Abstract
Problems arising during DNA replication require the activation of the ATR-CHK1 pathway to ensure the stabilization and repair of the forks, and to prevent the entry into mitosis with unreplicated genomes. Whereas the pathway is essential at the cellular level, limiting its activity is particularly detrimental for some cancer cells. Here we review the links between replication stress (RS) and cancer, which provide a rationale for the use of ATR and Chk1 inhibitors in chemotherapy. First, we describe how the activation of oncogene-induced RS promotes genome rearrangements and chromosome instability, both of which could potentially fuel carcinogenesis. Next, we review the various pathways that contribute to the suppression of RS, and how mutations in these components lead to increased cancer incidence and/or accelerated ageing. Finally, we summarize the evidence showing that tumours with high levels of RS are dependent on a proficient RS-response, and therefore vulnerable to ATR or Chk1 inhibitors.
Introduction
The faithful transmission of genetic information is challenged in every cell division due to DNA damage inflicted by both endogenous and exogenous sources that can hamper the correct replication of the DNA. The presence of DNA modifications, difficult to replicate structures or the depletion in the nucleotide pool, among others, all result in the stalling of replication forks and accumulation of single stranded DNA (ssDNA) that is rapidly coated by ssDNA-binding proteins such as RPA. The presence of RPA-bound to ssDNA next to a ssDNA-dsDNA junction leads to the activation of the ATR kinase, which subsequently phosphorylates the Chk1 kinase in the so-called replication stress (RS) response [1]. The activity of this pathway is necessary to stabilize stalled forks, and to activate checkpoints that limit the entry into mitosis in the presence of unreplicated DNA [2].
Both ATR and Chk1 are essential for development in mammals [3–6]. In their absence, stalled forks persist and additional backup origins are fired, leading to an exhaustion of dNTP pools and a further accumulation of non-progressive forks. When the amount of ssDNA exceeds the pool of available RPA, and by yet not fully understood mechanisms, forks collapse leading to the generation of DNA double strand breaks (DSB) [7]. The lack of checkpoint activation in these cells allows them to progress to mitosis with unreplicated chromosomes leading to death by mitotic catastrophe. Whether the collapse of the forks occurs already in S phase or is mediated by structure specific endonucleases that are activated later in mitosis remains to be fully solved. Whereas ATR deficiency is not viable, a synonymous mutation that compromises ATR mRNA splicing leads to a severe reduction of ATR levels and a human hereditary syndrome known as Seckel, associated to dwarfism and microcephaly [8]. A humanized mouse model of ATR-Seckel recapitulates the symptoms of the human patients, and revealed that ATR hypomorphism limits mammalian lifespan due to a generalized accumulation of RS during embryogenesis [9]. In contrast to other genomic instability syndromes that show cancer predisposition, ATR-Seckel mice do not develop spontaneous tumours. In fact, reduced levels of ATR prevent the onset of certain tumours such as Myc-induced B cell lymphomas or MLL-ENL driven acute myeloid leukemias [10,11]. We believe that the increased sensitivity of cancer cells towards limited activity of ATR is linked to the increased levels of RS that are associated with oncogene expression, which renders these cells highly dependant on a proficient RS-checkpoint. How RS arises and is dealt with, its impact on genomic instability during carcinogenesis, and the potential to exploit it for cancer treatment are the focus of the present review.
Sources of replication stress during transformation
Oncogene activation alters DNA replication leading to increased RS, DNA breaks and activation of the DNA damage response (DDR) [12]. There are several proposed mechanisms for the induction of RS upon oncogene activation. Two of the proposed mechanisms are related to an inappropriate – insufficient or excessive- usage of replication origins. Overexpression of cyclin E reduces the number of replication origins that are licensed during G1. As a consequence, RS increases in S-phase due to the shortage of back-up origins to cope with stalled forks [13]. This effect is similar to that observed in hypomorphic mutants of the components of the MCM helicase. The mutation of MCM4 or MCM2 reduces the activity of the MCM complex, limiting the amount of available back-up origins, which results in increased RS, genomic instability and a cancer-prone phenotype [14–16]. In contrast, the overexpression of certain oncogenes (Myc, Ras…) has the opposite effect and increases the firing of origins of replication, leading to a depletion of the cellular pool of nucleotides (dNTPs) [17–21]. The reduced level of dNTPs slows down the progression of the forks and increases the chance of fork stalling per se. Of note, other reports suggest that the nucleotide depletion during transformation is due to the reduction in the expression of the small subunit of the Ribonucleotide Reductase (RNR), the enzyme that catalyzes the last and rate-limiting step in the generation of dNTPs. The RNR enzyme is composed of the constitutively expressed RRM1 protein and the cell cycle regulated RRM2 [22]. Ras induced transformation of human fibroblasts decreases transcription of the RRM2 gene prior to the cell cycle exit [23]. However, these results are at odds with the fact that RRM2 is a cell-cycle regulated protein with maximum expression during S phase, and a target of E2F and Myc transcription factors. Moreover, overexpression of RNR, rather than its loss, is frequently found in human cancers [22]. Regardless of the mechanism, supplementation with nucleosides has been shown to limit oncogene-induced RS in vitro, supporting the idea that a limited availability of nucleotides can be a source of oncogene-driven RS [19].
Besides any effect on the nucleotide pool, the higher number of active origins generates more replication-transcription collisions that lead to stalled replication forks [18], expression of common fragile sites and genome instability [24]. There is increasing evidence showing that the formation of RNA-DNA hybrid called R-loops is central to the replication-transcription collisions [25]. R-loops are frequently observed in highly transcribed genes with GC rich promoters, linked to G-quadruplex structures and torsional constrains during transcription [26–28]. The generation of R-loops is limited by the THO complex that targets the nascent RNA to the RNA processing machinery and to the nuclear pore [29]. Further, several factors cooperate in the resolution of R-loops, including RNase H enzymes and RNA helicases such as senataxin [29–33]. Mutation of any of these factors increases genome instability secondary to replication-transcription collisions. In addition to RNA metabolizing enzymes, known oncogenes and tumour suppressors may also fuel genomic instability through the modulation of R-loops. For instance, BRCA2 deficiency leads to an increase in R-loops; it remains to be seen whether this is due to the known roles of BRCA2 at replication forks [34] or if BRCA2 also has an active role in dissolving the RNA-DNA hybrids [35]. In what regards to oncogenes, Next Generation Sequencing (NGS) of cancers has revealed that a large fraction of them are transcription factors or chromatin regulators that override the transcriptional control in transformed cells. Given that replication timings are cell-type specific, it is possible the firing of origins in each cells has been selected to minimize collisions with the transcriptional machinery. Thus, R-loops might be an important source of genomic instability in the presence of mutations that alter the transcriptional landscape.
