oocytes
Several groups have proposed that uridylation at the 3′ ends of mRNAs induces mRNA decay (Rissland et al. 2007; Rissland and Norbury 2009; Sement et al. 2013; Chang et al. 2014; Lim et al. 2014). If the oligo(U) tail of cyclin B mRNAs in starfish oocytes in the absence of hormonal stimulation is also involved in mRNA degradation, cyclin B mRNA with oligo(U) should be turned over more rapidly than the mRNA without oligo(U). Alternatively, given that many maternal mRNAs in oocytes are stored stably in the cytoplasm (Barckmann and Simonelig 2013), uridylated cyclin B mRNA may be stable in oocytes. To determine which hypothesis is correct, it is necessary to measure the stability of uridylated cyclin B mRNA in oocytes in the absence of hormonal stimulation.
To determine whether the 3′ oligo(U) tail of cyclin B mRNA plays a role in mRNA decay, we microinjected uniformly radiolabeled cycB-oligo(A) with or without cold U-tail [cycB-oligo(A) + U or cycB-oligo(A)] into oocytes, and analyzed the amount of injected RNA remaining after 0 or 20 h of incubation. After 20 h, the levels of both injected cycB-oligo(A) + U and cycB-oligo(A) were reduced (Fig. 3A, upper panel), and the remaining amount of radioactivity did not differ significantly between the two RNAs (Fig. 3A, lower panel). To determine the stability of 3′ termini of uridylated and nonuridylated RNAs, we synthesized cold cycB-oligo(A) with radiolabeled terminal oligo(U) or oligo(A) using hot UTP or ATP, and then performed an experiment similar to the one shown in Figure 3A. Again, the remaining radioactivity did not differ significantly between 3′ uridine and adenine residues after 20 h incubation (Fig. 3B). These results confirm that 3′ uridine modification does not induce mRNA decay in oocytes prior to hormonal stimulation.
FIGURE 3.
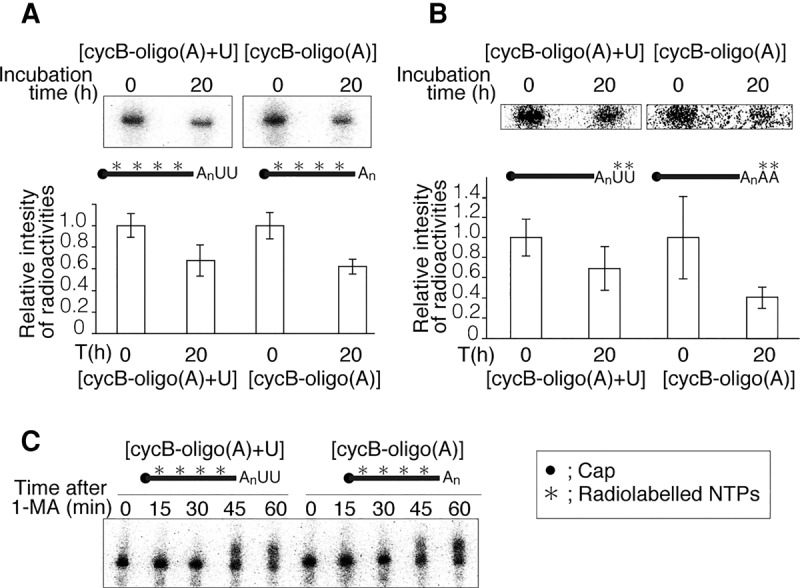
Uridylated and nonuridylated RNAs were similarly stable in starfish oocytes. Asterisks in the schematics denote locations of radiolabels. (A) Similar stability of uniformly labeled uridylated and nonuridylated RNAs in oocytes without hormonal stimulation. Upper panel shows representative autoradiography results. The intensity of each band was calculated using ImageQuant. Lower graphs show the remaining amounts of injected RNA after 20 h incubation. Error bars represent standard deviation from more than three experiments. (B) An experiment similar to the one shown in A was performed using 3′-end radiolabeled cycB-oligo(A) + U or cycB-oligo(A). Upper panel shows representative autoradiography results, and lower graphs show the remaining amounts of injected RNA after 20 h incubation. Error bars represent standard deviation (n = 3). (C) Similar band shifts of uniformly labeled uridylated and nonuridylated RNAs in oocytes after hormonal stimulation. Bands for both cycB-oligo(A) + U and cycB-oligo(A) were shifted up after 45–60 min.
