Abstract
Glial cell line-derived neurotrophic factor (GDNF) has been tested in clinical trials to treat Parkinson’s disease with promising but variable results. Improvement of therapeutic effectiveness requires solid understanding of the physiological role of GDNF in the maintenance of the adult brain catecholamine system. However, existing data on this issue is contradictory. Here we show with three complementary approaches that, independent of the time of reduction, Gdnf is not required for maintenance of catecholaminergic neurons in adult mice.
Neurotrophic factors hold promise as therapeutic tools to treat neurodegenerative diseases. GDNF was identified for its ability to support and maintain the midbrain dopamine and noradrenaline neurons1, 2, which are specifically affected in Parkinson’s disease3. Clinical trials to treat Parkinson’s disease using GDNF or its family member neurturin (NRTN), have yielded promising, yet conflicting results3. Improvement of therapeutic strategies requires substantial understanding of the physiological role of endogenous GDNF in the maintenance of brain catecholamine system in adult life. Knock-out mice for Gdnf, its binding receptor Gfrα1 or its main signaling receptor Ret die upon birth mainly due to the lack of kidneys but with intact catecholamine system, rendering postnatal analysis impossible4. However, conditional ablation using lox-Cre system (termed hereon as “floxed”) of GDNF main signaling receptor Ret and Gdnf have yielded conflicting results. Jain et al reported that Ret deletion from dopamine neurons using DAT-Cre has no effect on brain dopamine system until 9 months of age, when the study was concluded5. An independent study by Kramer et al using de novo generated Ret floxed allele demonstrated that the absence of Ret, either ubiquitously in the central nervous system (CNS) or in the dopamine neurons using Nestin-Cre or DAT-Cre respectively, results in modest, age-dependent dopamine system degeneration in substantia nigra, but not in ventral tegmental area or noradrenergic locus coeruleus, starting around 12 months of age6. In contrast to the above studies, Pascual et al reported that ubiquitous GDNF reduction in 2 months old adult mice using tamoxifen/Esr1-Cre system results in dramatic degeneration of dopamine neurons both in substantia nigra and ventral tegmental area, and a complete degenerative destruction of locus coeruleus 7 months after tamoxifen injection7. The existence of alternative GDNF receptors, NCAM and Syndecan-3, and/or onset of developmental compensation in Ret floxed mice have been suggested to explain the apparent discrepancy. However, due to the obvious clinical relevance of GDNF and the lack of follow-up studies on GDNF conditional deletion since the Pascual report in 2008, the above inconsistency has remained a matter of intensive debate.
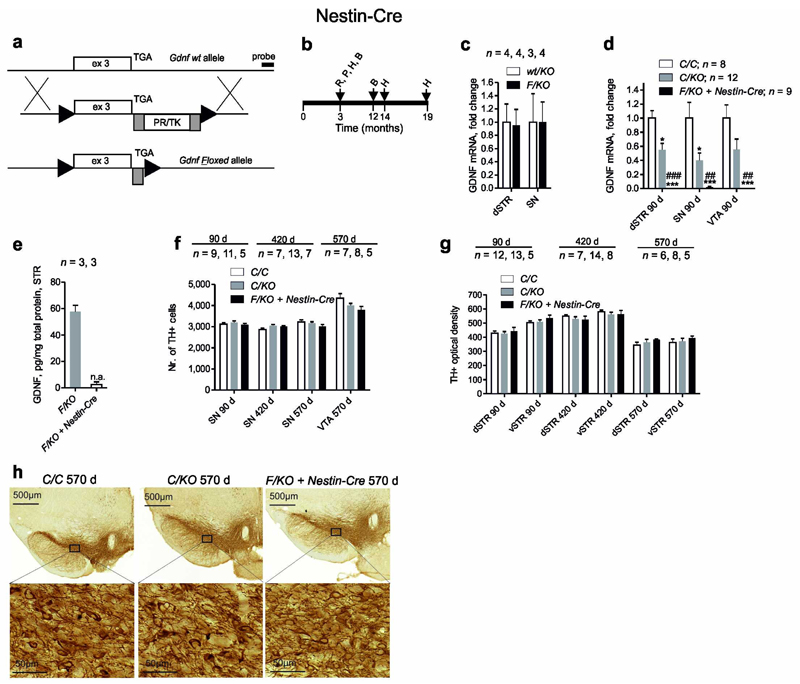