Chromosomal instability as a result of replication stress
In addition to the localized impact of RS at replication forks, several lines of evidence now support that RS can also lead to a wide range of chromosomal aberrations during malignant transformation. First, an in silico analysis of homozygous and heterozygous focal deletions in cancer samples and cell lines revealed that most of the heterozygous deletions in transformed cells are found in already defined common fragile sites (CFS) or in large genes [36]. Similar deletions can be induced by the treatment of cells with aphidicolin [36], a DNA polymerase inhibitor that induces replication stress. These data indicate that RS is likely the source of most of the passenger deletions during transformation and suggests a major role for RS in cancer genome evolution. Besides CFS, RS also induces the breakage of the recently identified early replicating fragile sites (ERFS), which are found at the breakpoints of some of the most common translocations observed in B-lymphoma [37]. Noteworthy, and in contrast to CFS, these sites are associated with “open” chromatin and present high levels of transcription, which suggests that the RS arising at ERFS might be linked to R-loops.
Second, RS might also lead to chromosomal instability (CIN) through an increase in defects on chromosome segregation. Consistently, the analysis of CIN+ versus CIN- colon adenocarcinoma cells revealed the presence of RS only in CIN+ cells, along with corresponding chromosome segregation defects [38]. This study identified a number of genes commonly deleted in CIN+ cell lines. Reduction of any of these genes in CIN- cells results in RS and alterations in chromosomes similar to those observed in CIN+ cells due to aberrant segregation during mitosis. Further proving the causal role of RS in the generation of CIN, the addition of nucleosides limited the segregation defects that arise upon deletion of CIN-suppressor genes [38]. At the molecular level, the link between RS and CIN might be intimately related to the observation that RS can promote the formation of anaphase bridges (see below).
Last, another mechanism by which RS could be involved on the generation of copy number alterations has also been recently described. This work revealed that break induced replication (BIR) is essential for cells to cope with RS induced by the overexpression of Cyclin E [39]. BIR is a specialized system of homologous recombination that can be used when the homology is limited to one side of the break, which is the case at a collapsed replication fork [40]. The use of BIR can generate amplifications with microhomology at the junctions, what resembles many of the copy number variations that are found on cancer cells. For DNA synthesis, BIR employs the basic DNA replication machinery, but in addition it specifically depends on the DNA Polymerase accessory subunits POLD3 and POLD4. The reduction in the levels of either of these proteins in cells overexpressing Cyclin E reduces fork speed, increases fork collapse, blocks the generation of small amplifications and ultimately limits the proliferation of these cells [39]. To what extent POLD3 or POLD4 are non-essential in the context of primary cells demands further study. Nevertheless, this work has opened the attractive possibility of targeting BIR in cancer.
RS as a fuel of tumorogenesis
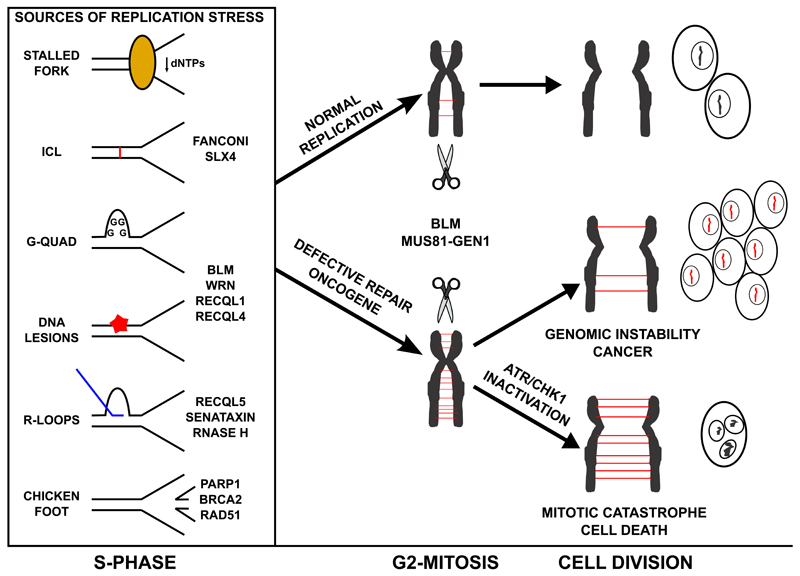
As mentioned above, RS has emerged as a major source of the deletions, translocations, amplifications and aneuploidies that are abundant in cancer. Evidence gathered in the last years shows that problems generated during replication, specially at fragile sites, can be carried on to mitosis and give rise to anaphase bridges [41]. After induction of RS, the number of anaphase bridges increases. If unresolved, these bridges can lead to chromosomal aberrations and micronuclei formation that are often found in cancer (Figure 1). In this context, several pathways have emerged to limit replication-borne genomic instability, mutations in which often derive in a higher incidence of tumours.
Figure 1. Replication Stress and Cancer.
Each S-phase, the replication machinery encounters different roadblocks and cells have specific systems to deal with each of them (left). Some of these alterations remain unresolved through G2 and into mitosis, generating DNA bridges during segregation. Under normal conditions the dissolution and resolution pathways take care of these joint DNA molecules (JM) during G2-Mitosis (centre, top), allowing for the completion of cell division (right, top). In the presence of oncogenes or when some of the mechanisms of repair are mutated or lost, the accumulation of JM in mitosis is too high (centre, bottom) and alterations are carried on through the cellular division, giving rise to genomic instability and cancer (right, middle). In the context of the high levels of RS that can be found on certain tumors, the inhibition of ATR and/or Chk1 is highly cytotoxic since it leads to (a) further amounts of RS and (b) it facilitates the entry into mitosis with unreplicated loci leading to mitotic catastrophe and cell death (right, bottom).