Finally, to determine whether uridylated mRNA is degraded after hormonal stimulation, we stimulated oocytes injected with uniformly radiolabeled cycB-oligo(A) with or without cold oligo(U) tails with 1-MA, and then analyzed the injected RNAs by denaturing acrylamide gel electrophoresis and autoradiography (Fig. 3C). Similar band shifts were observed in both cycB-oligo(A) + U and cycB-oligo(A) (Fig. 3C), indicating that polyadenylation per se does not require the 3′ uridine modification, and the oligo(U) tail does not induce remarkable mRNA decay during meiotic progression following hormonal stimulation (Fig. 3C).
In this study, we found that cyclin B mRNAs with oligo(U) tails were stored in starfish oocytes. After hormonal stimulation, cyclin B mRNAs could be trimmed in the 3′–5′ direction, and poly(A) elongation occurred. To our knowledge, this is the first report showing that uridylated mRNAs are stored in oocytes and that they can be activated translationally.
In somatic cells, factors involved in degradation of uridylated mRNAs localize to P-bodies (Sheth and Parker 2003; Decker and Parker 2012; Malecki et al. 2013); in some cases, stored mRNAs within P-bodies can reenter polysomes for translation (Brengues et al. 2005; Anderson and Kedersha 2006; Bhattacharyya et al. 2006). If storage of mRNAs in P-bodies is regulated by uridylation, deuridylation should be required for reentry of the stored mRNAs into polysomes, suggesting that uridylation is involved in translational inactivation. Indeed, uridylated poly(A)+ reporter luciferase RNA is translationally inactive in Xenopus oocytes (Lapointe and Wickens 2013). Therefore, modification of the 3′ oligo(U) tail in starfish oocytes may translationally inactivate certain mRNAs prior to hormonal stimulation.
MATERIALS AND METHODS
Animals and oocytes
Starfish (Asterina pectinifera) were collected on the Pacific coast of Honshu, Japan, and kept in laboratory aquaria supplied with circulating seawater. Immature oocytes were treated with cold calcium-free seawater (SW) to remove follicle cells, and then incubated in normal SW containing 1 μM 1-MA to induce germinal vesicle breakdown (GVBD). Usually, GVBD was induced 20–30 min after 1-MA treatment. All experiments were performed at 20°C unless otherwise stated.
Microinjection of in vitro synthesized RNA into immature oocytes
For rapid microinjection of synthesized RNA, oocytes were held between two coverslips separated by a double layer of Scotch tape, and then released after injection by peeling off the tape (Chiba et al. 1992). Microinjections were performed using a constricting pipet filled with in vitro synthesized RNA (80 ng/µL); the injection volume was 10–30 pL per oocyte. Injected oocytes were incubated for the indicated periods, and 100 oocytes were used for each experiment.
Extraction of RNA from oocytes
Total RNA from oocytes was extracted using the TRI reagent (Molecular Research Center, Inc.). In some experiments, 1 µg yeast tRNA was used to coprecipitate RNA. In oocytes stimulated with 1-MA, total RNA was extracted 30 min after GVBD.
Electrophoresis
RNA was analyzed by a 10% polyacrylamide gel in TBE (89 mM Tris borate [pH 8.0], 89 mM boric acid, 2 mM EDTA) containing 7.5 M urea.
RT-PCR with 3′ adaptor ligation and tail sequence
Synthesis of cDNA with biotinylated 3′ adaptor ligation was performed using small RNA Cloning Kit (Takara Bio) as reported previously (Wada et al. 2012), followed by PCR using a gene-specific primer and a 3′ adaptor primer (primer sets are listed below). In some experiments, in order to increase the amount of DNA available for cloning, secondary nested PCR was performed using internal (nested) PCR primers (primer sets for the second amplification are listed below). PCR products were purified by agarose gel electrophoresis and extracted from the gel using the Wizard SV Gel and PCR Clean-Up System (Promega). Purified PCR products were cloned into a pCR2.1-TOPO vector (Invitrogen), and the insert sequences were determined.
The following primer sets were used to target endogenous cyclin B mRNAs: for the first amplification, sfcycB Fwd primer 1 (5′-TTGTGCTAGATATCCGAGACC-3′) and 3′ adaptor Rev primer 1 (5′-GTCTCTAGCCTGCAGGA-3′); for the second amplification, sfcycB Fwd primer 2 (5′-TCCCACACAACTTCAGATGAAC-3′) and PCR-R&RT primer (5′-GTCTCTAGCCTGCAGGATCGATG-3′). In Figure 1E, PCR was performed using sfcycB short Fwd primer (5′-GCTTTTGTCTGTCAGTCTTTGT-3′) and 3′ adaptor Rev primer 1.
