Abstract
MPMV has great potential for development as a vector for gene therapy. In this respect, precisely defining the sequences and structural motifs that are important for dimerization and packaging of its genomic RNA (gRNA) are of utmost importance. A distinguishing feature of the MPMV gRNA packaging signal is two phylogenetically conserved long-range interactions (LRIs) between U5 and gag complementary sequences, LRI-I and LRI-II. To test their biological significance in the MPMV life cycle, we introduced mutations into these structural motifs and tested their effects on MPMV gRNA packaging and propagation. Furthermore, we probed the structure of key mutants using SHAPE (selective 2′hydroxyl acylation analyzed by primer extension). Disrupting base-pairing of the LRIs affected gRNA packaging and propagation, demonstrating their significance to the MPMV life cycle. A double mutant restoring a heterologous LRI-I was fully functional, whereas a similar LRI-II mutant failed to restore gRNA packaging and propagation. These results demonstrate that while LRI-I acts at the structural level, maintaining base-pairing is not sufficient for LRI-II function. In addition, in vitro RNA dimerization assays indicated that the loss of RNA packaging in LRI mutants could not be attributed to the defects in dimerization. Our findings suggest that U5-gag LRIs play an important architectural role in maintaining the structure of the 5′ region of the MPMV gRNA, expanding the crucial role of LRIs to the nonlentiviral group of retroviruses.
Keywords: retroviruses, Mason-Pfizer monkey virus (MPMV), RNA secondary structure, RNA packaging and dimerization, long-range interactions (LRI), SHAPE (selective 2′hydroxyl acylation analyzed by primer extension)
INTRODUCTION
One of the hallmarks of the retroviral life cycle is the efficient and specific packaging of the genomic RNA (gRNA) as dimers by the assembling virus particles (for review, see Paillart et al. 2004a). During this process, full-length, unspliced gRNA is preferentially packaged, whereas spliced viral and cellular RNAs are generally excluded from the nascent virus particles. The packaging specificity results from high-affinity interactions between the retroviral viral nucleocapsid domain (NC) within the Gag precursor and a specific sequence at the 5′ end of the viral genome called the packaging signal (psi, Ψ; for review, see D'Souza and Summers 2005; Lever 2007; Johnson and Telesnitsky 2010). Determinants of retroviral gRNA dimerization and packaging map to the same 100 to 400 nucleotides (nt) at the 5′ end of the gRNA; therefore, it is not surprising that packaging signals are located within the dimer linkage structure (DLS) that is formed when two gRNAs dimerize. A number of studies undertaken to identify the packaging determinants of retroviruses have shown that packaging sequences are generally present downstream from the major splice donor (mSD) (for review, see D'Souza and Summers 2005; Lever 2007; Johnson and Telesnitsky 2010). Therefore, a simplistic model would suggest that the full-length unspliced genomic RNA is preferentially packaged by virtue of the presence of the packaging determinant following interaction with the NC binding site (for review, see D'Souza and Summers 2005; Lever 2007; Johnson and Telesnitsky 2010). On the other hand, the packaging sequences are eliminated during splicing, thereby excluding the spliced RNAs from encapsidation into the nascent viral particles. However, in the case of HIV-1, the main determinant of Gag binding, stem–loop 1 (SL1) is located upstream of the major splice site (Abd El-Wahab et al. 2014; Smyth et al. 2015) and is thus present in both the spliced and unspliced viral mRNA, revealing a more complex mechanism of gRNA selection in which the gRNA structure plays a key role (Abd El-Wahab et al. 2014; Smyth et al. 2015). A similar situation also prevails in HIV-2 as both the spliced and unspliced viral mRNAs contain the packaging determinants, yet only the unspliced messages are encapsidated into the virus particle (for review, see Balvay et al. 2007).
Despite the fact that RNA packaging is a universal step in all retroviruses and the packaging signals are found at the 5′ end of the retroviral gRNAs, no sequence conservation has been found between the packaging signals of different retroviruses. Moreover, it is becoming evident from cross-packaging studies that switching the packaging sequences between two different retroviruses that have no primary sequence homology can maintain efficient packaging (Rizvi and Panganiban 1993; Yin and Hu 1997; White et al. 1999; Parveen et al. 2004; Moore et al. 2007; Al Dhaheri et al. 2009; Al Shamsi et al. 2011). The process of gRNA packaging thus seems to involve recognition of the structure rather than the sequence of the packaging signals. Consistent with these observations, packaging sequences of almost all retroviruses assume higher-order structures comprising several structural motifs (for review, see D'Souza and Summers 2005; Lever 2007; Johnson and Telesnitsky 2010). However, the significance of these structural motifs to gRNA packaging remains largely unclear. One such characteristic feature of retroviral gRNA packaging signals is the long-range interaction (LRI) involving sequences in R/U5 that are complementary to a sequence found downstream in the gag gene (Paillart et al. 2002, 2004b; Abbink and Berkhout 2003; Kenyon et al. 2008; Song et al. 2008; Jaballah et al. 2010; Aktar et al. 2013, 2014).
Genomic RNA packaging and dimerization among retroviruses are being intensively investigated, in part because of their use in human gene therapy. Among retroviruses, HIV-1-based vectors are being exploited currently; however, their use in humans raises critical safety concerns. Vectors from phylogenetically distant, nonhuman retroviruses such as Mason-Pfizer monkey virus (MPMV) may circumvent such safety concerns. The use of MPMV-based vectors are also being considered ideal since MPMV promoter is transcriptionally active in human cells and expression of the therapeutic genes of interest may require nuclear export signals like the MPMV constitutive transport element (CTE) (Bray et al. 1994) for their efficient expression. These unique features have led to increasing interest in developing MPMV vectors for human gene therapy. However, before MPMV-based vectors can be used in humans, it is crucial that the molecular mechanisms of pertinent aspects of their life cycle be fully understood. Due to these reasons, among β retroviruses, MPMV is the most extensively studied in terms of its gRNA packaging and dimerization.
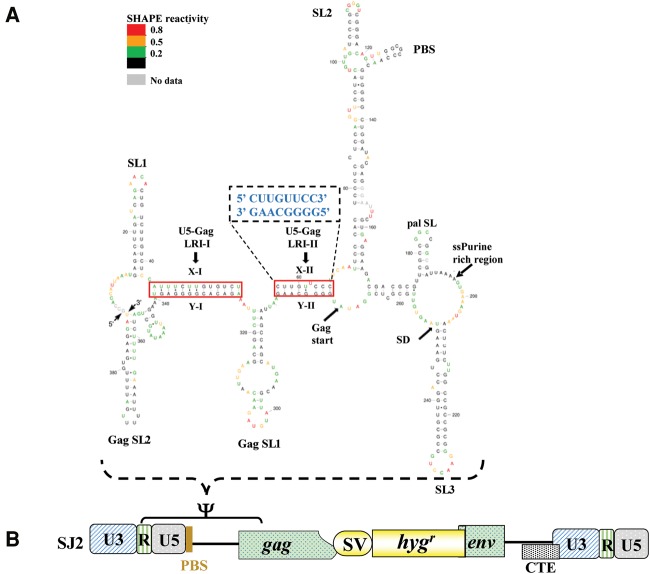
Initial RNA packaging studies have identified a stretch of nucleotides at the 5′ end of the MPMV genome important for incorporation of its RNA into the nascent virus particles (Vile et al. 1992; Harrison et al. 1995; Guesdon et al. 2001). Later, a more systematic mutational analysis of the 5′ end of the viral genome revealed that the packaging determinants of MPMV are multipartite and reside in two distinct regions: region “A” which includes the first 50 nts of the 5′ untranslated region (UTR); and region “B” that encompasses the last 23 nt of the 5′ UTR as well as the first 120 nt of gag (Schmidt et al. 2003; Jaballah et al. 2010). RNA secondary-structure predictions of this region suggested that it assumes several stable stem–loop structures (Jaballah et al. 2010). The predicted structure was later validated by using selective 2′ hydroxyl acylation by primer extension (SHAPE) (Fig. 1; Aktar et al. 2013).
FIGURE 1.
SHAPE-validated structure of MPMV packaging signal RNA. (A) RNA structure of MPMV packaging signal after applying SHAPE constraints. Nucleotides are color-coded as per the SHAPE reactivity key. Arrows point to important structural elements of the virus genome. The solid boxed regions show the two long-range interactions (LRIs), LRI-I and LRI-II, observed in the structure. The SHAPE-validated structure corroborates well with the predicted structure except for subtle differences in LRI-II (the predicted LRI-II structure shown in the enlarged dashed box). A uridine residue (U62) is unpaired and forms a bulge in the SHAPE-validated, but not in the predicted LRI-II structure, while C65 is base-paired in the SHAPE-validated structure, but unpaired in the predicted structure (figure adapted with permission from Aktar et al. 2013). (B) Schematic representation of MPMV subgenomic transfer vector, SJ2 (Jaballah et al. 2010), in which region between R and 120 nt of Gag that has been shown to be important for MPMV RNA packaging and dimerization is demarcated. The same region was used to predict and validate the RNA secondary structure.