Fanconi Anemia (FA) is a cancer-prone hereditary disease that is caused by mutations in proteins that repair DNA interstrand crosslinks (ICL) [42]. When a replication fork encounters a crosslink, the FA pathway is activated and the FANCD2-FANCI complex is ubiquitinated. This recruits the scaffold protein SLX4, which in turn brings several structure specific nucleases to the fork to free the linked strands by cleavage and allow the subsequent repair through homologous recombination [43,44]. Different mechanisms have been proposed to explain the predisposition to cancer observed upon FA protein mutation. On one hand, the absence of FA may favour DNA repair through non-homologous end joining, an error-prone mechanism. On the other hand, when FA activities are missing ICL or joint DNA molecules accumulate, which might be another source of genomic instability that drives carcinogenesis [45]. The latter idea is supported by the cancer prone phenotype observed in SLX4 or MUS81 deficient mice [44,46]. Besides its role in dealing with ICL, recent evidence has shown that the FA pathway might also be involved in safeguarding the integrity of replication forks. For instance, analysis of the composition of the replisome after treatment with hydroxyurea revealed the recruitment of FANCD2 through the interaction with phosphorylated MCM2, where it would slow down replication to allow fork stabilization [47]. FANCD2 has also been involved in the protection of reversed forks in cooperation with BRCA2 and Rad51 [34,48]. Obstacles such as bulky adducts might require the regression of the fork to give access to the appropriate repair machinery. This generates a chicken foot structure that is protected by Rad51 filaments, and is stabilized by BRCA1/2 and FANCD2. In the absence of these proteins a nuclease activity involving Mre11 degrades the regressed fork leading to the generation of broken ends and genomic instability. Finally, FA proteins are also localized to the anaphase bridges, where they cooperate with the DNA helicase BLM in the resolution of these structures [49,50]. All of the above indicate a key role for the FA pathway in the suppression of RS and replication-borne genomic instability.
In addition to BRCA1/2 and FA proteins, the repair of stalled forks demands a number of additional activities, some of which are also linked to human disease. One of such activities is that of DNA helicases that recognize and process specific toxic structures generated during replication. The RECQ family of helicases has been particularly linked to RS and disease [51]. The different members of the family show multiple roles in replication during initiation, elongation, lagging strand processing and fork repair. BLM and WRN mediate the dissolution of complex structures such as G-quadruplexes to facilitate replication and avoid fork stalling during S phase. In fact, the absence of these proteins increases the expression of common fragile sites associated with the presence of these structures. Besides preventing fork stalling, BLM, WRN, RECQL1 and RECQL4 are important in the repair and restart of stalled forks. First, they mediate the recombination steps required to skip bulky lesions that will be processed later by specific repair mechanisms [52]. Additionally, they work on fork regression and restoration of chicken foot structures to normal forks that may be generated by lesions such as ICL [53]. In fact, the absence of these proteins increases genome instability as reflected by the expression of common fragile sites, along with increased sister chromatid exchanges and anaphase bridges [49,50,54]. Defects in BLM, WRN and RECQL4 cause Bloom, Werner and Rothmund-Thomson syndromes, respectively, which are associated with cancer and accelerated ageing phenotypes. In contrast to the shared roles of BLM, WRN and RECQL4, RECQL5 is specifically associated with RNA polymerase II [55,56]. Yet, similar to the other family members, deletion of RECQL5 also results in a cancer prone phenotype [57]. A recent paper has shown that RECQL5 slows down RNA polymerase II elongation and increases the robustness of transcription. Depletion of RECQL5 increases polymerase pausing and leads to genomic instability with gains and losses in common fragile sites and long genes, further supporting that stalled forks, regardless of the cause, can ultimately derive into pro-tumoral mutations [58]. These data suggest that RECQL5 may be limiting the generation of R-loops, yet further analyses should address this point.
Apart from the RECQ family, SNF2 helicases also present roles in the RS response. First, the PICH helicase has a role in the processing of anaphase bridges [59,60]. Although PICH might not be directly involved in the dissolution of the bridge, it is required to stabilize the DNA and recruit BLM to allow their repair [61]. Second, the SMARCAL1 helicase is recruited to stalled forks, interacts with WRN and RPA, and is essential for the restart of forks after stress [62–64]. Defects in SMARCAL1 are the cause of the Schimke immunoosseous dysplasia (Siod) that is characterized by growth retardation, skeletal abnormalities and a severe immunodeficiency. Whereas it has been proposed that the patients suffering from Siod have increased cancer susceptibility, the severity of the phenotype precludes a deeper analysis, as most of the patients succumb to infections at a very young age.
RS is a direct source of joint DNA molecules (JM), where sister chromatids or even different chromosomes remain linked by DNA. JM can be generated at stalled replication forks through unreplicated DNA linking sister chromatids [65], or by recombinational repair of forks that generate Holliday Junction structures and further promote inter-sister bridges. The persistence of JM into mitosis can account for the segregation problems derived from the accumulation of RS. The action of DNA helicases such as BLM is particularly relevant for the dissolution of JM through a pathway that does not involve the generation of chromosome breaks. In the absence of BLM-dependent dissolution, a backup genotoxic pathway (resolution) is activated which involves the cleavage of the JM by structure-specific endonucleases and recombinational repair. In contrast to the action of helicases, resolution by the nucleases generates a strong increase in the number of chromosomal aberrations observed in mitosis, such as sister chromatid exchanges [66]. SLX1-SLX4, MUS81-EME1 and GEN1 are the main structure-specific nucleases involved in the processing of stalled replication forks through the resolution pathway [67–69]. Their activity is restricted to the G2/M phases of the cell cycle where they are in charge of the resolution of persistent Holliday junctions that have not been dissolved by the helicase system (Figure 1). The resolution by endonucleases implies the generation of double strand breaks and favours their repair through non-homologous end joining, an error prone pathway that may increase genome instability [70]. Interestingly, a recent report indicates that the inactivation of these endonucleases also reduces replication fork speed, highlighting their important mechanism as a back-up repair system [71]. Besides their mitotic roles, MUS81-EME2 has been described as a new S-phase specific nuclease with a role in fork restart after damage [72], indicating that helicases and nucleases might work together to minimize the detrimental effects of damaged forks during DNA replication. Regardless of their temporal regulation, the JM-resolution and/or repair activities of helicases are necessary to prevent carcinogenesis, further supporting the role of RS as a driver of tumorigenesis.