Here we report de novo generation and analysis of Gdnf floxed mice. We used the same strategy as Pascual et al7 in that Gdnf exon three encoding for GDNF protein was “floxed (Fig. 1a). We studied the effect of Gdnf deletion on the catecholamine system with a focus on the dopamine system using three different gene deletion methods: Nestin-Cre; AAV5-Cre and Esr1-Cre. Nestin-Cre deletes Gdnf from the CNS during embryonic development, while intrastriatal injection of adeno-associated virus (AAV) encoding for Cre deletes Gdnf from the innervation target of substantia nigra dopamine neurons in adult mice. Finally, we used the same tamoxifen inducible Esr1-Cre mouse line and experimental procedures as in Pascual et al study to replicate their experiments. Gdnf floxed (GdnfF/F) mice were generated, genotyped and validated using routine methods (Supplementary Fig. 1a, b; Online Methods). Nestin-Cre mice were analyzed as indicated in Fig. 1b. Gdnf mRNA expression from Gdnfwt and GdnfF alleles was comparable (Fig. 1c). As expected from earlier work6, 8, cross to Nestin-Cre resulted in CNS-specific Gdnf ablation (Fig. 1d,e Supplementary Fig. 1b,c). As in Pascual et al7, compound heterozygous GdnfF/KO mice were analyzed (Fig. 1d). Morphological analysis of the catecholamine system using immunostaining for tyrosine hydroxylase, stereological quantification of tyrosine hydroxylase expressing cells in the midbrain, and optical density (OD) measurements of dorsal striatum (dSTR) and ventral striatum (vSTR) reflecting density of catecholaminergic innervation revealed no difference between the genotypes at 3, 14 and 19 months of age (Fig. 1f–h). Locus coeruleus appeared morphologically normal at 19 months of age (Supplementary Fig. 1d). Analysis of motor function with open field and rotarod tests revealed no difference in young (2 months) and old (12 months) mice (Supplementary Table 1). Since dopamine system dysfunction is involved in schizophrenia and anxiety mice were analyzed with pre-pulse inhibition, light-dark and elevated plus-maze tests at both ages. No difference between the genotypes was found (Supplementary Table 1).
Figure 1.
Generation of Gdnf floxed mice and catecholamine system analysis after crosses to Nestin-Cre. (a) Gdnf wt (Gdnfwt) allele, targeting construct and floxed allele (GdnfF), triangles, loxP; grey rectangles, FRT sequences; PR/TK-selectable marker. (b) Experimental timeline for analysis of GdnfF/KO + Nestin-Cre animals, time points for RNA (R); protein (P); histology (H); and behavioral (B) analysis are indicated. (c) Gdnf mRNA expression from GdnfF and Gdnfwt allele, determined by qPCR. (d) Relative levels of Gdnf mRNA after cross to Nestin-Cre line, determined by qPCR (P < 0.0001 for dorsal striatum (dSTR) and substantia nigra (SN), P = 0,001 for ventral tegmental area (VTA); Kruskal-Wallis test). (e) GDNF protein levels in the striatum (STR) after cross to Nestin-Cre line. (f) Unbiased stereological cell counts of SN and VTA tyrosine hydroxylase (TH) immunoreactive neurons at indicated time points. (g) Unbiased measurement of striatal TH+ optical density reflecting TH+ fiber density. (h) Representative TH+ immunostaining of coronal midbrain slices at 19 months of age. wt-wild-type; F-floxed; KO-knock-out; C-control, containing both wt and F alleles; n = number of animals analyzed in each experiment; n.a. = not applicable; *, *** p > 0.05 or 0.001 relative to GdnfC/C, ##, ### p > 0.01 or 0.001 relative to Gdnfwt/KO. Error bars are mean ± s.e.m.
