Abstract
The production of limitless carbon-free energy is a long-sought dream of scientists and politicians alike. One strategy for achieving this aim is the production of hydrogen by photosynthetic microorganisms – harnessing the effectively limitless power of the sun to power our cars, toasters and PCR machines. It may be tempting to think of host expression systems as miniature factories given over entirely to the production our molecule of interest. However, the biological nature of the host must be taken into account if we are to maximize productivity. The circadian rhythm, an organism’s entrainable oscillation of biological processes with a period of around 24 hours, is one such aspect that has received scant attention but is likely to be of particular importance to photosynthetic host systems. In this issue of current biology Xu et al. describe how our knowledge of the Synechococcus elongatus circadian clock can be leveraged to improve the production of exogeneous proteins, including those involved in the production of hydrogen [1].
Co-Opting the KaiABC Oscillator
S. elongatus has been a useful model for understanding circadian regulation of gene expression, independently of its potential utility in biotechnology [2]. The S. elongatus circadian clockwork is based upon rhythms of phosphorylation in the protein KaiC and its regulatory binding partners KaiA and KaiB. In its simplest form the KaiABC complex will undergo circadian rhythms in autophosphorylation and dephosphorylation when its constituents are mixed in vitro in the presence of ATP [3]. In vivo this basic oscillator is elaborated by autoregulation of the KaiABC gene cluster via accessory transcription factors [4]. The KaiABC oscillator also feeds into the genome-wide regulation of transcription to generate rhythmic outputs from the circadian clock [5, 6]. Genes which accumulate during the day and peak at dusk are known as Class I genes. Those with an opposing peak at dawn are termed Class II. Overexpression of the KaiA protein enhances gene expression from the KaiBC promoter [4], giving the cells an apparently continuous dusk signal. Conversely, expression of KaiC has been claimed to globally repress gene expression [7]. With this in mind, the production of potentially useful gene products might be enhanced throughout the night by exogenous overexpression of KaiA – effectively holding the cells in a permanent dusk-like state. The goal: to turn the circadian S. elongatus protein factory into a round-the-clock production line for exogenous protein.
In their efforts to stably enhance gene expression, Xu et al found that overexpression of KaiA (KaiA-OX) in constant light conditions causes about 20% of genes to be up-regulated at the transcript level. Around 12% were down-regulated by KaiA-OX. Interestingly the over-expression of KaiC has the opposite effect on individual genes. The authors describe this reciprocal regulation by KaiA and KaiC as a Yin-Yang model of regulation. Circadian rhythms of transcript abundance are produced by induction and repression of specific loci at opposing phases of the circadian cycle – presumably enhancing the amplitude of rhythmic gene expression in wild type cells.
The endogenous NAD/NADP-utilizing hydrogenase displays a peak-at-dusk (Class I) profile and has enhanced expression during KaiA-OX. This raises the tantalizing possibility of increasing hydrogenase activity by pausing the circadian oscillator at subjective dusk when these genes are most prolifically transcribed.
When is a “Neutral Site” not a neutral site?
Hydrogenases have evolved naturally in cyanobacteria to catalyze the reversible reaction 2H+ + 2e- → H2, using, NADH, NADPH and Ferrodoxin as electron donors. The direct production of H2 circumvents the inefficient Calvin cycle and offers the tantalizing possibility of highly efficient fuel production. Currently yield is poor due to the oxygen sensitivity of hydrogenases and energetic favorability of H2 uptake over production [8].
The RC41 S.elongatus strain was developed by Weyman et al in an effort to optimize H2 production from S.elongatus [9]. The endogenous bidirectional hydrogenase HoxYH was deleted and the HynSL hydrogenase and accessory gene cluster from Alteromonas macleodii expressed under the control of trcp at neutral site NS I. Unfortunately, RC41 has rather disappointing levels of hydrogenase activity owing to poor expression of hydrogenase and its accessory proteins [9].
An intriguing observation by Xu et al revealed that several constitutive promoters from Escherichia coli behave in a circadian manner when expressed in S.elongatus from “Neutral Sites” NS I and NS II. Neutral Sites are so called because their loci can be disrupted by incorporation of exogenous DNA with no aberrant phenotype in the host cell. As expression varies over the circadian cycle, it perhaps confusing to label them neutral sites [10]. In particular the conIIp constitutive promoter drives strong rhythmic expression of the luxAB reporter from NS I or NS II in S.elongatus, which has a peak-at-dusk expression profile similar to that observed in KaiA-OX induced genes. KaiA-OX potentiates luxAB activity both in constant light and constant darkness, which suggests that NS I and NS II are circadian-regulated loci.
Towards round-the-clock hydrogen production
The observed increase in endogenous hydrogenase activity during KaiA overexpression in constant light conditions, coupled with the possibility of enhanced NS I activity during KaiA-OX prompted Xu et al to optimize HynSL production in RC41.
Overexpression of KaiA in RC41 improved the expression of the HynL and increased H2 production two-fold. This dramatic improvement in the production of H2 is unlikely to spark an immediate clean energy revolution however, as the hydrogenase activity is still lower than that observed in native S.elongatus. Clearly, there exist other limiting factors which need to be overcome before S.elongatus becomes a viable system for H2 production.
Accordingly, when the KaiA-overexpressing S.elongatus was used to produce Human Pro-insulin fused to glutathione S-transferase the results were more impressive – achieving up to a five-fold increase relative to cells not overexpressing KaiA. This demonstrates that pausing the circadian clock in a locus’s most productive time window is a viable strategy for maximizing the production of exogenous proteins. Interestingly the potentiation of gene expression by KaiA-overexpression was maximal during the dark phase of the Light:Dark culture cycle which is the unexpected given that NS I integrated genes normally behave like class I genes. It may be that the general inactivity of S. elongatus gene expression machinery during the dark phase generates spare capacity for exogenous protein expression and thus allows it to occur more efficiently.