Finally, we can also mention the emerging roles of poly(ADP)ribosylation (PAR) mediated by PARP proteins, that are receiving increasing attention as targets for cancer therapy. PARP1 is rapidly recruited to damage sites to initiate PARylation, a phenomenon that is also observed at stalled replication forks. The main role proposed for PARylation in genome stability is linked to the recruitment of specific proteins through PAR binding domains such as the macro domain. At replication forks, the establishment of a PAR scaffold is necessary for the reversal of forks generated by TopI inhibition [73,74]. In addition, PAR protects reversed forks from MRE11 mediated degradation by either physically impeding the access to the fork or by cooperating in the recruitment of BRCA2 and RAD51. Additionally, PARylation also blocks the access of the RECQL1 helicase and prevents the restart of the fork before it has been repaired [53]. Finally, PARylation at stalled forks also transiently recruits the scaffold protein SAFB1. This recruitment is essential to amplify the ATR signal and elicit a strong phosphorylation of its main substrate H2AX. Importantly, SAFB1 is recurrently loss in human tumours, underscoring the role of this pathway in human cancer [75].
RS as a brake for tumorigenesis
All of the observations mentioned above suggest that mild levels of RS allow the accumulation of genome instability that can drive tumorigenesis. However, whether ATR and Chk1, the main kinases of the RS-response, operate as tumour suppressors in human cancer remains unclear. First, heterozygous deletion of ATR or Chk1 in mice only mildly increases tumour incidence [3,5]. Second, comprehensive analysis of cancer genomes has failed to find a significant incidence of mutations in ATR or Chk1 in human tumours (http://cancergenome.broadinstitute.org), and no examples of homozygous mutations has been ever found. On the contrary, ATR and Chk1 expression are frequently upregulated in cancer, in part because they are cell-cycle regulated genes and tumours have high proliferative indexes. Moreover, the expression of Chk1 is under the control of oncogenes such as Myc or E2F [76,77], which might be responsible for its enhanced levels in certain tumours. Why would tumours select for higher ATR and Chk1 levels? We believe that excessive levels RS are deleterious even in the context of a transformed cell. Cells cannot complete mitosis with unreplicated regions of their genome, and will undergo mitotic catastrophe or polyploidization if RS persists into mitosis (Figure 1). In this context, tumours need to maintain a proficient RS-response to cope with the high levels of RS that are induced by oncogenes and ensure their survival. In support for these ideas, a mild increase of Chk1 levels facilitates transformation with Ras and E1A by reducing the amount of RS-induced by the oncogenes and thus limiting the toxicity of the transformation process [78]. In this context, we believe that oncogenes that promote DNA replication, such as E2F or Myc, will upregulate factors that are necessary to cope with the increased replication rates (RS-buffers) as part of their transcriptional response. For instance, and in addition to the control of Chk1 by Myc and E2F, Myc also promotes the expression of RRM2, which would buffer RS by increasing dNTP pools [19]. Along these lines, we have recently showed that Chk1 expression is also under the control of the oncogene c-fos, which might facilitate transformation by limiting fos-induced RS [79]. Altogether, these results support that, rather than losing ATR and Chk1, tumours bearing high levels of RS become addicted to a proficient ATR-Chk1 response, which they need to limit the deleterious consequences of RS and constitutes the basis for the use of ATR and Chk1 inhibitors in cancer chemotherapy.
Targeting RS in cancer treatment
The rationale for targeting ATR in cancer stems from the study of mouse models with reduced ATR activity that showed a synthetic lethal effect between ATR deletion or hypomorphism and the loss of p53 [80,81] suggesting that the inhibition of ATR could be particularly useful for the treatment of p53-deficient tumours. The analysis of tissues deficient in ATR and p53 revealed that the absence of p53 further promoted the accumulation of RS in ATR mutant cells, likely due to a more promiscuous S-phase entry driven by p53 deficiency. These results suggested that low levels of ATR could also be particularly toxic in the context of other mutations that promote DNA replication. Accordingly, subsequent studies showed that low levels of ATR prevented the onset of Myc-induced Burkitt lymphomas in mice [10], a tumour type that presents high level of RS. Besides Myc, reduced levels of ATR were also shown to be particularly limiting for leukemias initiated by an MLL-ENL translocation or for H-Ras driven p53-null fibrosarcomas [11]. All of the above indicated that targeting ATR could be particularly toxic for certain tumours, particularly those with high levels of RS, and prompted the development of ATR inhibitors. One limitation of this strategy is the lack of appropriate biomarkers of RS that work on clinically relevant samples. So far, pan-nuclear histone H2AX phoshorylation has been used as a surrogate marker, given that it is restricted to S-phase cells and is detected in cells that show RPA foci [10,81–83]. An interesting alternative is the detection of 53BP1 bodies, large nuclear 53BP1 foci which accumulate preferentially at fragile sites in G1 cells after exposure to RS [84,85]. However, both strategies have their limitations and better biomarkers are needed for an accurate identification of RS.