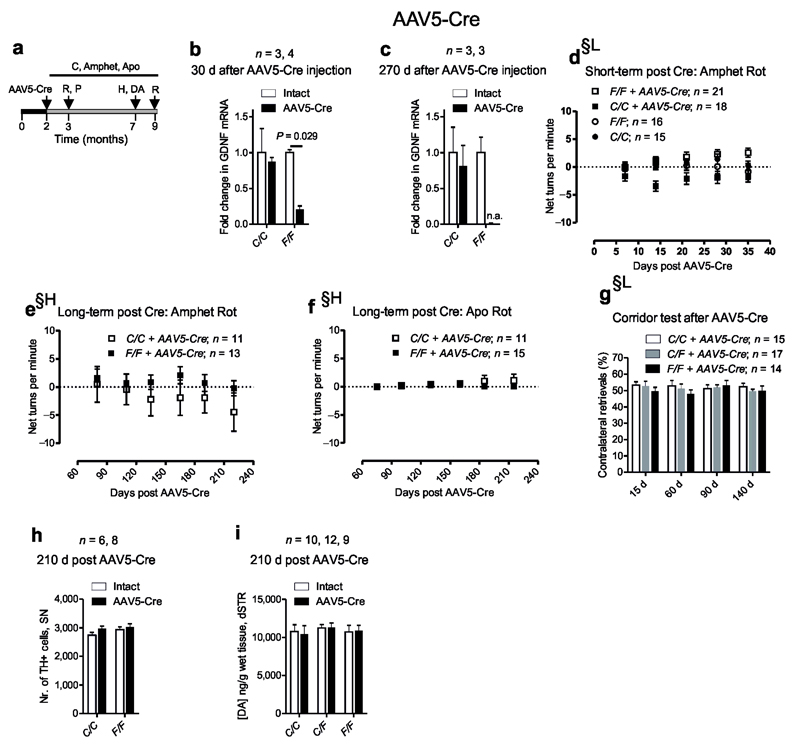
Next we tested, whether the absence of GDNF is compensated during the development by deleting Gdnf in adult animals. Mice were unilaterally striatally injected with AAV5-Cre (Supplementary Fig. 2a) and analyzed as indicated on Fig. 2a. To strengthen the conclusions, behavioral experiments were performed in parallel in two independent laboratories, in Lund University, Sweden and University of Helsinki, Finland. Analysis of striatal Gdnf mRNA levels revealed five-fold reduction at one month, and almost complete ablation at nine months post AAV5-Cre injection (Fig. 2b,c). We later measured Gdnf mRNA and GDNF protein levels using a separate batch of animals and a separate stock of the virus to confirm the acute reduction in striatal GDNF. In this batch we detected a 40 % drop in GDNF protein levels accompanied by a 30 % reduction in Gdnf mRNA 30 days post injection (Supplementary Fig. 2b,c). Unilateral decline in dopamine system function is associated with side bias in corridor test9, 10 and by amphetamine (ipsilateral), or apomorphine (contralateral) induced rotational bias11. During the seven month follow-up unilateral GDNF reduction had no impact on rotational behavior in either laboratory (Fig. 2d-g, Supplementary Fig. 2d-g), no effect on catecholaminergic cell cell survival in substantia nigra (Fig. 2h, Supplementary Fig. 2h) and no effect on striatal tissue dopamine (Fig. 2i) or its metabolite levels (Supplementary Fig. 2i,j).
Figure 2.
Dopamine system analysis after striatal Gdnf reduction by AAV5- Cre injection in adult mice. (a) Experimental timeline for analysis, time points for AAV5 encoding for synapsin promoter driven Cre injection (AAV5-Cre), RNA (R); protein (P); histology (H); striatal dopamine level measurements (DA) and behavior by corridor test (C) and after amphetamine (Amphet) or apomorphine (Apo) injections are indicated. (b–c) Relative levels of Gdnf mRNA determined by qPCR in the intact and AAV5 -Cre injected striata. (d–f) Measurements of rotations (Rot) after amphetamine (Amphet) and apomorphine (Apo) injection at the indicated time-points after AAV5 -Cre injection. (g) Corridor test at the indicated timepoints after AAV5 -Cre injection. (h) Unbiased stereological cell counts of substanti nigra tyrosine hydroxylase (TH) immunoreactive neurons 7 months (210 days) after Gdnf reduction. (i) Striatal tissue dopamine (DA) levels 7 months after Gdnf reduction. §-experiments were independently performed in Lund University, Sweden and University of Helsinki, Finland; §H-experiment performed in Helsinki; §L-experiment performed in Lund. F-floxed; C-control containing only wt alleles; n = number of animals analyzed in each experiment; n.a. = not applicable. Error bars are mean ± s.e.m.