The SHAPE-validated secondary structure of MPMV gRNA packaging signal revealed the presence of two LRIs (LRI-I and LRI-II) (Fig. 1) between U5 and gag complementary sequences in the region that has been shown to be important for MPMV gRNA packaging (Schmidt et al. 2003; Jaballah et al. 2010). These LRIs are phylogenetically conserved among different strains of MPMV both at the sequence and secondary structural levels (Aktar et al. 2013), suggesting that these two LRIs could play a role in gRNA packaging, potentially by maintaining the overall secondary structure of the gRNA packaging signal. Therefore, to provide functional and structural evidence for the existence of U5-gag LRIs and establish their biological significance during the MPMV life cycle, we introduced a series of mutations and tested their effects on MPMV gRNA packaging and propagation using a biologically relevant trans-complementation assay (Supplemental Fig. 1), while the structure of selected key mutants was analyzed by SHAPE.
RESULTS
Experimental approach
The sequences involved in formation of the U5-gag LRIs fall in a region that has been reported to be important for both MPMV RNA packaging and dimerization and have been shown to extend into gag (Schmidt et al. 2003; Jaballah et al. 2010; Aktar et al. 2013). Therefore, it is difficult to test the effects of mutations in U5-gag LRIs in the full-length genomic context as mutations introduced in this region might affect Gag/Pol protein synthesis. To overcome such a caveat, we used the three-plasmid trans-complementation assay developed in our laboratory for MPMV (Browning et al. 2001). It provides the necessary biological components to generate virus particles containing the packaged RNA, the replication of which is limited to a single round because reinfection of the target cells cannot take place (Supplemental Fig. 1; Browning et al. 2001). The Gag and Gag/Pol proteins were provided in trans from a separate expression plasmid. Briefly, the wild-type (SJ2) or mutant transfer vectors, the Gag/Pol packaging construct (TR301), and the envelope expressing plasmid (MD.G), along with a firefly luciferase expression plasmid (pGL3C), were cotransfected into the producer 293T cells. The resulting virus particles were used to isolate and quantify the packaged RNA and to monitor its propagation in the infected HeLa T4 cells following their transduction with the marker hygromycin-resistant gene. The number of resulting hygromycin-resistant colonies should be directly proportional to the packaged RNA content, providing an indirect estimate of RNA packaging efficiency (Supplemental Fig. 1).
A custom-made qPCR assay was developed for quantitation of wild-type or mutant MPMV RNA both in the cytoplasm as well as in the virions (see Materials and Methods) that employed a probe along with primers that bound within the U5/PBS region of MPMV (nucleotides 702–770). This region is common to the wild-type and mutant transfer vector RNAs and away from the site of the mutations, ensuring 100% complementary binding efficiency of the primers and probe to the target sites. A human β-actin assay was used as an endogenous control, as described previously (Mustafa et al. 2012; Aktar et al. 2014).
Possible role of the wobble guanine–uracil (G•U) base pairs in LRI-I and LRI-II during MPMV RNA packaging and propagation
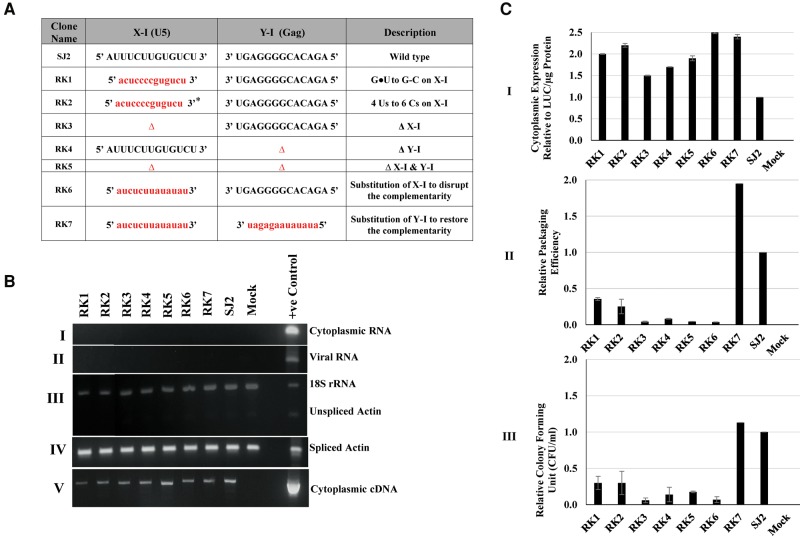
Sequence analysis of the U5-gag LRI-I revealed that out of the 13 pairs of complementary nucleotides, four are wobble guanine–uracil (G•U) base pairs (Fig. 1). Since the LRI sequences play a role in anchoring the RNA structure (Jaballah et al. 2010; Aktar et al. 2013), we reasoned that further strengthening the base-pairing between the U5 and gag sequences of the LRI-I could stabilize its structure, resulting in enhanced RNA packaging. Thus, mutant RK1was created in which the four G•U base pairs of LRI-I were substituted by more stable G = C base pairs (Fig. 2A). Similarly, mutant RK2 was created in which all the six G•U base pairs in both LRIs were converted into G = C base pairs (Fig. 2A).
FIGURE 2.
Effect of substitution/deletion mutations in U5-gag LRIs on MPMV gRNA packaging and propagation. (A) Table outlining the substitution/deletion mutations introduced into the U5-gag sequences of LRI-I and LRI-II. Sequences of the mutations introduced are represented in lower case and in red. In RK2, (*) denotes that only part of the U5 sequence for both LRIs is shown due to space limitations. (B) Representative gel images of the controls needed for validating different aspects of the three-plasmid in vivo packaging and propagation assay: (I and II) PCR amplification of DNase-treated RNA from the cytoplasmic and viral RNA preparations, respectively, with MPMV-specific vector primers; (III) multiplex amplification of unspliced β-actin mRNA and 18S rRNA; (IV) PCR amplification of spliced β-actin mRNA to check for the nucleocytoplasmic fractionation technique; and (V) PCR amplification of transfer vector cytoplasmic cDNAs using MPMV vector-specific primers. (C) Bar graphs represent: (I) the relative cytoplasmic expression of transfer vector RNAs in 293T cells relative to the wild-type (SJ2 vector) after normalization with the β-actin endogenous control and luciferase expression; (II) packaging efficiency of the mutant transfer vector RNAs relative to SJ2 after normalization with β-actin and luciferase expression; and (III) the relative propagation of MMTV transfer vector RNAs as measured by the luciferase-normalized hygromycin-resistant colony-forming units (CFU)/mL for mutant transfer vectors compared to the wild-type SJ2 vector. The data represented in the histograms correspond to the mean of samples tested in triplicates (±SD) following transfection and infection experiments.
The three-plasmid trans-complementation assay was used to test the effects of the introduced mutations on vector RNA packaging and propagation. Following transfection, cells were fractionated into cytoplasmic and nuclear fractions to assess vector RNA expression and nuclear export. RNA isolated from both the cytoplasmic fractions and pelleted viral particles was DNase-treated and PCR-amplified using MPMV vector-specific primers. The absence of amplification product confirmed the absence of DNA contamination in these RNA preparations (Fig. 2B.I and II). The integrity of the nucleocytoplasmic fractionation was confirmed by demonstrating the absence of unspliced β-actin mRNA in the cytoplasmic fractions (Fig. 2B.III) and the presence of spliced β-actin mRNA in these fractions (Fig. 2B.IV). The unspliced β-actin amplifications were performed as a multiplex PCR in the presence of primers/competimer for 18S ribosomal RNAs and successful amplification of 18S ribosomal RNAs across all the samples validated the presence of amplifiable cDNAs (Fig. 2B.III). Finally, cDNAs prepared from cytoplasmic RNA fractions were amplified using MPMV transfer vector-specific primers to ensure that the transfer vector RNAs were efficiently expressed and properly transported from the nucleus to the cytoplasm (Fig. 2B.V).
Next, we analyzed the packaging efficiencies of RK1 and RK2 mutant RNAs in virus particles using the custom-designed qPCR real-time assay, as described in Materials and Methods. The viral RNA packaging results obtained were further normalized to their cytoplasmic expression (Fig. 2C.I), the transfection efficiency (data not shown), and the packaging of the wild-type (SJ2) RNA to determine the relative packaging efficiency (RPE) of each mutant RNA (Fig. 2C.II). In contrast to our expectations, the RPE of the RK1 and RK2 mutants designed to stabilize the LRIs was actually reduced (2.8-fold and fourfold, respectively; P < 0.01; Fig. 2C.II). In agreement with the RNA packaging data, these mutants were also defective for RNA propagation (Fig. 2C.III). These effects on RNA packaging and propagation were observed despite the fact that the transfection efficiencies of the RK1 and RK2 mutants and wild-type (SJ2) in multiple experiments were within twofold of each other (data not shown), while real-time qPCR analysis of mutant transfer vector RNAs showed similar levels of expression in the cytoplasm to the wild-type, SJ2 (Fig. 2C.I).
Role of the LRI-I structure in MPMV RNA packaging and propagation
To investigate the complementary nature of the U5-gag LRI-I sequences, we deleted either the U5 (X-I; RK3) or gag (Y-I; RK4), or both U5 and gag sequences (RK5) while maintaining LRI-II (Fig. 2A). The dual mutation in RK5 was introduced to determine whether MPMV could package its RNA in the absence of LRI-I while maintaining only LRI-II.