The attempt to leverage our knowledge of the circadian cycle to enhance exogenous gene expression has yielded interesting but perhaps unexpected results. The efficient production of Human Pro-Insulin during the subjective night may hint that other exogenous proteins might be better suited to expression at this time. Perhaps HynSL production would be more efficiently produced from a KaiC-OX induced locus or when present at multiple temporally divergent genetic loci – allowing an even production of protein throughout the circadian cycle without the need for the permanent ‘on’ switch of KaiA overexpression.
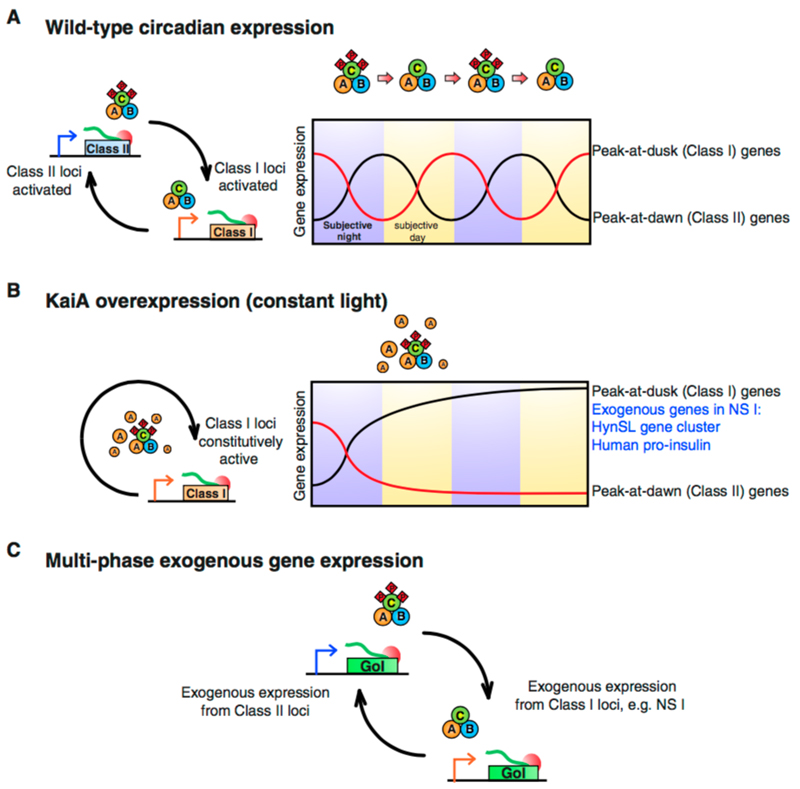
Circadian oscillations in gene expression are linked to the phosphorylation – dephosphorylation cycle of the KaiABC complex (A). Class I and Class II genes oscillate out of phase with one another, and maintain their rhythm during constant light. When KaiA is overexpressed (B) a subset of genes including Class I genes are expressed stably. This strategy was successfully employed to improve expression of exogenous HynSL hydrogenase and Human Pro-Insulin. Another potential strategy for round-the-clock expression of exogenous genes would be to incorporate genes at multiple sites with varying Class I and Class II character (C) – enabling stable expression of any gene of interest (GoI) without the need to disrupt the circadian rhythm of the host by KaiA-overexpression.
References
- 1.Xu Y, Weyman PD, Umetani M, Xiong J, Qi X, Xu Q, Iwasaki H, Johnson CH. Circadian Yin-Yang regulation and its manipulation to globally reprogram gene expression. Current BIology. 2013 doi: 10.1016/j.cub.2013.10.011. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson CH, Stewart PL, Egli M. The cyanobacterial circadian system: from biophysics to bioevolution. Annual review of biophysics. 2011;40:143–167. doi: 10.1146/annurev-biophys-042910-155317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 4.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, Golden SS, Johnson CH, Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, Tsinoremas NF, Johnson CH, Lebedeva NV, Golden SS, Ishiura M, Kondo T. Circadian orchestration of gene expression in cyanobacteria. Genes & development. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 6.Nair U, Ditty JL, Min H, Golden SS. Roles for sigma factors in global circadian regulation of the cyanobacterial genome. Journal of bacteriology. 2002;184:3530–3538. doi: 10.1128/JB.184.13.3530-3538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakahira Y, Katayama M, Miyashita H, Kutsuna S, Iwasaki H, Oyama T, Kondo T. Global gene repression by KaiC as a master process of prokaryotic circadian system. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:881–885. doi: 10.1073/pnas.0307411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrieri D, Wawrousek K, Eckert C, Yu J, Maness PC. The role of the bidirectional hydrogenase in cyanobacteria. Bioresource technology. 2011;102:8368–8377. doi: 10.1016/j.biortech.2011.03.103. [DOI] [PubMed] [Google Scholar]
- 9.Weyman PD, Vargas WA, Tong Y, Yu J, Maness PC, Smith HO, Xu Q. Heterologous expression of Alteromonas macleodii and Thiocapsa roseopersicina [NiFe] hydrogenases in Synechococcus elongatus. PloS one. 2011;6:e20126. doi: 10.1371/journal.pone.0020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson CR, Tsinoremas NF, Shelton J, Lebedeva NV, Yarrow J, Min H, Golden SS. Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods in enzymology. 2000;305:527–542. doi: 10.1016/s0076-6879(00)05511-7. [DOI] [PubMed] [Google Scholar]