Selective ATR inhibitors only started to emerge in 2011 and were derived by independent strategies [81,86–88]. In our case, we employed a screening based on a cellular model of “at will” ATR activation upon the addition of an inert derivative of tamoxifen [81]. In agreement with the previous mouse data, ATR inhibitors were particularly toxic for cells expressing S-phase promoting oncogenes such as cyclin E, specially in the context of p53-deficiency [81]. Additionally, ATR inhibition was toxic for ATM deficient cells [87], a synthetic lethal effect that had already been described with mouse models [9]. The availability of these compounds has allowed the discovery of new synthetic lethal interactions that confer sensitivity to ATR inhibitors and which so far include mutations in XRCC1, ERCC1-XPF or BRCA1 [89–91]. These studies will help to direct the use of ATR inhibitors towards specific mutations. It is expected that most mutations that sensitize to ATR inhibitors will be related to the metabolism of replication forks and these studies will help in the design of combined drug treatments that would benefit from the use of these compounds. In fact, our early works revealed that ATR inhibitors enhance the toxicity of Chk1 inhibitors [81]. Given that drugs never achieve a full inhibition of their targets, we think that the addition of ATR inhibitors further decreases Chk1 activity in Chk1 inhibitor-treated cells, resulting in a higher accumulation of replicative stress than with any of the individual treatments. Whereas some studies report a radiosensitization effect of these drugs, genetic data do not support a major role of this pathway in the sensitization against ionizing radiation, and the potential off-target effects of the inhibitors on other DNA repair pathways might be accountable for these effects. In contrast, both ATR and Chk1 inhibitors strongly synergize with drugs that promote replication fork stalling such as gemcitabine, etoposide or cisplatin [91]. While most data with ATR inhibitors so far relates to in vitro studies due to the limited availability of ATR inhibitors that can work in vivo, some of the observations from ATR mutant mouse models could be validated with Chk1 inhibitors. First, Chk1 inhibitors showed efficacy on the treatment of Myc-lymphoma xenografts [10,76]. In addition, high levels of Chk1 are induced by N-Myc in a xenograft model of neuroblastoma, making the tumours highly sensitive to the treatment with Chk1 inhibitors [92,93]. Promising results with ATR inhibitors in vivo are also starting to emerge. For instance, these compounds limit the growth of colon cancer xenograft models with high basal levels of RS [86]. It should be noted that a recent work has identified that ATR is also essential for the activation of a checkpoint that responds to mechanical and osmotic stress, and which coordinates chromatin condensation with the breakdown of the nuclear envelope [94]. To what extent the effect of ATR inhibitors in cancer cells is also influenced by this novel checkpoint remains unknown. In any case, with the first series of ATR inhibitors now entering into early clinical trials, the next years should give a clearer picture of the potential to use RS-response targeting compounds in cancer treatment (Figure 1).
Besides ATR or Chk1 inhibitors, an alternative strategy could be to target the availability of nucleotides that, as mentioned, becomes particularly limiting for cells suffering from RS. Several compounds targeting the ribonucletide reductase have been used for cancer treatment or are currently in clinical trials [95]. Hydroxyurea is the best-known of the RRM2 inhibitors although its effectiveness is reduced in the clinic due to its poor pharmacokinetic properties [96]. Gemcitabine is a nucleoside analogue that affects DNA replication and inhibits RRM1, and which is widely used in the clinic [97]. The contribution of the depletion of the dNTP pool to the action of gemcitabine is not clear, but recent reports show that the employment of CHK1 inhibitors can enhance the effects of gemcitabine treatment [92,98–100]. The disruption of the interaction of RRM1 with RRM2 also depletes the dNTP pool, inhibiting cancer cell growth in vitro and in vivo, and overcoming resistance to gemcitabine [101]. However, it is important to note that cells not only synthesize nucleotides but can also scavenge them from the environment. In fact, a recent study revealed that the efficiency of chemotherapeutic strategies targeting nucleotide biosynthesis is greatly enhanced by simultaneously targeting the nucleotide savage pathway [102]. The need of cancer cells to maintain a healthy pool of dNTPs is highlighted by the recent discovery of inhibitors of the MTH1 enzyme, responsible for sanitizing oxidized nucleotides in the cells. MTH1 inhibition leads to an increase in DNA damage, and is preferentially toxic for cancer cells due to the increased levels of oxidation stress [103,104]. Interestingly, studies in yeast suggest that ATR might suppress RS by modulating nucleotide metabolic pathways [105,106], further linking the connection between the RS-targeting strategies. In 1948, a young Sidney Farber showed that antifolates could promote remissions in children with leukemia [107], providing one of the first examples of cancer chemotherapy. More than half a century of research later it seems that targeting nucleotide metabolism might still provide one of the most beneficial routes for the sensitization of cancer cells, and ATR inhibitors now bring a promising new route to further exploit this avenue.
Acknowledgements
Work in the OF laboratory is supported by the Spanish Ministry of Science (SAF2011-23753), Association for International Cancer Research (12-0229), Fundació LaMarató de TV3 (33/C (2013)), Howard Hughes Medical Institute and the European Research Council (ERC-210520).
References
- 1.López-Contreras AJ, Fernandez-Capetillo O. The ATR barrier to replication-born DNA damage. DNA Repair (Amst.) 2010;9:1249–1255. doi: 10.1016/j.dnarep.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eykelenboom JK, Harte EC, Canavan L, Pastor-Peidro A, Calvo-Asensio I, Llorens-Agost M, et al. ATR activates the S-M checkpoint during unperturbed growth to ensure sufficient replication prior to mitotic onset. Cell Rep. 2013;5:1095–1107. doi: 10.1016/j.celrep.2013.10.027. [DOI] [PubMed] [Google Scholar]
- 3.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 4.de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, Carr AM, et al. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol. 2000;10:479–482. doi: 10.1016/s0960-9822(00)00447-4. [DOI] [PubMed] [Google Scholar]
- 5.Q Liu, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 6.Takai H, Tominaga K, Motoyama N, Minamishima YA, Nagahama H, Tsukiyama T, et al. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(-/-) mice. Genes Dev. 2000;14:1439–1447. [PMC free article] [PubMed] [Google Scholar]