These mutant transfer vectors (RK3-RK5) were tested in the in vivo packaging and propagation assays. Figure 2B,C.I details the necessary controls (DNase treatment of RNA, nucleocytoplasmic fractionation control, presence of amplifiable cDNA, and successful expression and cytoplasmic transport of vector RNAs) followed by quantitation of the packaging potential of RNAs into nascent virions. These analyses demonstrated that deletion of either one (U5-X-I in RK3 or gag-Y-I in RK4) or both complementary sequences (RK5) involved in LRI-I almost abolished RNA packaging (>10-fold compared to the wild-type; Fig. 2C.II). Consistent with RPEs, the propagation efficiencies of these mutant RNAs were also dramatically reduced (Fig. 2C.III).
To establish whether base-pairing between the complementary U5 and gag sequences forming LRI-I is sufficient to promote gRNA packaging or whether the native viral sequences (U5 and gag) are required, two additional mutants were generated. We substituted either the U5 (X-I) sequence to disrupt LRI-I base-pairing (Fig. 2A, mutant RK6), or both the U5 (X-I) and gag (Y-I) sequences to restore it (Fig. 2A, mutant RK7). Consistent with the deletion mutants RK3-RK5, disruption of the LRI-I complementarity in RK6 [due to the substitution of U5 (X-1) sequence with a heterologous one] almost abolished RNA packaging (>20-fold reduction; P < 0.01; Fig. 2C.II) and dramatically compromised RNA propagation (25-fold reduction; P < 0.01; Fig. 2C.III). Restoration of LRI-I with heterologous sequences in RK7 not only restored RNA packaging, but in fact improved it (Fig. 2C.II), and also restored RNA propagation to wild-type levels (Fig. 2C.III).
Altogether, data obtained from the test of these mutations in biological assays demonstrate that complementarity between the U5 and gag sequences forming LRI-I is crucial for MPMV RNA packaging and propagation. These results also show that the mere presence of LRI-II is not sufficient to encapsidate MPMV RNA into the budding virus particles.
Role of LRI-II in MPMV RNA packaging and propagation
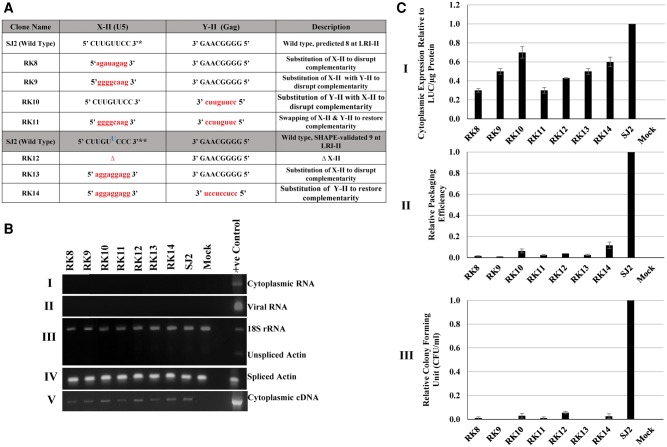
A similar mutational approach was used toward investigating the role of U5-gag complementary sequences forming LRI-II (Fig. 3A). LRI-II was disrupted in mutants RK8, RK9, RK10, RK12, and RK13 as a result of deletion of U5 (X-II: mutant RK12) or various substitutions of either U5 (X-II: RK8, RK9, RK13) or gag (Y-II: RK10). After performing all the necessary controls (Fig. 3B,C.I), these substitution and deletion mutants (RK8-RK10, RK12, and RK13) were tested in the in vivo packaging and propagation assays. Similar to our observations with regard to LRI-I, RNA packaging and propagation of these mutants were nearly ablated (Fig. 3C.II,C.III).
FIGURE 3.
Effect of deletion/substitution mutations introduced into U5-gag LRI-II on MPMV gRNA packaging and propagation. (A) Table outlining the deletion/substitution mutations introduced into the U5-gag sequences of LRI-II. Sequences of the introduced mutations are represented in lower case and in red. Two sets of mutants are outlined in the table. The first set of mutations were introduced into the Mfold-predicted 8-nt LRI-II structure, while the second set describes mutations introduced into the SHAPE-validated 9-nt LRI-II structure. (*) Denotes the 8-nt U5 sequence of the predicted LRI-II. (**) Denotes the 9-nt U5 sequence of the SHAPE-validated LRI-II. (B) Representative gel images of the controls needed for validating different aspects of the three-plasmid in vivo packaging and propagation assay: (I and II) PCR amplification of DNase-treated RNA from the cytoplasmic and viral RNA preparations, respectively, with MPMV-specific vector primers; (III) multiplex amplification of unspliced β-actin mRNA and 18S rRNA; (IV) PCR amplification of spliced β-actin mRNA to check for the nucleocytoplasmic fractionation technique; and (V) PCR amplification of transfer vector cytoplasmic cDNAs using MPMV vector-specific primers. (C) Bar graphs represent: (I) the relative cytoplasmic expression of transfer vector RNAs in 293T cells relative to the wild-type (SJ2 vector) after normalization with the β-actin endogenous control and luciferase expression; (II) packaging efficiency of the mutant transfer vector RNAs relative to SJ2 after normalization with β-actin and luciferase expression; and (III) the relative propagation of MMTV transfer vector RNAs as measured by the luciferase-normalized hygromycin resistant colony-forming units (CFU)/mL for mutant transfer vectors compared to the wild-type SJ2 vector. The data represented in the histograms correspond to the mean of samples tested in triplicates (±SD) following transfection and infection experiments.
We next attempted to design mutants that would restore a heterologous wild-type-like LRI-II structure by simultaneously substituting the U5 (X-II) and gag (Y-II) sequences with each other to restore complementarity using native sequences in reverse. However, the U5 and gag sequences forming LRI-II differed somewhat between the predicted and SHAPE-validated secondary structure models of MPMV packaging signal RNA (Aktar et al. 2013). Specifically, the uridine (U) residue at position 62 (U62) forms a G•U wobble base pair in the predicted structure, whereas it forms a bulge in the SHAPE-validated structure (Fig. 1; Jaballah et al. 2010; Aktar et al. 2013). In addition, cytosine at position 65 (C65) that was not originally predicted to be part of the U5 (X-II) complementary sequences was included in the SHAPE-validated LRI-II structure. As a result, the U5 (X-II) sequence became 9 nt long instead of 8 nt as predicted earlier (Fig. 1; Jaballah et al. 2010; Aktar et al. 2013). Thus, we designed two complementary mutants, RK11, which should restore the predicted LRI-II structure, and RK14, which should restore the SHAPE-validated LRI-II structure (Fig. 3A).
Quite surprisingly, packaging of both RK11 and RK14 RNAs was severely affected (Fig. 3C.II). In line with these results, propagation of RK11 and RK14 RNAs was also dramatically reduced (Fig. 3C.III). These results suggest that either the wild-type complementary sequences of U5 (X-II) and gag (Y-II) are required for LRI-II function, or that the LRI-II structure was not restored in these mutants, contrary to our expectations. It was therefore important to experimentally test the structure of these mutants.
Structural analyses of the mutant transfer vector RNAs
In an attempt to perform a structure/function relationship of the MPMV gRNA packaging signal, all mutants were analyzed using the Mfold software (Mathews et al. 1999; Zuker 2003) to predict their thermodynamically most stable structures. While mutants predicted to destabilize the LRI structures are likely to do so, it is more difficult to correctly predict the structures of mutants designed to stabilize or to restore the LRI-I/II structures, as sequence alterations offer alternative base-pairing possibilities. Therefore, we analyzed the structures of key LRI-I and II mutant transfer vector RNAs using SHAPE.