The following primer sets were used to target in vitro synthesized RNAs encoding the 3′ region of cyclin B mRNA: for the first amplification, exo tag Fwd primer 1 (5′-CCCCTCGAGAAAGATCTGC-3′) and 3′adaptor Rev primer 1; for the second amplification, exo tag Fwd primer2 (5′-GAAAGATCTGCAGCGTGCGTCA-3′) and 3′adaptor Rev primer 2 (5′-AGCCTGCAGGATCGATG-3′).
DNA cloning and plasmids for in vitro RNA synthesis
To create pcDNAsfcycB3′UTR1, the 3′ UTR of starfish cyclin B cDNA was amplified by RT-PCR using a sfcycB Fwd primer 1 and a (dT)12-anchor primer (5′-GCGAGCTCCGCGGCCGCGTTTTTTTTTTTT-3′) (Wada et al. 2012), which was TA-cloned into vector pCR2.1-TOPO (Invitrogen).
Plasmids encoding the T7 promoter, exogenous sequence tag (5′-AAAGATCTGCAGCGTGCGTCA-3′, 21 nt), cyclin B 3′ UTR of starfish oocyte (Supplemental Fig. S1), short poly(A) tail, and a FokI site (see also Supplemental Fig. S4-[1]) were generated by the following two-step procedure: (i) A DNA fragment encoding the exogenous sequence tag and the portion of the 3′ UTR of cyclin B (nucleotides 1946–2179, Supplemental Fig. S1) was PCR amplified from pcDNAsfcycB3′UTR1 using a forward primer (5′-GCTCGAG AAAGATCTGCAGCGTGCGTCATTGTGCTAGATATCCGAGACC-3′; exogenous sequence tag underlined and XhoI site double underlined) and reverse primer (5′-CGAATTC GTATACAAAGACTGACAGACAAAAGC-3′; Bst1107I [SnaI] site underlined and EcoRI site double-underlined). The PCR product was cloned into the XhoI and EcoRI sites of pBluescript SK II (Stratagene), and the resultant plasmid was named pcDNAsfcycB3′UTR2. (ii) A synthetic double-stranded DNA (5′-CTGTCAGTCTTTGTAAATAAAAGTTAAATGGAAATAAAAAAAAAAAACGCATGTTCCATCCTACGAATTCCTGCAG-3′/5′-CTGCAGGAATTCGTAGGATGGAACATGCGTTTTTTTTTTTTATTTCCATTTAACTTTTATTTACAAAGACTGACAG) encoding the portion of the cyclin B 3′ UTR (nucleotides 2180–2199, Supplemental Fig. S1), a short poly(A) tail, and a FokI site were cloned into the SnaI site of pcDNAsfcycB3′UTR2 by the In-Fusion (Takara Bio) method, and the resultant plasmid was named pcDNAsfcycB3′UTRFokI.
In vitro transcription
A DNA template for in vitro transcription of cycB-oligo(A) was PCR amplified from pcDNAsfcycB3′UTRFokI using a forward primer (5′-GCGTAATACGACTCACTATAGG-3′; T7 promoter sequence underlined) and a universal M13 reverse primer (5′-CAGGAAACAGCTATGAC-3′). The PCR product was digested with FokI, and the objective fragment was purified by agarose gel electrophoresis using the Wizard SV Gel and PCR Clean-Up System, followed by phenol/chloroform extraction and ethanol precipitation, and the resultant DNA pellet was subsequently dissolved in sterile water. Using this DNA as a template, in vitro transcription was performed using the mMESSAGE mMACHINE T7 kit (Invitrogen) with the following modified NTP concentrations: 18.75 mM ATP, 3.75 mM CTP, 3.75 mM CTP, 0.75 mM GTP, and 3 mM cap analog unless otherwise indicated (see also text). Transcripts were purified by phenol/chloroform extraction and ethanol precipitation and were subsequently separated by electrophoresis on a 5% polyacrylamide gel in TBE (89 mM Tris [pH 8.0], 89 mM boric acid, 2 mM EDTA) containing 8 M urea. The gel was stained with SYBR Green I, and ∼318-nt RNA fragments were cut out, crushed, and eluted from gel slices in four volumes of TE buffer (10 mM Tris [pH 8.0] 0.1 mM EDTA) at 37°C for 4–5 h. Eluted RNA was recovered by phenol/chloroform extraction and ethanol precipitation, and the resultant RNA pellet was dissolved in sterile water.