- 7.Toledo LI, Altmeyer M, Rask M-B, Lukas C, Larsen DH, Povlsen LK, et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 8.O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 9.Murga M, Bunting S, Montaña MF, Soria R, Mulero F, Cañamero M, et al. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat Genet. 2009;41:891–898. doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murga M, Campaner S, López-Contreras AJ, Toledo LI, Soria R, Montaña MF, et al. Exploiting oncogene-induced replicative stress for the selective killing of Mycdriven tumors. Nat Struct Mol Biol. 2011;18:1331–1335. doi: 10.1038/nsmb.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoppy DW, Ragland RL, Gilad O, Shastri N, Peters AA, Murga M, et al. Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR. The Journal of Clinical Investigation. 2012;122:241–252. doi: 10.1172/JCI58928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 13.Ekholm-Reed S, Mendez J, Tedesco D, Zetterberg A, Stillman B, Reed SI. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. J Cell Biol. 2004;165:789–800. doi: 10.1083/jcb.200404092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shima N, Alcaraz A, Liachko I, Buske TR, Andrews CA, Munroe RJ, et al. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet. 2007;39:93–98. doi: 10.1038/ng1936. [DOI] [PubMed] [Google Scholar]
- 15.Pruitt SC, Bailey KJ, Freeland A. Reduced Mcm2 expression results in severe stem/progenitor cell deficiency and cancer. Stem Cells. 2007;25:3121–3132. doi: 10.1634/stemcells.2007-0483. [DOI] [PubMed] [Google Scholar]
- 16.Bagley BN, Keane TM, Maklakova VI, Marshall JG, Lester RA, Cancel MM, et al. A dominantly acting murine allele of Mcm4 causes chromosomal abnormalities and promotes tumorigenesis. PLoS Genet. 2012;8:e1003034. doi: 10.1371/journal.pgen.1003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyperreplication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 18.Jones RM, Mortusewicz O, Afzal I, Lorvellec M, García P, Helleday T, et al. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene. 2013;32:3744–3753. doi: 10.1038/onc.2012.387. [DOI] [PubMed] [Google Scholar]
- 19.Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 21.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 22.Y Aye, M Li, Long MJC, Weiss RS. Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies. Oncogene. 2014 doi: 10.1038/onc.2014.155. [DOI] [PubMed] [Google Scholar]
- 23.Aird KM, Zhang G, Li H, Z Tu, Bitler BG, Garipov A, et al. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Rep. 2013;3:1252–1265. doi: 10.1016/j.celrep.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helmrich A, Ballarino M, Nudler E, Tora L. Transcription-replication encounters, consequences and genomic instability. Nat Struct Mol Biol. 2013;20:412–418. doi: 10.1038/nsmb.2543. [DOI] [PubMed] [Google Scholar]
- 25.Aguilera A, García-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 26.Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roy D, Lieber MR. G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol Cell Biol. 2009;29:3124–3133. doi: 10.1128/MCB.00139-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helmrich A, Ballarino M, Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell. 2011;44:966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 29.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 30.Alzu A, Bermejo R, Begnis M, Lucca C, Piccini D, Carotenuto W, et al. Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell. 2012;151:835–846. doi: 10.1016/j.cell.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yüce Ö, West SC. Senataxin, defective in the neurodegenerative disorder ataxia with oculomotor apraxia 2, lies at the interface of transcription and the DNA damage response. Mol Cell Biol. 2013;33:406–417. doi: 10.1128/MCB.01195-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuduri S, Crabbé L, Conti C, Tourrière H, Holtgreve-Grez H, Jauch A, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11:1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-Strand Break Repair-Independent Role for BRCA2 in Blocking Stalled Replication Fork Degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhatia V, Barroso SI, García-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014 doi: 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]
- 36.Dereli-Öz A, Versini G, Halazonetis TD. Studies of genomic copy number changes in human cancers reveal signatures of DNA replication stress. Mol Oncol. 2011;5:308–314. doi: 10.1016/j.molonc.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barlow JH, Faryabi RB, Callén E, Wong N, Malhowski A, Chen H-T, et al. Identification of early replicating fragile sites that contribute to genome instability. Cell. 2013;152:620–632. doi: 10.1016/j.cell.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller M-C, Shaikh N, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, et al. Break-Induced Replication Repair of Damaged Forks Induces Genomic Duplications in Human Cells. Science. 2013 doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malkova A, Ira G. Break-induced replication: functions and molecular mechanism. Curr Opin Genet Dev. 2013;23:271–279. doi: 10.1016/j.gde.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan KL, Hickson ID. New insights into the formation and resolution of ultra-fine anaphase bridges. Semin Cell Dev Biol. 2011;22:906–912. doi: 10.1016/j.semcdb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 42.Clauson C, Schärer OD, Niedernhofer L. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harb Perspect Med. 2013;3:a012732. doi: 10.1101/cshperspect.a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klein Douwel D, Boonen RACM, Long DT, Szypowska AA, Räschle M, Walter JC, et al. XPF-ERCC1 acts in Unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell. 2014;54:460–471. doi: 10.1016/j.molcel.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hodskinson MRG, Silhan J, Crossan GP, Garaycoechea JI, Mukherjee S, Johnson CM, et al. Mouse SLX4 is a tumor suppressor that stimulates the activity of the nuclease XPF-ERCC1 in DNA crosslink repair. Mol Cell. 2014;54:472–484. doi: 10.1016/j.molcel.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pamidi A, Cardoso R, Hakem A, Matysiak-Zablocki E, Poonepalli A, Tamblyn L, et al. Functional interplay of p53 and Mus81 in DNA damage responses and cancer. Cancer Res. 2007;67:8527–8535. doi: 10.1158/0008-5472.CAN-07-1161. [DOI] [PubMed] [Google Scholar]