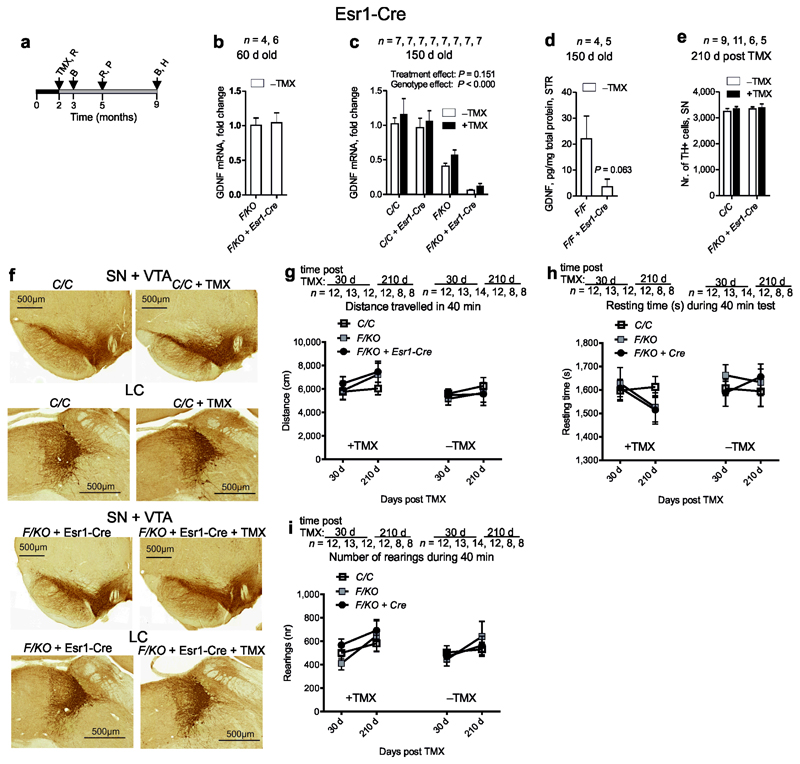
Finally, we repeated the experiments reported by Pascual et al using the same Esr1-Cre line and experimental conditions (Fig. 3a). At two months of age, Gdnf mRNA levels in GdnfF/KO mice and GdnfF/KO mice expressing Esr1-Cre were comparable (Fig. 3b). Compared to the controls, mice hemizygous for Gdnf allele displayed about 50% reduction in Gdnf mRNA levels (Fig. 3c). However, in line with a previous report on “leakiness” of Esr1-Cre12 we noted that Gdnf exon 3 is spontaneously deleted independently of tamoxifen injection between 2 and 5 months of age, resulting in an 85 % drop in Gdnf mRNA (Fig. 3c). We later measured striatal GDNF protein levels in the presence and absence of Esr1-Cre using a separate batch of GdnfF/F animals and detected an 80 % drop in GDNF protein levels at 5 months of age (Fig. 3d). Unlike Pascual et al7 who reported close to 100% destruction of locus coeruleus and around 70 % loss of dopamine cells in substantia nigra and ventral tegmental area with concomitant decline in motor activity, we found no morphological changes in any of these structures (Fig. 3e,f), , and no changes in motor activity in GdnfF/KO + Esr1-Cre animals with reduced GDNF independent of tamoxifen application (Fig. 3g-i).
Figure 3.
Dopamine system analyses after Gdnf reduction in adult mice. (a) Experimental timeline for analysis of tamoxifen (TMX) treated mice; time-points for TMX injection and brain harvesting for RNA (R); protein (P); histology (H); and behavioral analysis (B) were selected as in Pascual et al7. (b) Relative levels of Gdnf mRNA determined by qPCR in the whole brain extract at 2 months of age. (c) Relative levels of Gdnf mRNA determined by qPCR in the whole brain extract at 5 months (150 days) of age with (+) and without (−) TMX injection at 2 months. (d) GDNF protein levels in striatum (STR) at 5 months of age determined by ELISA. (e) Unbiased stereological cell counts of substantia nigra (SN) tyrosine hydroxylase (TH) immunoreactive neurons 7 months after Gdnf ablation. (f) Representative TH immunostaining of coronal midbrain displaying intact SN, ventral tegmental area (VTA) and locus coeruleus (LC) 7 months post Gdnf reduction. (g–i) Open field activity at 1 and 7 months after Gdnf reduction is not affected. F-floxed; KO-knock-out; C-control containing only wt alleles; n = number of animals analyzed in each experiment. Error bars are mean ± s.e.m.