Structure–function relationship of the stabilizing mutations in U5-gag LRI-I and LRI-II
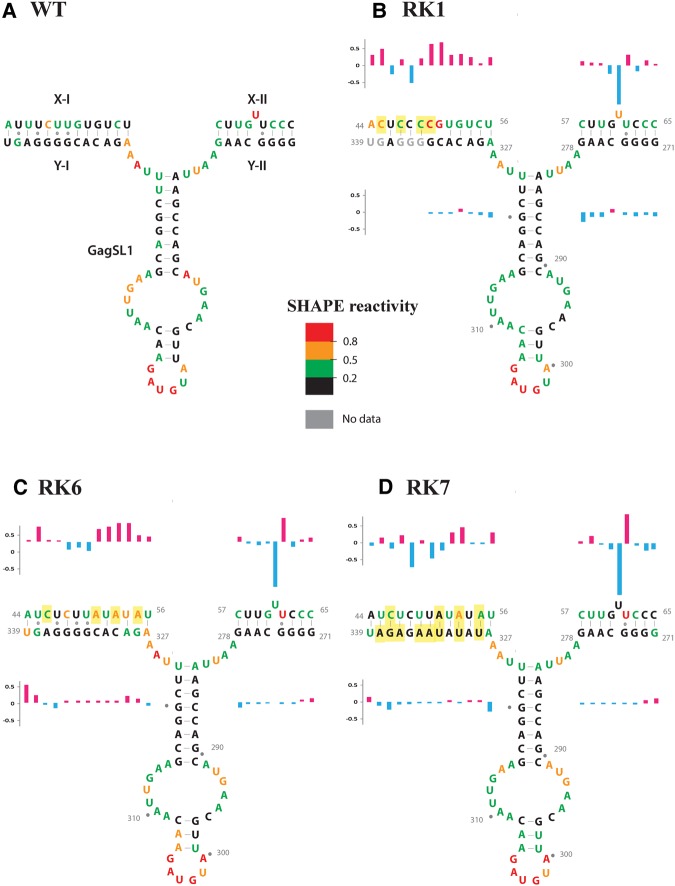
The Mfold predictions of the secondary structures of U5-gag LRI-I and LRI-II mutants with G•U base pairs, RK1 and RK2, did not reveal any disruptions of LRIs when compared to the wild-type, even though there was a significant decrease (three- to fourfold) in RPE of these mutants (Supplemental Fig. 2). As expected, the substitution of G•U base pairs with G = C pairs stabilized the predicted secondary structures by 10 and 16 kcal/mol, respectively (Supplemental Fig. 2B,C). To experimentally validate these predictions, we performed SHAPE on an in vitro transcribed RNA corresponding to the first 550 nt of RK1 gRNA, and the SHAPE data (Supplemental Table 1) were applied as constraints in the structure prediction program RNAstructure (version 5.3) (Reuter and Mathews 2010). Of the five most likely RK1 structures, after taking into account the SHAPE data, four predicted the existence of both LRI-I and LRI-II, and one predicted the existence of LRI-I, but not LRI-II. The second of these SHAPE-validated structures was similar to the wild-type SHAPE-validated structure (Supplemental Fig. 2D). However, careful inspection of the SHAPE reactivity of the nucleotides predicted to be involved in LRI-I and LRI-II strongly argues against the existence of LRI-I in mutant RK1. Several nucleotides in the X-I (U5) sequences (i.e., A44, C45, C50, G51, U52, and G53) were more reactive in RK1 than in the wild-type MMPV strain, whereas only 1 nt (C48) was less reactive in the RK1 mutant (Fig. 4A,B). The observed reactivity pattern of RK1 was thus at odds with the predicted stabilization of LRI-I. Besides that, the reactivity of the Y-I (gag) sequence was only marginally affected in mutant RK1, except for strong reverse transcription pausing at nucleotides G334, G335, G336, G338, and U339, which prevented determination of the reactivity of these residues (Fig. 4A,B). These pausing sites, which were not observed with the wild-type gRNA, suggest that the Y-I (gag) sequence is involved in different structures in the wild-type and RK1 gRNAs. Altogether, these data strongly suggest that LRI-I does not exist in mutant RK1 even though predicted otherwise. By comparison, little difference was found between wild-type and RK1 RNA when looking at the X-II and Y-II sequences forming LRI-II (Fig. 4A,B). The only significant difference was a decreased reactivity of U61 and a concomitant reactivity increase of U62. Similar changes were observed with the other mutants (see below); they suggest that either U61 or U62 can base-pair with G274 and that unlike LRI-I, LRI-II is formed in mutant RK1. Considering this structural information, the gRNA packaging and propagation defects of RK1 are most likely due to the disruption of LRI-I, in line with the results we obtained with mutants RK3, RK4, RK5, and RK6 (see below).
FIGURE 4.
SHAPE reactivity of the region corresponding to LRI-I/II and Gag SL1 of wild-type and mutant (RK1, RK6, and RK7) gRNAs. The SHAPE reactivity is color-coded on the SHAPE-validated structure of wild-type and mutant RNAs. (A–D) The SHAPE reactivity is reported on the wild-type structure for the sake of comparison. For mutants RK1, RK6, and RK7, a structure similar to the wild-type structure is drawn to facilitate comparison of the SHAPE reactivity, but this structure does not necessarily reflect the actual structure (see text and Fig. 5 for details). The differences in reactivity in LRI-I and LRI-II in the mutants compared to wild-type are indicated by histograms (red and blue correspond to increase and decrease in reactivity, respectively). Mutations are highlighted in yellow.
Structure–function relationship of U5-gag LRI-I
The predicted structures of mutants RK3 and RK5 showed that whenever the U5 (X-I) sequence was deleted, both LRI-I and LRI-II structures were lost, in addition to the gag stem–loop 1, SL1 (Supplemental Fig. 3B,D). In RK4, deletion of the gag (Y-I) sequence affected LRI-I and gag SL1, while LRI-II remained intact (Supplemental Fig. 3C). All three mutations resulted in a complete loss of RNA packaging and propagation, suggesting that the maintenance of LRI-I structure is critical for these functions.
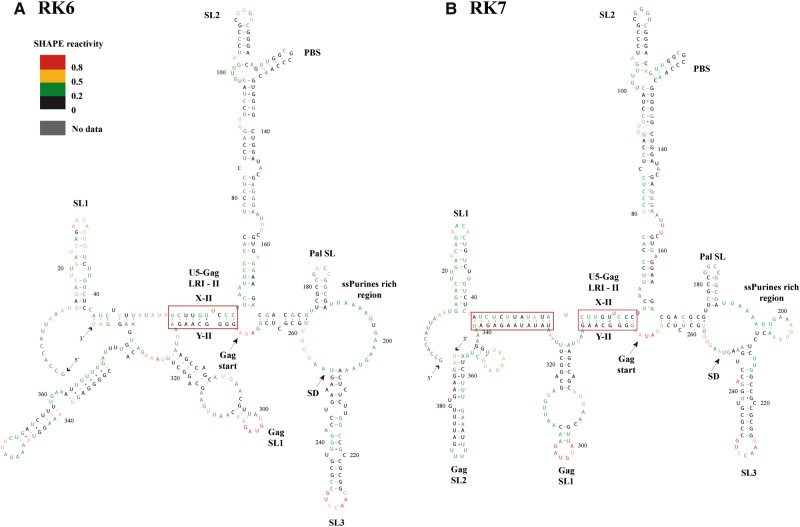
Mutants RK6 and RK7 were created to destabilize and reestablish the LRI-I structure, respectively. As shown in Supplemental Figure 4B, the predicted structure of RK6 revealed loss of the LRI-I stem, but maintenance of all the other important structural motifs. The mutation introduced in RK7, on the other hand, predicted restoration of the LRI-I structure (Supplemental Fig. 4C). We performed SHAPE on mutants RK6 and RK7 to experimentally validate these predicted structures (Fig. 4C,D; Supplemental Table 1). Among the five most likely RK6 structures incorporating the SHAPE data, none predicted the existence of LRI-I, while two predicted disruption of LRI-II. Examination of the SHAPE reactivity of the sequences potentially involved in LRI-I/II supports the existence of LRI-II (Fig. 4C). Several nucleotides in U5 (X-1) were more reactive in RK6 than in the wild-type RNA (i.e., U45, A51, U52, A53, U54, and A55), consistent with disruption of LRI-I. The gag (Y-I) sequence remained weakly reactive, with the exception of U339, indicating that this sequence is base-paired elsewhere in the RK6 structure. The only significant reactivity change in LRI-II was a decrease in reactivity of U61 and a concomitant increase in reactivity of U62 in RK6 compared to the wild-type (Fig. 4C). This change is compatible with the existence of LRI-II, except that U62, rather than U61 is bulging out in the mutant structure. Importantly, the SHAPE-validated RK6 structure maintained the SL1, LRI-II, SL2/PBS, pal SL, ssPurine rich region, SL3, and GagSL1 structural motifs (Fig. 5A), strongly suggesting that the essentially complete loss of RNA packaging and propagation of this mutant was a direct result of the disruption of LRI-I.
FIGURE 5.
SHAPE-validated RNA secondary structural models of mutant RK6 designed to disrupt LRI-I (A) and RK7 designed to restore LRI-I (B). Nucleotides are color-coded as per the SHAPE reactivity key. Major structural elements that also exist in wild-type gRNA are labeled as in Figure 1.
The SHAPE data of mutant RK7 (Fig. 4D) reinforced the predicted structure (Supplemental Fig. 4C) in which the substituted, heterologous yet complementary U5 (X-I) and gag (Y-I) sequences restored the LRI-I interaction (Fig. 5B). The SHAPE reactivity of the sequences involved in LRI-I and LRI-II also supported the existence of these two long-range interactions in mutant RK7 (Fig. 4D). While some nucleotides in U5 (X-I) are slightly more reactive in RK7 than in the wild-type RNA, others were less reactive, keeping the overall reactivity of the substituted U5 (X-I) similar to the wild-type sequence. Meanwhile, the reactivity of gag (Y-I) was not found to be significantly different in the wild-type and RK7 mutant, except for the now familiar strong reactivity decrease of U61 and the concomitant reactivity increase of U62 (Fig. 4D). Altogether, our data show that the double substitution introduced in RK7 restored the wild-type structure (Figs. 4D, 5B; Supplemental Fig. 4C) and that a heterologous LRI-I interaction can efficiently support gRNA packaging and propagation (Fig. 2C.II,III).