In vitro uridylation/adenylation at the 3′ ends of RNAs
To produce cycB-oligo(A) + U RNAs, the uridylation reaction shown in Supplemental Figure S3C was carried out in reaction mixture A [40 unit/mL of poly(U) polymerase (NEB: M0337S), 1.6 unit/µL RNase inhibitor (Promega:N2611), 50 mM NaCl, 10 mM Tris–HCl (pH 7.9), 10 mM MgCl2, 1 mM DTT, 450 nM of a 42-nt synthetic RNA 5′-end labeled with Alexa Fluor 488 (5′-Alexa488UGUCAGUCUUUGUAAAUAAAAGUUAAAUGGAAAUAAAAAAAA-3′, marker substrate) and 50 nM cycB-oligo(A) RNA] with 1 mM UTP. When the uridylation reaction was performed without cycB-oligo(A) in Supplemental Figure S3B, 500 nM 42-nt synthetic RNA 5′-end labeled with Alexa Fluor 488 was used. The reaction mixture was incubated at 37°C for 60 sec or as indicated. The reaction was stopped by addition of EDTA (28 mM final concentration). Reaction products were purified by phenol/chloroform extraction and ethanol precipitation and separated on a 10% polyacrylamide gel in TBE containing 7.5 M urea. The gel was scanned on a Fuji-imager LAS 4000 mini with or without SYBR-Green I staining.
In vitro synthesis of radiolabeled RNA
To produce uniformly radiolabeled cycB-oligo(A) + U and cycB-oligo(A), hot cycB-oligo(A) was produced and purified by the in vitro transcription procedure with 111 KBq/µL [α-32P]UTP in T7 polymerase reaction mixture, and the 3′ end of hot cycB-oligo(A) was uridylated or adenylated in reaction mixture A with 1 mM cold UTP or 1 mM cold ATP, respectively. Reaction mixtures containing UTP or ATP were incubated at 37°C for 60 sec or 25°C for 90 sec, respectively. The reaction was stopped by addition of EDTA (28 mM final concentration). The resultant uniformly radiolabeled cycB-oligo(A) + U and cycB-oligo(A) RNAs were purified using the RNA Clean-Up and Concentration Kit (Norgen Biotek Corporation).
To radiolabel the 3′ end of cycB-oligo(A) RNA with [α-32P]UTP or [α-32P]ATP in Figure 3B, 11.1 KBq/µL [α-32P] UTP + 2.5 mM UTP or 11.1 KBq/µL [α-32P]ATP + 2.5 mM ATP were added to reaction mixture A, respectively. Reaction mixtures containing [α-32P]UTP or [α-32P] ATP were incubated at 37°C for 60 sec or at 25°C for 90 sec, respectively, followed by addition of EDTA (28 mM final concentration) to stop the reaction. 3′ radiolabeled cycB-oligo(A) + U and cycB-oligo(A) were purified using the RNA Clean-Up and Concentration Kit (Norgen Biotek Corporation).
RNA stability assay
Radiolabeled cycB-oligo(A) + U or cyclinB-oligo(A) was injected into oocytes without hormonal stimulation, and total RNA was extracted before or after 20 h incubation (Fig. 3A,B). In addition, injected oocytes were treated with 1−MA and extracted every 15 min after hormonal stimulation (Fig. 3C). Purified RNAs were separated by electrophoresis on a 10% polyacrylamide gel in TBE containing 8 M urea, followed by autoradiography (Typhoon FLA 7000, GE Healthcare). The bands of labeled RNAs incubated in oocytes were quantitated using the Image Quant TL software (GE Healthcare).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by JSPS KAKENHI grant numbers 25440102 and 25.10787. H.O. is a research fellow of the Japan Society for the Promotion of Science.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.054882.115.