Ret deficient central dopamine neurons do not respond to GDNF13, suggesting that RET is the main signaling receptor for GDNF in those neurons. We have with three complementary approaches shown that GDNF reduction independent of the time of deletion does not lead to any significant changes in the number and function of central monoaminergic neurons. Since GDNF is one of the four ligands activating RET signaling in the CNS4, our results are well in line with those of Ret deletion. We conclude that GDNF reduction is dispensable for maintenance of central catecholaminergic neurons in mice.
Materials and Methods
Generation and housing of Gdnf floxed mice
Mice were housed under temperature controlled conditions at 20–22°C in 12h light/dark cycle with “lights-on” at 06.00h, 2–5 animals per cage with ad libitum access to standard chow and water. All animal experiments were authorized by the national Animal Experiment Board of Finland and the Ethical Committee for the use of laboratory animals at Lund University. Mice carrying GdnfF allele were generated using routine methods. Briefly, 5668bp 5’ and 6055bp 3’ homologous arms and Gdnf exon 3 until and including the stop codon were amplified with PCR from Gdnf containing PAC (PAC ID: RP21-583-K20, CHORI) and cloned into PmeI, NotI and HindIII sites in pFlexible respectively (GenBank: CR847878.1.) to generate Gdnf allele as depicted on Fig.1a. Exon 3 of Gdnf gene encodes for GDNF protein. ES (IB10) clones that had undergone homologous recombination (17 %) were identified using standard Southern blotting with probes located outside the homologous arms. Standard karyotyping was performed prior morula aggregations. Following germ-line transmission selectable marker PR/TK in pFlexible (GenBank: CR847878.1.) was removed by cross to Deleter-FLP line resulting in GdnfF allele depicted on Fig. 1a. GdnfF mice were maintained and used in experiments in mixed genetic background (129Ola/ICR/C57bl6). Both males and females were used in all the experiments. Littermate controls were used in all the experiments with Nestin-Cre line. For the other mouse lines age and gender matched wt controls in the comparable genetic background were bred separately.
Brain dissection
For biochemistry and qPCR analyses, samples were obtained from sub-dissected brain regions using a mouse brain matrix. Mice underwent cervical-dislocation before the whole brain was washed in ice-cold saline to remove blood, and then placed in a mouse brain matrix (Stoelting, Wood Dale, IL). Slices were taken, from structures such as striatum (STR) and ventral midbrain. STR, substantia nigra (SN) and ventral tegmental area (VTA) samples were dissected using either a sample corer (inner diameter, 2 mm) or a scalpel. Samples were snap-frozen on dry-ice and stored at –80 °C until assayed, except for the ELISA analysis samples were kept on regular ice and assayed immediately or stored at –80°C after homogenization in the lysis buffer, see section for ELISA analysis below. For HPLC analysis, all samples were weighed prior to snap-freezing to estimate wet-weight.
RNA isolation
RNA from total brain, dorsal striatum, substantia nigra and ventral tegmental area was isolated using RNeasy Lipid Tissue Mini Kit (74804, Qiagen) or TRIzol (15596-026, Ambion/Life Technologies) according to manufacturer’s recommendations. Briefly, the tissue was mechanically homogenized in QIAzol Lysis Reagent or TRIzol Reagent and incubated at room temperature for 5 min. Homogenates were then thoroughly mixed with chloroform, incubated at room temperature for 3 minutes and centrifuged at 12,000 g for 15 min at 4 °C. When using RNeasy Lipid Tissue Mini Kit, the upper aqueous phase was then mixed with 1 volume of 70 % ethanol and mixed thoroughly by vortexing. RNA was collected on spin columns by centrifugation. To ensure removal of genomic DNA from the sample, on-column DNase digestion was performed using DNase I (79254, Qiagen), as recommended by the manufacturer. Columns were then washed twice with Buffer RPE and RNA was eluted using RNase-free water. RNA yield was 4–5.5 µg (dorsal striatum), 2.2–3.3 µg (substantia nigra), and 2.3–4.7 µg (ventral tegmental area). Alternatively, the upper aqueous phase obtained after phase separation with TRIzol Reagent was mixed with isopropanol, incubated for 10 min at room temperature and centrifuged at 12,000 g for 10 min at 4 °C. RNA pellet was then washed with 70 % ethanol and centrifuged at 7500 g for 5 min at 4 °C. The pellet was allowed to air dry for 5–10 min and eluted with RNase-free water. To avoid contamination with genomic DNA, samples were then treated with DNA-free DNase treatment and removal reagents (AM1906, Invitrogen/Life Technologies). RNA yield was 40–100 µg (total brain) 2.3–5 µg (dorsal striatum), 2.6–3.8 µg (substantia nigra), and 4.6–7.2 µg (ventral tegmental area). RNA was stored at –80 °C or used immediately for cDNA synthesis.