Structure–function relationship of the sequences involved in U5-gag LRI-II
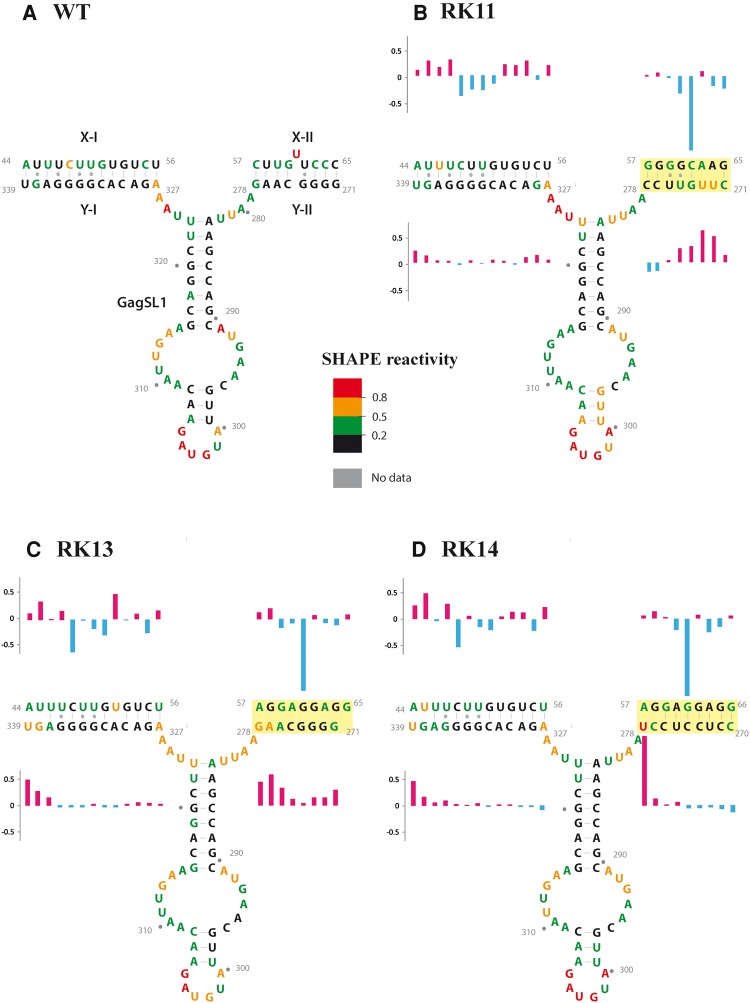
Substitution mutants RK8-RK10 were designed to disrupt LRI-II, while RK11 was designed to restore LRI-II by flipping the U5 (X-II) and gag (Y-II) complementary sequences. Mutants RK8 and RK10 were predicted to adopt structures that led to the creation of stem structures that somewhat resembled LRI-II but involved different sequences; however, RNA packaging and propagation of these mutants were still found to be completely abrogated (Fig. 3C.II,III; Supplemental Fig. 5). Similarly, mutant RK9 and RK11 were predicted to create a LRI-II-like stem separated from LRI-I by a 2-nt bulge, but both resulted in a complete abrogation of packaging and propagation (Fig. 3C.II,III; Supplemental Fig. 6). Since “compensatory” mutant RK11 did not restore RNA packaging, we performed SHAPE on this mutant. None of the four most likely structures incorporating the SHAPE constraints actually restored LRI-II, while LRI-I was always present. The SHAPE reactivity of the sequences potentially involved in forming LRI-I and II also supports this conclusion (Fig. 6A,B). The nucleotides at both ends of U5 (X-I) were slightly more reactive in mutant RK11 than in the wild-type, while the reactivity of the nucleotides in the center was weaker in the mutant RNA. At the same time, the reactivity of the gag (Y-I) sequence was only marginally affected, supporting the existence of LRI-I. In contrast, U5 (X-II) and gag (Y-II) had different reactivity profiles. Nucleotide C61, which is the equivalent of the bulging out U61 in the wild-type structure, had a dramatically reduced reactivity, indicating that it is likely base-paired. Indeed the reactivity of the entire U5 (X-II) was low, strongly suggesting that this 8-nt sequence is base-paired. On the other hand, the reactivity of nucleotides U272, U273, and to a lesser extent G274 and U275, increased in RK11, indicating that the 5′ end of gag (Y-II) is not base-paired, thus ruling out the existence of the native LRI-II structure (Fig. 6B). Since LRI-I was maintained in RK11, its lack of functionality can therefore most likely be attributed to disruption of LRI-II.
FIGURE 6.
SHAPE reactivity of the region corresponding to LRI-I/II and Gag SL1 of wild-type gRNA and mutants RK11, RK13, and RK14. The SHAPE reactivity is color-coded on the SHAPE-validated structure of wild-type and mutant RNAs. (A–D) The SHAPE reactivity is reported on the wild-type structure for the sake of comparison. For these mutants, RK11, RK13, and RK14, a structure similar to the wild-type structure is drawn to facilitate comparison of the SHAPE reactivity, but this structure does not necessarily reflect the actual structure (see text and Fig. 7 for details). The differences in reactivity in LRI-I and LRI-II in the mutants compared to wild-type are indicated by histograms (red and blue correspond to increase and decrease in reactivity, respectively). Mutations are highlighted in yellow.
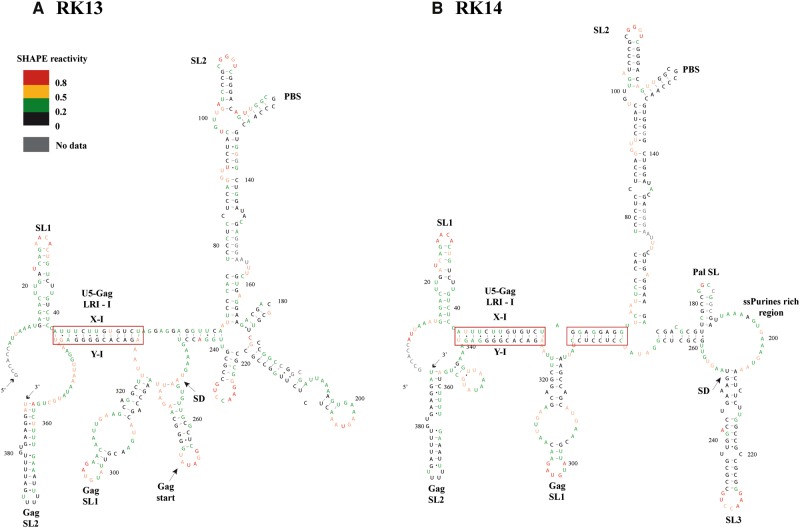
Mutants RK12 and RK13 were designed to disrupt U5-gag complementarity in LRI-II, while RK14 was designed to restore that complementarity using heterologous sequences. Even though these mutations resulted in a complete abrogation of packaging and propagation (Fig. 3C.II,III), Mfold predicted that LRI-II should be disrupted in RK12 and RK13, but restored in RK14 (Supplemental Fig. 7). As function was not restored in mutant RK14 despite recreating a heterologous LRI-II, we used SHAPE to experimentally analyze the RNA structure of mutants RK13 and RK14. In the five most likely structures of RK13 that incorporated the structural information provided by SHAPE, LRI-I was always formed, and the gag (Y-II) sequence always formed a hairpin, while the U5 (X-II) sequence base-paired with various sequences in gag (Fig. 7A and data not shown). The SHAPE reactivity of U5 (X-I), gag (Y-I), U5 (X-II), and gag (Y-II) sequences supports these models. The probing data were consistent with the existence of LRI-I, even though it was slightly destabilized at the “left” end (Fig. 6C). U5 (X-II) was weakly or not reactive, indicating it is base-paired, whereas the reactivity of the 3′ end of gag (Y-II) indicated that nucleotides A277 and G278 were single-stranded (Fig. 6C). The reactivity pattern of LRI-I in mutant RK14 was the same as in RK13, again showing a slightly destabilized “left” end (Fig. 6D). The reactivity of U5 (X-II) and gag (Y-II) was compatible with the existence of a heterologous LRI-II, with the exception of the A57-U278 base pair, which probably does not exist, as U278 was highly reactive (Fig. 6D). It should also be mentioned that the heterologous LRI-II in mutant RK14 does not contain any bulge, in contrast with the SHAPE-validated wild-type LRI-II structure (Fig. 6A,D). Thus, the overall SHAPE-validated structure of RK14 is very similar, but not identical, to the wild-type structure (Fig. 7B and data not shown). However, this mutant still failed to restore RNA packaging and propagation (Fig. 3C.II,III), indicating that maintaining LRI-II base-pairing was not sufficient for function.
FIGURE 7.
SHAPE-validated RNA secondary structure models of mutant RK13 designed to disrupt the complementarity of LRI-II (A) and RK14 designed to restore a heterologous LRI-II (B). Nucleotides are color-coded as per the SHAPE reactivity key. Structural elements that also exist in wild-type gRNA are labeled as in Figure 1.
In vitro RNA dimerization on SHAPE-interrogated LRI mutants
As packaging of retroviral RNA is tightly coupled to its dimerization, we next checked if introduced mutations in LRIs affected in vitro RNA dimerization. Wild-type or mutant in vitro transcribed RNAs were incubated in monomer or dimer buffer and analyzed on agarose gel in TBM buffer at 4°C or in TB buffer at 20°C (see Materials and Methods). While electrophoresis at 4°C in TBM buffer stabilizes the dimer species, only the most stable dimers survive electrophoresis at 20°C in the absence of Mg2+ ions (TB buffer). Dimerization of RK1 (LRI-I stabilizing mutant) and RK13 (LRI-II destabilizing mutant) RNAs was significantly reduced in TB buffer (Fig. 8, top panel), but this reduction (2.2- and 2.5-fold, respectively) is rather modest. Dimerization of the other mutant RNAs was only slightly affected (<25%). The electrophoresis profile of mutants RK1, RK11 (mutant with heterologous LRI-II), and RK13 is qualitatively similar to the wild-type profile, with one predominant dimer species. There is, however, a faint band corresponding to a second dimer species that migrated between the monomer and the main dimer species; this intermediate band is more visible with mutant RK7 (mutant with heterologous LRI-I), and especially with RK6 (LRI-I destabilizing mutant). Interestingly, this intermediate band is more predominant when electrophoresis was performed at 20°C in TB (Fig. 8, lower panel), indicating that the dimer in this intermediate band is more stable than the dimer in the upper band. Our data also reveal that these two dimer species are in equilibrium with each other, as the intermediate band is usually more pronounced in TB than in TBM buffer. However, the relative intensity of the top and intermediate bands does not correlate with RNA packaging: on the one hand the intermediate band is more pronounced in mutant RK7 (which restored RNA packaging and propagation; Fig. 2C.II,III) than in wild-type RNA; on the other hand, mutant RK14 (mutant with heterologous LRI-II), whose dimerization profile is qualitatively and quantitatively very similar to wild-type, is defective at the RNA packaging and propagation levels (Fig. 3C.II,III). Altogether, these results indicate that functional loss of RNA packaging of the LRI mutants cannot be directly correlated to defects in RNA dimerization.