REFERENCES
- Anderson P, Kedersha N. 2006. RNA granules. J Cell Biol 172: 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barckmann B, Simonelig M. 2013. Control of maternal mRNA stability in germ cells and early embryos. Biochim Biophys Acta 1829: 714–724. [DOI] [PubMed] [Google Scholar]
- Benoit P, Papin C, Kwak JE, Wickens M, Simonelig M. 2008. PAP- and GLD-2-type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila. Development 135: 1969–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125: 1111–1124. [DOI] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310: 486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HM, Triboulet R, Thornton JE, Gregory RI. 2013. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28–let-7 pathway. Nature 497: 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Lim J, Ha M, Kim VN. 2014. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol Cell 53: 1044–1052. [DOI] [PubMed] [Google Scholar]
- Chiba K, Tadenuma J, Matsumoto M, Takahashi K, Katada T, Hoshi M. 1992. The primary structure of the α subunit of a starfish guanosine-nucleotide-binding regulatory protein involved in 1-methyladenine-induced oocyte maturation. Eur J Biochem 207: 833–838. [DOI] [PubMed] [Google Scholar]
- Chiba K, Kontani K, Tadenuma H, Katada T, Hoshi M. 1993. Induction of starfish oocyte maturation by the βγ-subunit of starfish G protein and possible existence of the subsequent effector in cytoplasm. Mol Biol Cell 4: 1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland PR, Wormington M. 2001. The mechanism and regulation of deadenylation: identification and characterization of Xenopus PARN. RNA 7: 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R. 2012. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 4: a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox CA, Wickens M. 1990. Poly(A) removal during oocyte maturation: a default reaction selectively prevented by specific sequences in the 3′ UTR of certain maternal mRNAs. Genes Dev 4: 2287–2298. [DOI] [PubMed] [Google Scholar]
- Fox CA, Sheets MD, Wickens MP. 1989. Poly(A) addition during maturation of frog oocytes: distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes Dev 3: 2151–2162. [DOI] [PubMed] [Google Scholar]
- Hake LE, Richter JD. 1994. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell 79: 617–627. [DOI] [PubMed] [Google Scholar]
- Hara M, Mori M, Wada T, Tachibana K, Kishimoto T. 2009. Start of the embryonic cell cycle is dually locked in unfertilized starfish eggs. Development 136: 1687–1696. [DOI] [PubMed] [Google Scholar]
- Hiraoka D, Hori-Oshima S, Fukuhara T, Tachibana K, Okumura E, Kishimoto T. 2004. PDK1 is required for the hormonal signaling pathway leading to meiotic resumption in starfish oocytes. Dev Biol 276: 330–336. [DOI] [PubMed] [Google Scholar]
- Hoefig KP, Rath N, Heinz GA, Wolf C, Dameris J, Schepers A, Kremmer E, Ansel KM, Heissmeyer V. 2013. Eri1 degrades the stem-loop of oligouridylated histone mRNAs to induce replication-dependent decay. Nat Struct Mol Biol 20: 73–81. [DOI] [PubMed] [Google Scholar]
- Jaffe LA, Gallo CJ, Lee RH, Ho YK, Jones TL. 1993. Oocyte maturation in starfish is mediated by the βγ-subunit complex of a G-protein. J Cell Biol 121: 775–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatani H, Shirai H, Nakanishi K, Kurokawa T. 1969. Isolation and identification of meiosis inducing substance in starfish Asterias amurensis. Nature 221: 273–274. [DOI] [PubMed] [Google Scholar]
- Kim JH, Richter JD. 2006. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol Cell 24: 173–183. [DOI] [PubMed] [Google Scholar]
- Kishimoto T. 2015. Entry into mitosis: a solution to the decades-long enigma of MPF. Chromosoma 124: 417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp G. 1988. RNA synthesis: strategies for the use of bacteriophage RNA polymerases. Gene 72: 75–89. [DOI] [PubMed] [Google Scholar]
- Kwak JE, Wang L, Ballantyne S, Kimble J, Wickens M. 2004. Mammalian GLD-2 homologs are poly(A) polymerases. Proc Natl Acad Sci 101: 4407–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe CP, Wickens M. 2013. The nucleic acid-binding domain and translational repression activity of a Xenopus terminal uridylyltransferase. J Biol Chem 288: 20723–20733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Kim B, Kim VN. 2014. Emerging roles of RNA modification: m6A and U-tail. Cell 158: 980–987. [DOI] [PubMed] [Google Scholar]
- Lim J, Ha M, Chang H, Kwon SC, Simanshu DK, Patel DJ, Kim VN. 2014. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 159: 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecki M, Viegas SC, Carneiro T, Golik P, Dressaire C, Ferreira MG, Arraiano CM. 2013. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J 32: 1842–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Murthy KG, Ryan K, Manley JL, Richter JD. 2000. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol Cell 6: 1253–1259. [DOI] [PubMed] [Google Scholar]
- Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res 15: 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N, Walker J, Dale M, Standart N. 1999. Dual roles of p82, the clam CPEB homolog, in cytoplasmic polyadenylation and translational masking. RNA 5: 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF. 2008. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev 22: 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norbury CJ. 2013. Cytoplasmic RNA: a case of the tail wagging the dog. Nat Rev Mol Cell Biol 14: 643–653. [DOI] [PubMed] [Google Scholar]
- Okumura E, Sekiai T, Hisanaga SI, Tachibana K, Kishimoto T. 1996. Initial triggering of M-phase in starfish oocytes: a possible novel component of maturation-promoting factor besides cdc2 kinase. J Cell Biol 132: 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura E, Fukuhara T, Yoshida H, Hanada S, Kozutsumi R, Mori M, Tachibana K, Kishimoto T. 2002. Akt inhibits Myt1 in the signalling pathway that leads to meiotic G2/M-phase transition. Nat Cell Biol 4: 111–116. [DOI] [PubMed] [Google Scholar]
- Paris J, Swenson K, Piwnica-Worms H, Richter JD. 1991. Maturation-specific polyadenylation: in vitro activation by p34cdc2 and phosphorylation of a 58-kD CPE-binding protein. Genes Dev 5: 1697–1708. [DOI] [PubMed] [Google Scholar]
- Radford HE, Meijer HA, de Moor CH. 2008. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim Biophys Acta 1779: 217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD. 2007. CPEB: a life in translation. Trends Biochem Sci 32: 279–285. [DOI] [PubMed] [Google Scholar]
- Rissland OS, Norbury CJ. 2009. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol 16: 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland OS, Mikulasova A, Norbury CJ. 2007. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol Cell Biol 27: 3612–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallés FJ, Strickland S. 1995. Rapid and sensitive analysis of mRNA polyadenylation states by PCR. PCR Methods Appl 4: 317–321. [DOI] [PubMed] [Google Scholar]
- Sement FM, Ferrier E, Zuber H, Merret R, Alioua M, Deragon JM, Bousquet-Antonelli C, Lange H, Gagliardi D. 2013. Uridylation prevents 3′ trimming of oligoadenylated mRNAs. Nucleic Acids Res 41: 7115–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets MD, Fox CA, Hunt T, Vande Woude G, Wickens M. 1994. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev 8: 926–938. [DOI] [PubMed] [Google Scholar]
- Sheth U, Parker R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slevin MK, Meaux S, Welch JD, Bigler R, Miliani de Marval P, Su W, Rhoads RE, Prins JF, Marzluff WF. 2014. Deep sequencing shows multiple oligouridylations are required for 3′ to 5′ degradation of histone mRNAs on polyribosomes. Mol Cell 53: 1020–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MG, Kiledjian M. 2007. 3′ Terminal oligo U-tract-mediated stimulation of decapping. RNA 13: 2356–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadenuma H, Chiba K, Takahashi K, Hoshi M, Katada T. 1991. Purification and characterization of a GTP-binding protein serving as pertussis toxin substrate in starfish oocytes. Arch Biochem Biophys 290: 411–417. [DOI] [PubMed] [Google Scholar]
- Tadenuma H, Takahashi K, Chiba K, Hoshi M, Katada T. 1992. Properties of 1-methyladenine receptors in starfish oocyte membranes: involvement of pertussis toxin-sensitive GTP-binding protein in receptor mediated signal transduction. Biochem Biophys Res Commun 186: 114–121. [DOI] [PubMed] [Google Scholar]
- Tay J, Hodgman R, Richter JD. 2000. The control of cyclin B1 mRNA translation during mouse oocyte maturation. Dev Biol 221: 1–9. [DOI] [PubMed] [Google Scholar]
- Ustianenko D, Hrossova D, Potesil D, Chalupnikova K, Hrazdilova K, Pachernik J, Cetkovska K, Uldrijan S, Zdrahal Z, Vanacova S. 2013. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA 19: 1632–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Hara M, Taneda T, Qingfu C, Takata R, Moro K, Takeda K, Kishimoto T, Handa H. 2012. Antisense morpholino targeting just upstream from a poly(A) tail junction of maternal mRNA removes the tail and inhibits translation. Nucleic Acids Res 40: e173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, Wilusz J. 2008. New ways to meet your (3′) end oligouridylation as a step on the path to destruction. Genes Dev 22: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K, Kotani T, Ota R, Yamashita M. 2010. Transgenic zebrafish reveals novel mechanisms of translational control of cyclin B1 mRNA in oocytes. Dev Biol 348: 76–86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.