- 47.Lossaint G, Larroque M, Ribeyre C, Bec N, Larroque C, Décaillet C, et al. FANCD2 binds MCM proteins and controls replisome function upon activation of s phase checkpoint signaling. Mol Cell. 2013;51:678–690. doi: 10.1016/j.molcel.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 48.Schlacher K, Wu H, Jasin M. A Distinct Replication Fork Protection Pathway Connects Fanconi Anemia Tumor Suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 50.Naim V, Rosselli F. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol. 2009;11:761–768. doi: 10.1038/ncb1883. [DOI] [PubMed] [Google Scholar]
- 51.Croteau DL, Popuri V, Opresko PL, Bohr VA. Human RecQ Helicases in DNA Repair, Recombination, and Replication. Annu Rev Biochem. 2014;83:519–552. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamath-Loeb AS, Lan L, Nakajima S, Yasui A, Loeb LA. Werner syndrome protein interacts functionally with translesion DNA polymerases. Proc Natl Acad Sci USA. 2007;104:10394–10399. doi: 10.1073/pnas.0702513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berti M, Ray Chaudhuri A, Thangavel S, Gomathinayagam S, Kenig S, Vujanovic M, et al. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat Struct Mol Biol. 2013;20:347–354. doi: 10.1038/nsmb.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pirzio LM, Pichierri P, Bignami M, Franchitto A. Werner syndrome helicase activity is essential in maintaining fragile site stability. J Cell Biol. 2008;180:305–314. doi: 10.1083/jcb.200705126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Islam MN, Fox D, Guo R, Enomoto T, Wang W. RecQL5 promotes genome stabilization through two parallel mechanisms--interacting with RNA polymerase II and acting as a helicase. Mol Cell Biol. 2010;30:2460–2472. doi: 10.1128/MCB.01583-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aygün O, Xu X, Liu Y, Takahashi H, Kong SE, Conaway RC, et al. Direct inhibition of RNA polymerase II transcription by RECQL5. J Biol Chem. 2009;284:23197–23203. doi: 10.1074/jbc.M109.015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Y Hu, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saponaro M, Kantidakis T, Mitter R, Kelly GP, Heron M, Williams H, et al. RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell. 2014;157:1037–1049. doi: 10.1016/j.cell.2014.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. Embo J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baumann C, Körner R, Hofmann K, Nigg EA. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell. 2007;128:101–114. doi: 10.1016/j.cell.2006.11.041. [DOI] [PubMed] [Google Scholar]
- 61.Biebricher A, Hirano S, Enzlin JH, Wiechens N, Streicher WW, Huttner D, et al. PICH: a DNA translocase specially adapted for processing anaphase bridge DNA. Mol Cell. 2013;51:691–701. doi: 10.1016/j.molcel.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Couch FB, Bansbach CE, Driscoll R, Luzwick JW, Glick GG, Bétous R, et al. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 2013;27:1610–1623. doi: 10.1101/gad.214080.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ciccia A, Bredemeyer AL, Sowa ME, Terret M-E, Jallepalli PV, Harper JW, et al. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 2009;23:2415–2425. doi: 10.1101/gad.1832309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan J, Ghosal G, Chen J. The annealing helicase HARP protects stalled replication forks. Genes Dev. 2009;23:2394–2399. doi: 10.1101/gad.1836409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Y Liu, Nielsen CF, Yao Q, Hickson ID. The origins and processing of ultra fine anaphase DNA bridges. Curr Opin Genet Dev. 2014;26C:1–5. doi: 10.1016/j.gde.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 66.Wechsler T, Newman S, West SC. Aberrant chromosome morphology in human cells defective for Holliday junction resolution. Nature. 2011;471:642–646. doi: 10.1038/nature09790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wyatt HDM, Sarbajna S, Matos J, West SC. Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells. Mol Cell. 2013;52:234–247. doi: 10.1016/j.molcel.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 68.Castor D, Nair N, Déclais A-C, Lachaud C, Toth R, Macartney TJ, et al. Cooperative control of holliday junction resolution and DNA repair by the SLX1 and MUS81-EME1 nucleases. Mol Cell. 2013;52:221–233. doi: 10.1016/j.molcel.2013.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garner E, Kim Y, Lach FP, Kottemann MC, Smogorzewska A. Human GEN1 and the SLX4-associated nucleases MUS81 and SLX1 are essential for the resolution of replication-induced Holliday junctions. Cell Rep. 2013;5:207–215. doi: 10.1016/j.celrep.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.LaRocque JR, Stark JM, Oh J, Bojilova E, Yusa K, Horie K, et al. Interhomolog recombination and loss of heterozygosity in wild-type and Bloom syndrome helicase (BLM)-deficient mammalian cells. Proc Natl Acad Sci USA. 2011;108:11971–11976. doi: 10.1073/pnas.1104421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sarbajna S, Davies D, West SC. Roles of SLX1-SLX4, MUS81-EME1, and GEN1 in avoiding genome instability and mitotic catastrophe. Genes Dev. 2014;28:1124–1136. doi: 10.1101/gad.238303.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pepe A, West SC. MUS81-EME2 promotes replication fork restart. Cell Rep. 2014;7:1048–1055. doi: 10.1016/j.celrep.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bryant HE, Petermann E, Schultz N, Jemth A-S, Loseva O, Issaeva N, et al. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. Embo J. 2009;28:2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, Bermejo R, et al. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat Struct Mol Biol. 2012;19:417–423. doi: 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- 75.Altmeyer M, Toledo L, Gudjonsson T, Grøfte M, Rask M-B, Lukas C, et al. The Chromatin Scaffold Protein SAFB1 Renders Chromatin Permissive for DNA Damage Signaling. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.08.025. [DOI] [PubMed] [Google Scholar]
- 76.Höglund A, Nilsson LM, Muralidharan SV, Hasvold LA, Merta P, Rudelius M, et al. Therapeutic implications for the induced levels of Chk1 in Myc-expressing cancer cells. Clin Cancer Res. 2011;17:7067–7079. doi: 10.1158/1078-0432.CCR-11-1198. [DOI] [PubMed] [Google Scholar]
- 77.Verlinden L, Vanden Bempt I, Eelen G, Drijkoningen M, Verlinden I, Marchal K, et al. The E2F-regulated gene Chk1 is highly expressed in triple-negative estrogen receptor /progesterone receptor /HER-2 breast carcinomas. Cancer Res. 2007;67:6574–6581. doi: 10.1158/0008-5472.CAN-06-3545. [DOI] [PubMed] [Google Scholar]