FIGURE 8.
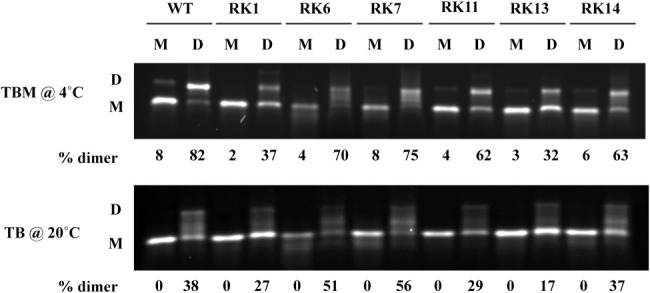
In vitro RNA dimerization on SHAPE-interrogated LRI mutants. Wild-type and mutant RNAs were incubated in monomer (M) or dimer (D) buffer and analyzed by electrophoresis on 1% agarose gel either in TBM buffer at 4°C (top panel) or in TB buffer at 20°C (lower panel). Experiments were performed in duplicates and the mean dimerization percent is indicated below each lane.
DISCUSSION
One of the distinguishing features of the MPMV packaging signal RNA secondary structure model based on SHAPE data is the presence of two LRIs involving 5′ (U5) and 3′ (gag) complementary sequences that are believed to anchor the structure of the 5′ UTR (Fig. 1; Jaballah et al. 2010; Aktar et al. 2013). LRIs have been found to be conserved in several retroviruses, including HIV-1 and 2, simian immunodeficiency virus (SIV), feline immunodeficiency virus (FIV), and mouse mammary tumor virus (MMTV) (Paillart et al. 2002, 2004b; Abbink and Berkhout 2003; Kenyon et al. 2008, 2011; Aktar et al. 2014; Siegfried et al. 2014; Keane et al. 2015; Smyth et al. 2015; Tran et al. 2015). Despite the fact that there are substantial sequence heterogeneities among human, simian, and feline lentiviruses, the preservation of such LRIs in these lentiviruses suggest they play important function(s) in the retroviral life cycle (Paillart et al. 2002; Kenyon et al. 2008, 2011). A pivotal role of LRIs is further evidenced by the fact that mutations that destabilize these interactions affect several important steps in the retroviral life cycle, including RNA packaging and dimerization (Paillart et al. 2002; Abbink and Berkhout 2003; Song et al. 2008; Rizvi et al. 2010; Lu et al. 2011). Similarly, an earlier study on MPMV has shown the importance of gag sequences (involved in maintaining U5-gag LRIs) in RNA packaging, further arguing in favor of the existence of LRIs in vivo and their functional role in the MPMV life cycle (Schmidt et al. 2003).
Unlike other retroviruses with one identified LRI, the SHAPE-validated MPMV secondary structure model contains two LRIs (LRI-I and LRI-II; Fig. 1), both of which maintain the interaction between U5 and gag sequences (Aktar et al. 2013). The functional and SHAPE results we obtained on a number of LRI mutants validate the existence and biological significance of LRIs and show that LRI-I could be functionally replaced by a heterologous LRI, while LRI-II could not.
All mutations that were designed to disrupt LRI-I (RK3, RK4, RK5, and RK6) dramatically reduced RNA packaging and propagation (Figs. 2C, 4C, 5A; Supplemental Figs. 3, 4), demonstrating the functional role of LRI-I. This was especially true as other motifs, such as pal SL, ssPurine-rich region, SL3, and Region B, previously shown to be critical for RNA packaging and dimerization (Jaballah et al. 2010; Aktar et al. 2014), were preserved in these mutants. Mutant RK7 was designed to restore a heterologous LRI-I (Supplemental Fig. 4C) and SHAPE data supported our structural predictions (Figs. 4D, 5B). This heterologous LRI-I was fully functional (Fig. 2C); hence the 13 complementary nucleotides of U5-gag LRI-I of MPMV functionally resembles the R/U5-gag heptanucleotide of FIV LRI, where a structural motif rather than a particular sequence has been shown to be essential for RNA packaging (Rizvi et al. 2010).
The wild-type LRI-I and the heterologous LRI-I of mutant RK7 are moderately stable, as predicted by the predominance of A–U and G•U base pairs and shown by a significant level of SHAPE reactivity (Fig. 4A,D). Indeed, it is well known that the gRNA is an active player in many critical stages in the retroviral life cycle, including processes that are driven mainly by the proper three-dimensional folding of multiple RNA domains to recruit viral and host factors required for activity. This structural flexibility is of great functional importance to the RNA as it is a dynamic structure that undergoes several intramolecular and intermolecular interactions during RNA dimerization and packaging processes (for review, see D'Souza and Summers 2005; Lever 2007; Johnson and Telesnitsky 2010). Therefore, even though the LRI-I structure is important, a limited stability might be required for function. The RK1 and RK2 mutants designed to test this possibility did not allow us to answer this question, as careful inspection of the SHAPE data revealed that LRI-I was not formed in RK1 (Fig. 4B). Therefore, the strong decrease in RNA packaging and propagation observed with this mutant is likely due to disruption of LRI-I, in line with the results obtained with mutants RK3, RK4, RK5, and RK6 discussed above. Unexpectedly, when SHAPE data were introduced as constraints in RNAstructure (Reuter and Mathews 2010) for RK1, this software still proposed structures in which U5 (X-I) and gag (Y-1) were base-paired, despite the inconsistency with SHAPE data for this region (Supplemental Fig. 2D). There are several plausible reasons why RNAstructure is unable to propose a secondary structure model that correctly accounts for the reactivity of U5 (X-I) and gag (Y-1) sequences. One possibility is that the actual RK1 RNA structure is stabilized by a pseudoknot or another higher-order structural motif that cannot be predicted by the software. An alternative explanation could be that the RK1 RNA structure corresponds to a kinetically trapped conformation during RNA folding. Such a phenomenon has recently been reported in the case of the gRNA of a protease defective Moloney mouse leukemia virus mutant (Grohman et al. 2014). Even though SHAPE most often does not always propose correct secondary structure models (Kladwang et al. 2011), our data show that it is very important to verify experimentally that “compensatory mutations” actually restore the wild-type structure. In the absence of such verification, a large proportion of “structure–function relationships” might indeed turn out to be inaccurate.
LRI-II is also required for function, as all mutants designed to disrupt it (Figs. 6, 7; Supplemental Figs. 5, 6, 7) displayed dramatically altered RNA packaging and propagation efficiencies (Fig. 3). Two mutants, RK11 and RK14, were designed to restore a heterologous LRI-II, but according to structural predictions and SHAPE analysis, LRI-II was formed in RK14, but not in RK11 (Figs. 6, 7; Supplemental Figs. 6, 7). Nevertheless, RNA packaging and propagation of mutant RK14 were severely affected (Fig. 3) despite its wild-type-like structure (Fig. 7B). Thus, in the case of LRI-II, base-pairing is not sufficient for function, suggesting some sequence specificity. This could be the case for instance if the MPMV Gag precursor directly interacts with LRI-II. Notably, the heterologous LRI-II does not contain any bulge, in contrast with the wild-type LRI-II (Figs. 6, 7B). However, deleting U61 (or U62) in the wild-type sequence did not affect viral propagation (data not shown), indicating that a bulge is not required for LRI-II function. Finally, SHAPE data of RK14 designed to restore heterologous LRI-II revealed that nucleotide U278 was highly reactive, precluding the existence of the A57–U278 base pair (Fig. 6D). As a consequence, the geometry of the junction formed by LRI-I, Gag SL1, and LRI-II (Fig. 1) was most likely compromised, and the overall 3D-structure might be significantly altered. The loss of the native RNA 3D structure may in turn alter, directly or indirectly, recognition by the Gag precursor.
Indeed, since LRI-I, Gag SL1, and LRI-II involve sequences within gag downstream from the mSD, and are thus restricted to the full-length unspliced gRNA, this junction represents a putative Gag binding site. In addition, similar to the proposed role of the HIV-1 U5-AUG LRI in the regulation of Gag translation (Abbink and Berkhout 2003; Lu et al. 2011), the MPMV LRIs may also have a role in the translation of the Gag protein since the guanine (G) residue of the gag AUG start codon is within the gag (Y-II) sequence of LRI-II. Thus, the MPMV LRIs plausibly could allow the regulation of a temporal switch between viral protein synthesis and packaging, conferring high specificity of packaging that is restricted to the full-length unspliced RNA.
MATERIALS AND METHODS
Genome nucleotide numbering system
The MPMV nucleotide numbering system refers to the genome sequence deposited in the Genbank (accession number M12349) by Sonigo et al. 1986.
Plasmid construction
The packaging construct, TR301, expresses the MPMV gag/pol genes under the transcriptional control of the human cytomegalovirus (hCMV) intron A promoter/enhancer described earlier (Browning et al. 2001; Supplemental Fig. 1A). The vesicular stomatitis virus glycoprotein G (VSV-G) expression vector, MD.G, was used to produce pseudotyped viruses to infect target cells since its receptors are present on many cell types (Naldini et al. 1996; Supplemental Fig. 1A).
Substitution and deletion mutations were cloned into the MPMV sub-genomic transfer vector, SJ2, described previously (Jaballah et al. 2010), which expresses the hygromycin B phosphotransferase selectable marker from an internal simian virus 40 early promoter (SV-Hygr) to monitor the effect of the mutations on propagation of the packaged viral RNA (Fig. 1B).
The mutations into the LRIs (Figs. 2A, 3A) were introduced using splice overlap extension (SOE) PCR, requiring two rounds of amplifications, using SJ2 as a template. Round 1 consisted of two independent PCRs, A and B in which PCR (A) was performed using the universal outer forward (sense; S) primer OTR787 (5′-ccctcgagTGTCCGGAGCCGTGCTGCCCG-3′) along with the inner reverse (antisense; AS) primer that varied depending upon the mutation introduced. In PCR (B), the AS outer primer OTR788 (5′-cccggatccTTCTTTCTTATCTATCAATTCTTTAATTAAG-3′) was used along with the inner S primer which also varied depending upon the mutations introduced. The resulting PCR products from these two separate reactions (PCR A and PCR B) contained overlapping complementary sequences that allowed them to anneal, forming a template with the introduced mutation for round 2 PCR, which was performed using outer S and AS primers (OTR787/OTR788), generating a final product harboring the mutation. The PCR-amplified mutant products were cleaved with XhoI and BamHI restriction endonucleases and cloned into similarly digested wild-type transfer vector, SJ2. This cloning strategy resulted in replacement of the wild-type sequences, starting from the 5′ end of U3 to 282 bp of gag (Fig. 1), with the PCR-amplified fragments containing the desired mutations. Sequences of primers used for the PCR and cloning are listed in Supplemental Table 2. The sequence of all mutant clones was verified by DNA sequencing.
Nucleocytoplasmic fractionation, virion and RNA isolation, and cDNA preparation
Transfected cells were fractionated into cytoplasmic and nuclear fractions and RNA extracted as described earlier (Mustafa et al. 2005; Ghazawi et al. 2006; Jaballah et al. 2010; Rizvi et al. 2010). Packaged viral RNAs were prepared from clarified supernatants from transfected cultures after filtration through 0.2 µm filters as described previously (Mustafa et al. 2005; Ghazawi et al. 2006; Jaballah et al. 2010; Rizvi et al. 2010). RNA preparations were DNase-treated and subjected to 30-cycle PCR to test for the presence of any residual contaminating plasmid DNA using MPMV specific primer pair: OTR 1161 (S; 5′-GATCAGAACACTGTCTTGTC-3′) and OTR 1163 (AS; 5′-CTT TCTTATCTATCAATTCTTTAA-3′) as described previously (Jaballah et al. 2010). After confirming the absence of any plasmid DNA contamination in RNA preparations by PCR, RNAs were reverse transcribed and PCR amplified to determine the quality of cDNA preparations. Cytoplasmic cDNAs were amplified for both unspliced as well as spliced β-actin mRNA to ensure the integrity of the fractionation process, as described previously (Mustafa et al. 2005; Ghazawi et al. 2006; Jaballah et al. 2010; Rizvi et al. 2010). Finally, cytoplasmic cDNA preparations were additionally amplified using MPMV-specific vector primer pair OTR 1161/1163 to ascertain the stability as well as the proper nuclear export of the mutant transfer vector RNAs.
Real-time quantitative PCR (qPCR) of transfer vector RNA expression
The relative expression of transfer vector RNAs in the cytoplasm and their packaging efficiency into the nascently produced virions was quantified by developing a custom-made Taqman gene expression assay (Applied Biosystems Inc., ABI). This assay employed a FAM/MGB-labeled probe along with primers within the U5/PBS region of MPMV, a region common to the wild-type and the mutant transfer vector RNAs and away from the site of the mutations analyzed in this study. Although this region is also upstream of the major splice donor of MPMV, there are no known splice acceptor sites in the wild-type as well as mutant transfer vectors, including the SV-Hygro cassette. Therefore, it is unlikely that any splicing will take place resulting in the generation of spliced RNAs. Such a vector design has been used successfully in a number of other retroviral systems for studying genomic RNA packaging efficiencies. In addition, RNA packaging results obtained from using such assays have correlated well in the past with the RNA propagation data obtained as measured by the counts of hygromycin-resistant colonies (CFU/mL) following transduction of the target cells with virus particles containing packaged RNA (Rizvi et al. 2010; Jaballah et al. 2010; Mustafa et al. 2012; Aktar et al. 2014).
The assay specifically consisted of a sense primer (5′-CTCCTCCAGGTTCCTACTGTTGA-3′; nucleotides 702–724), an antisense primer (5′-TCGTATCCAGCCCCACGTT-3′; nucleotides 770–752), and a FAM/MGB-labeled probe (5′-TCGGGACAGTTGGC-3′; nucleotides 734–747). The PCR efficiency of the MPMV assay was calculated using the online PCR efficiency calculator (http://srvgen.upct.es/index.html; Mallona et al. 2011), which gave a value of 1.86. This value fell slightly below the level suggested for the use of the ΔΔCT method (≥1.9). To ensure that we could use this method for the relative quantification purposes, the assay was empirically tested against the standard curve method (data not shown) using test samples with varying amounts of MPMV plasmid DNA. This comparative analysis validated the use of the ΔΔCT method for quantitating relative packaging efficiencies.
As an endogenous control, a predesigned VIC/MGB-labeled human β-actin assay (ABI #4326315E) was used as described previously (Mustafa et al. 2012; Aktar et al. 2014). Briefly, equal amounts of cDNAs from the wild-type and mutant samples were amplified for 50 cycles in triplicates for both MPMV and β-actin using the ABI thermocycler 7500 (ABI). The relative quantification (RQ) values thus obtained for MPMV expression in both the cytoplasmic and viral samples were normalized to the luciferase values obtained per μg protein to control for differences in transfection efficiencies. Finally, to estimate the packaging efficiency, results obtained for MPMV expression in the virions were divided by those for the cytoplasmic expression and then reported relative to the wild-type levels.
In silico and SHAPE analyses of MPMV RNA secondary structure
The 5′ end of the wild-type and mutant MPMV genomes (region between R and the first 120 nt of gag) were folded using Mfold (Mathews et al. 1999; Zuker 2003) to correlate the effects of the introduced mutations on MPMV packaging signal RNA secondary structure in an attempt to establish structure–function relationship of this region during MPMV RNA packaging. SHAPE was performed on purified in vitro transcribed wild-type and mutant MPMV RNAs as previously described (Aktar et al. 2013) using primers complementary to nucleotides 483–502. Electropherograms were analyzed with the SHAPEfinder program (Vasa et al. 2008; Wilkinson et al. 2008). The SHAPE reactivity data were then applied as constraints in the structure prediction program RNAstructure (version 5.3; Reuter and Mathews 2010) in order to obtain the SHAPE-derived RNA structure of the wild-type and mutant MPMV packaging signal RNAs.
In vitro RNA dimerization assays
In vitro RNA dimerization was performed as previously described (Aktar et al. 2013). Briefly, 300 nM of wild-type or mutant RNA was incubated in monomer (50 mM sodium cacodylate at pH 7.5, 40 mM KCl, 0.1 mM MgCl2) or dimer buffer (50 mM sodium cacodylate at pH 7.5, 300 mM KCl, 5 mM MgCl2) for 30 min at 37°C. After addition of loading buffer, the samples were analyzed on 1% agarose gels either at 4°C in a TBM buffer (50 mM Tris base, 45 mM boric acid, 0.1 mM MgCl2) or at 20°C TB buffer (50 mM Tris base, 45 mM boric acid), visualized using ethidium bromide, and quantified.
Statistical analysis
Statistical analysis was performed employing the standard paired two-tailed Student's t-test between the wild-type and the mutant clones to establish statistical significance. A P-value of 0.01 was considered to be significant.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded primarily by a grant from the United Arab Emirates University (UAEU Program for Advanced Research-UPAR; 31M131) to T.A.R. and R.M., in part by a grant from the College of Medicine and Health Sciences (NP/13/26) to T.A.R., and UAEU-NRF (31M101) and Start-Up (31M122) grants to F.M.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.055731.115.