- 78.López-Contreras AJ, Gutierrez-Martinez P, Specks J, Rodrigo-Perez S, Fernandez-Capetillo O. An extra allele of Chk1 limits oncogene-induced replicative stress and promotes transformation. J Exp Med. 2012;209:455–461. doi: 10.1084/jem.20112147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schulze J, López-Contreras AJ, Uluçkan O, Graña-Castro O, Fernandez-Capetillo O, Wagner EF. Fos-dependent induction of Chk1 protects osteoblasts from replication stress. Cell Cycle. 2014;13:1980–1986. doi: 10.4161/cc.28923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ruzankina Y, Schoppy DW, Asare A, Clark CE, Vonderheide RH, Brown EJ. Tissue regenerative delays and synthetic lethality in adult mice after combined deletion of Atr and Trp53. Nat Genet. 2009;41:1144–1149. doi: 10.1038/ng.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, et al. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol. 2011;18:721–727. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gagou ME, Zuazua-Villar P, Meuth M. Enhanced H2AX phosphorylation, DNA replication fork arrest, and cell death in the absence of Chk1. Mol Biol Cell. 2010;21:739–752. doi: 10.1091/mbc.E09-07-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Syljuåsen RG, Sørensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25:3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Harrigan JA, Belotserkovskaya R, Coates J, Dimitrova DS, Polo SE, Bradshaw CR, et al. Replication stress induces 53BP1-containing OPT domains in G1 cells. J Cell Biol. 2011;193:97–108. doi: 10.1083/jcb.201011083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243–253. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- 86.Foote KM, Blades K, Cronin A, Fillery S, Guichard SS, Hassall L, et al. Discovery of 4-{4-[(3R)-3-Methylmorpholin-4-yl]-6-[1-(methylsulfonyl)cyclopropyl]pyrimidin-2-yl}-1H-indole (AZ20): a potent and selective inhibitor of ATR protein kinase with monotherapy in vivo antitumor activity. J Med Chem. 2013;56:2125–2138. doi: 10.1021/jm301859s. [DOI] [PubMed] [Google Scholar]
- 87.Reaper PM, Griffiths MR, Long JM, Charrier J-D, Maccormick S, Charlton PA, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol. 2011;7:428–430. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 88.Charrier J-D, Durrant SJ, Golec JMC, Kay DP, Knegtel RMA, Maccormick S, et al. Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J Med Chem. 2011;54:2320–2330. doi: 10.1021/jm101488z. [DOI] [PubMed] [Google Scholar]
- 89.Mohni KN, Kavanaugh GM, Cortez D. ATR pathway inhibition is synthetically lethal in cancer cells with ERCC1 deficiency. Cancer Res. 2014 doi: 10.1158/0008-5472.CAN-13-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sultana R, Abdel-Fatah T, Perry C, Moseley P, Albarakti N, Mohan V, et al. Ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase inhibition is synthetically lethal in XRCC1 deficient ovarian cancer cells. PLoS ONE. 2013;8:e57098. doi: 10.1371/journal.pone.0057098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huntoon CJ, Flatten KS, Wahner Hendrickson AE, Huehls AM, Sutor SL, Kaufmann SH, et al. ATR inhibition broadly sensitizes ovarian cancer cells to chemotherapy independent of BRCA status. Cancer Res. 2013;73:3683–3691. doi: 10.1158/0008-5472.CAN-13-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Walton MI, Eve PD, Hayes A, Valenti MR, De Haven Brandon AK, Box G, et al. CCT244747 is a novel potent and selective CHK1 inhibitor with oral efficacy alone and in combination with genotoxic anticancer drugs. Clin Cancer Res. 2012;18:5650–5661. doi: 10.1158/1078-0432.CCR-12-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cole KA, Huggins J, Laquaglia M, Hulderman CE, Russell MR, Bosse K, et al. RNAi screen of the protein kinome identifies checkpoint kinase 1 (CHK1) as a therapeutic target in neuroblastoma. Proc Natl Acad Sci USA. 2011;108:3336–3341. doi: 10.1073/pnas.1012351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kumar A, Mazzanti M, Mistrik M, Kosar M, Beznoussenko GV, Mironov AA, et al. ATR Mediates a Checkpoint at the Nuclear Envelope in Response to Mechanical Stress. Cell. 2014;158:633–646. doi: 10.1016/j.cell.2014.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wijerathna SR, Ahmad MF, Xu H, Fairman JW, Zhang A, Kaushal PS, et al. Targeting the Large Subunit of Human Ribonucleotide Reductase for Cancer Chemotherapy. Pharmaceuticals (Basel) 2011;4:1328–1354. doi: 10.3390/ph4101328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Donehower RC. An overview of the clinical experience with hydroxyurea. Semin Oncol. 1992;19:11–19. [PubMed] [Google Scholar]
- 97.Moysan E, Bastiat G, Benoit J-P. Gemcitabine versus Modified Gemcitabine: a review of several promising chemical modifications. Mol Pharm. 2013;10:430–444. doi: 10.1021/mp300370t. [DOI] [PubMed] [Google Scholar]
- 98.Xiao Y, Ramiscal J, Kowanetz K, Del Nagro C, Malek S, Evangelista M, et al. Identification of preferred chemotherapeutics for combining with a CHK1 inhibitor. Mol Cancer Ther. 2013;12:2285–2295. doi: 10.1158/1535-7163.MCT-13-0404. [DOI] [PubMed] [Google Scholar]
- 99.Montano R, Thompson R, Chung I, Hou H, Khan N, Eastman A. Sensitization of human cancer cells to gemcitabine by the Chk1 inhibitor MK-8776: cell cycle perturbation and impact of administration schedule in vitro and in vivo. BMC Cancer. 2013;13:604. doi: 10.1186/1471-2407-13-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Blackwood E, Epler J, Yen I, Flagella M, O'Brien T, Evangelista M, et al. Combination drug scheduling defines a “window of opportunity” for chemopotentiation of gemcitabine by an orally bioavailable, selective ChK1 inhibitor. GNE-900, Mol Cancer Ther. 2013;12:1968–1980. doi: 10.1158/1535-7163.MCT-12-1218. [DOI] [PubMed] [Google Scholar]
- 101.Zhou B, Su L, Hu S, Hu W, Yip MLR, Wu J, et al. A small-molecule blocking ribonucleotide reductase holoenzyme formation inhibits cancer cell growth and overcomes drug resistance. Cancer Res. 2013;73:6484–6493. doi: 10.1158/0008-5472.CAN-13-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nathanson DA, Armijo AL, Tom M, Z Li, Dimitrova E, Austin WR, et al. Co-targeting of convergent nucleotide biosynthetic pathways for leukemia eradication. J Exp Med. 2014;211:473–486. doi: 10.1084/jem.20131738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.H Gad, Koolmeister T, Jemth A-S, Eshtad S, Jacques SA, Ström CE, et al. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature. 2014;508:215–221. doi: 10.1038/nature13181. [DOI] [PubMed] [Google Scholar]
- 104.Huber KVM, Salah E, Radic B, Gridling M, Elkins JM, Stukalov A, et al. Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature. 2014;508:222–227. doi: 10.1038/nature13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Andreson BL, Gupta A, Georgieva BP, Rothstein R. The ribonucleotide reductase inhibitor, Sml1, is sequentially phosphorylated, ubiquitylated and degraded in response to DNA damage. Nucleic Acids Res. 2010;38:6490–6501. doi: 10.1093/nar/gkq552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. Embo J. 2001;20:3544–3553. doi: 10.1093/emboj/20.13.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.FARBER S, DIAMOND LK. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N Engl J Med. 1948;238:787–793. doi: 10.1056/NEJM194806032382301. [DOI] [PubMed] [Google Scholar]