REFERENCES
- Abbink TE, Berkhout B. 2003. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J Biol Chem 278: 11601–11611. [DOI] [PubMed] [Google Scholar]
- Abd El-Wahab EW, Smyth RP, Mailler E, Bernacchi S, Vivet-Boudou V, Hijnen M, Jossinet F, Mak J, Paillart JC, Marquet R. 2014. Specific recognition of the HIV-1 genomic RNA by the Gag precursor. Nat Commun 5: 4304. [DOI] [PubMed] [Google Scholar]
- Aktar SJ, Jabeen A, Ali LM, Vivet-Boudou V, Marquet R, Rizvi TA. 2013. SHAPE analysis of the 5′ end of the Mason-Pfizer monkey virus (MPMV) genomic RNA reveals structural elements required for genome dimerization. RNA 19: 1648–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktar SJ, Vivet-Boudou V, Ali LM, Jabeen A, Kalloush RM, Richer D, Mustafa F, Marquet R, Rizvi TA. 2014. Structural basis of genomic RNA (gRNA) dimerization and packaging determinants of mouse mammary tumor virus (MMTV). Retrovirology 11: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Dhaheri NS, Phillip PS, Ghazawi A, Ali J, Beebi E, Jaballah SA, Rizvi TA. 2009. Cross-packaging of genetically distinct mouse and primate retroviral RNAs. Retrovirology 6: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Shamsi IR, Al Dhaheri NS, Phillip PS, Mustafa F, Rizvi TA. 2011. Reciprocal cross-packaging of primate lentiviral (HIV-1 and SIV) RNAs by heterologous non-lentiviral MPMV proteins. Virus Res 155: 352–357. [DOI] [PubMed] [Google Scholar]
- Balvay L, Lopez Lastra M, Sargueil B, Darlix JL, Ohlmann T. 2007. Translational control of retroviruses. Nat Rev Microbiol 5: 128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M, Prasad S, Dubay JW, Hunter E, Jeang KT, Rekosh D, Hammarskjöld ML. 1994. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci 91: 1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning MT, Schmidt RD, Lew KA, Rizvi TA. 2001. Primate and feline lentivirus vector RNA packaging and propagation by heterologous lentivirus virions. J Virol 75: 5129–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza V, Summers MF. 2005. How retroviruses select their genomes. Nat Rev Microbiol 3: 643–655. [DOI] [PubMed] [Google Scholar]
- Ghazawi A, Mustafa F, Phillip PS, Jayanth P, Ali J, Rizvi TA. 2006. Both the 5′ and 3′ LTRs of FIV contain minor RNA encapsidation determinants compared to the two core packaging determinants within the 5′ untranslated region and gag. Microbes Infect 8: 767–778. [DOI] [PubMed] [Google Scholar]
- Grohman JK, Gorelick RJ, Kottegoda S, Allbritton NL, Rein A, Weeks KM. 2014. An immature retroviral RNA genome resembles a kinetically trapped intermediate state. J Virol 88: 6061–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guesdon FM, Greatorex J, Rhee SR, Fisher R, Hunter E, Lever AM. 2001. Sequences in the 5′ leader of Mason-Pfizer monkey virus which affect viral particle production and genomic RNA packaging: development of MPMV packaging cell lines. Virology 288: 81–88. [DOI] [PubMed] [Google Scholar]
- Harrison GP, Hunter E, Lever AM. 1995. Secondary structure model of the Mason-Pfizer monkey virus 5′ leader sequence: identification of a structural motif common to a variety of retroviruses. J Virol 69: 2175–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaballah SA, Aktar SJ, Ali J, Phillip PS, Al Dhaheri NS, Jabeen A, Rizvi TA. 2010. A G-C-rich palindromic structural motif and a stretch of single-stranded purines are required for optimal packaging of Mason-Pfizer monkey virus (MPMV) genomic RNA. J Mol Biol 401: 996–1014. [DOI] [PubMed] [Google Scholar]
- Johnson SF, Telesnitsky A. 2010. Retroviral RNA dimerization and packaging: the what, how, when, where, and why. PLoS Pathog 6: e1001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane SC, Heng X, Lu K, Kharytonchyk S, Ramakrishnan V, Carter G, Barton S, Hosic A, Florwick A, Santos J, et al. 2015. RNA structure. Structure of the HIV-1 RNA packaging signal. Science 348: 917–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon JC, Ghazawi A, Cheung WK, Phillip PS, Rizvi TA, Lever AM. 2008. The secondary structure of the 5′ end of the FIV genome reveals a long-range interaction between R/U5 and gag sequences, and a large, stable stem-loop. RNA 14: 2597–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon JC, Tanner SJ, Legiewicz M, Phillip PS, Rizvi TA, Le Grice SF, Lever AM. 2011. SHAPE analysis of the FIV Leader RNA reveals a structural switch potentially controlling viral packaging and genome dimerization. Nucleic Acids Res 39: 6692–6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kladwang W, VanLang CC, Cordero P, Das R. 2011. Understanding the errors of SHAPE-directed RNA structure modeling. Biochemistry 50: 8049–8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever AM. 2007. HIV-1 RNA packaging. Adv Pharmacol 55: 1–32. [DOI] [PubMed] [Google Scholar]
- Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, et al. 2011. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science 334: 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallona I, Weiss J, Egea-Cortines M. 2011. pcrEfficiency: a Web tool for PCR amplification efficiency prediction. BMC Bioinformatics 12: 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol 288: 911–940. [DOI] [PubMed] [Google Scholar]
- Moore MD, Fu W, Nikolaitchik O, Chen J, Ptak RG, Hu WS. 2007. Dimer initiation signal of human immunodeficiency virus type 1: its role in partner selection during RNA copackaging and its effects on recombination. J Virol 81: 4002–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa F, Ghazawi A, Jayanth P, Phillip PS, Ali J, Rizvi TA. 2005. Sequences intervening between the core packaging determinants are dispensable for maintaining the packaging potential and propagation of feline immunodeficiency virus transfer vector RNAs. J Virol 79: 13817–13821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa F, Al Amri D, Al Ali F, Al Sari N, Al Suwaidi S, Jayanth P, Philips PS, Rizvi TA. 2012. Sequences within both the 5′ UTR and Gag are required for optimal in vivo packaging and propagation of mouse mammary tumor virus (MMTV) genomic RNA. PLoS One 7: e47088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272: 263–267. [DOI] [PubMed] [Google Scholar]
- Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. 2002. In vitro evidence for a long range pseudoknot in the 5′-untranslated and matrix coding regions of HIV-1 genomic RNA. J Biol Chem 277: 5995–6004. [DOI] [PubMed] [Google Scholar]
- Paillart JC, Shehu-Xhilaga M, Marquet R, Mak J. 2004a. Dimerization of retroviral RNA genomes: an inseparable pair. Nat Rev Microbiol 2: 461–472. [DOI] [PubMed] [Google Scholar]
- Paillart JC, Dettenhofer M, Yu XF, Ehresmann C, Ehresmann B, Marquet R. 2004b. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J Biol Chem 279: 48397–48403. [DOI] [PubMed] [Google Scholar]
- Parveen Z, Mukhtar M, Goodrich A, Acheampong E, Dornburg R, Pomerantz RJ. 2004. Cross-packaging of human immunodeficiency virus type 1 vector RNA by spleen necrosis virus proteins: construction of a new generation of spleen necrosis virus-derived retroviral vectors. J Virol 78: 6480–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter JS, Mathews DH. 2010. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi TA, Panganiban AT. 1993. Simian immunodeficiency virus RNA is efficiently encapsidated by human immunodeficiency virus type 1 particles. J Virol 67: 2681–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi TA, Kenyon JC, Ali J, Aktar SJ, Phillip PS, Ghazawi A, Mustafa F, Lever AM. 2010. Optimal packaging of FIV genomic RNA depends upon a conserved long-range interaction and a palindromic sequence within gag. J Mol Biol 403: 103–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RD, Mustafa F, Lew KA, Browning MT, Rizvi TA. 2003. Sequences within both the 5′ untranslated region and the gag gene are important for efficient encapsidation of Mason-Pfizer monkey virus RNA. Virology 309: 166–178. [DOI] [PubMed] [Google Scholar]
- Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. 2014. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat Methods 11: 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth RP, Despons L, Huili G, Bernacchi S, Hijnen M, Mak J, Jossinet F, Weixi L, Paillart JC, von Kleist M, et al. 2015. Mutational interference mapping experiment (MIME) for studying RNA structure and function. Nat Methods 12: 866–872. [DOI] [PubMed] [Google Scholar]
- Song R, Kafaie J, Laughrea M. 2008. Role of the 5′ TAR stem-loop and the U5-AUG duplex in dimerization of HIV-1 genomic RNA. Biochemistry 47: 3283–3293. [DOI] [PubMed] [Google Scholar]
- Sonigo P, Barker C, Hunter E, Wain-Hobson S. 1986. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell 45: 375–385. [DOI] [PubMed] [Google Scholar]
- Tran T, Liu Y, Marchant J, Monti S, Seu M, Zaki J, Yang AL, Bohn J, Ramakrishnan V, Singh R, et al. 2015. Conserved determinants of lentiviral genome dimerization. Retrovirology 12: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. 2008. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA 14: 1979–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vile RG, Ali M, Hunter E, McClure MO. 1992. Identification of a generalised packaging sequence for D-type retroviruses and generation of a D-type retroviral vector. Virology 189: 786–791. [DOI] [PubMed] [Google Scholar]
- White SM, Renda M, Nam NY, Klimatcheva E, Zhu Y, Fisk J, Halterman M, Rimel BJ, Federoff H, Pandya S, et al. 1999. Lentivirus vectors using human and simian immunodeficiency virus elements. J Virol 73: 2832–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol 6: e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin PD, Hu WS. 1997. RNAs from genetically distinct retroviruses can copackage and exchange genetic information in vivo. J Virol 71: 6237–6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.