cDNA synthesis
cDNA was synthesized using Transcriptor First Strand cDNA Synthesis Kit (04379012001, Roche) according to manufacturer’s protocol. Briefly, 150–500 ng of RNA was mixed with random hexamer primers and incubated at 65 °C for 10 min. Transcriptor RT Reaction Buffer, Protector RNase inhibitor, dNTP mix and Transcriptor Reverse Transcriptase were then added and samples were incubated for 10 min at 25 °C, 30 min at 55 °C and 5 min at 85 °C. cDNA was diluted 1:10 and 2.5 µl was used for quantitative real-time PCR.
Real-time quantitative PCR (qPCR)
Real-time quantitative PCR (qPCR) was performed on a Lightcycler®480 real-time PCR system (Roche Diagnostics) using Lightcycler®480 SYBR Green I Master complemented with 2.5 pmol of primers in the final volume of 10 µl on white 384-well plates sealed with adhesive plate sealer (04729749001, Roche). 2.5 µl of the diluted cDNA product was used for each reaction. Reactions were performed in duplicate. The following qPCR program was used: (1) pre-incubation 10 min at 95 °C, (2) amplification 10 s at 95 °C, 15 s at 60 °C, 15 s at 72 °C for 45 cycles, (3) melting curve 5 s at 95 °C, 30 s at 55 °C, continuous acquisition mode at 95 °C with 2 acquisitions per °C, and (4) cooling 10 s at 40 °C. The results were analyzed with Lightcycler® 480 Software Release 1.5.0 SP1 using the Absolute Quantification / 2nd Derivative Max calculation. Results for a biological repeat were discarded when the Cq value for one or more of the replicates was 40 (or 0) or when the Cq difference between replicates was >1. For each primer pair, primer efficiencies (the ratio of amplified products if average Cq difference is 1) were determined. Gene expression was normalized to β-actin. The following intron-spanning primers were used: Gdnf F: CGCTGACCAGTGACTCCAATATGC, Gdnf R: CGCTGACCAGTGACTCCAATATGC, β-actin F: CCAGTTCGCCATGGATGAC, β-actin R: GAGCCGTTGTCGACGACC. Primer efficiencies were 2.428 (Gdnf) and 1.763 (β-actin).
ELISA analysis
Brain tissues were kept briefly on regular ice after harvesting, and sonicated immediately in ice cold lysis buffer, pH 7.4 containing 8.1 mM Na2PO4, 137 mM NaCl, 2.7 mM KCl and supplemented with 1 % Igepal CA-630 (I-3021 Sigma), 10 % Glycerol and protease inhibitor cocktail tablet Complete EDTA-free Mini (04693159001, Roche). Lysate was centrifuged for 5 min at 5000 rpm at 4 ºC and supernatant was used directly on ELISA or aliquoted and snap frozen on dry ice and stored at −80 ºC. Aliquots were thawed only once for ELISA analysis. To assess GDNF expression in brain tissue, GDNF Emax® ImmunoAssay System (G7621 Promega) was used as recommended by the manufacturer. Samples were acid treated on room temperature for 15 min, centrifuged few seconds at a microcentrifuge and stored on ice for 3 min. Samples were neutralized on ice by gentle pipetting and immediately loaded to the ELISA plate. Absorbance was recorded immediately after neutralizing the color development with Victor3 plate reader (Wallac-PerkinElmer). Absorbance values at 450 nm were converted to concentrations of GDNF. GDNF levels were normalized to total protein concentration using DC Protein Assay (#500-0116, Bio-Rad) as recommended by the manufacturer, 100–200 µg total protein was loaded per well. The signal from brain lysate from Gdnf full knockout mice (Supplementary Fig. 1b) and GdnfF/KO + Nestin-Cre mice at embryonic day 18 was the same and was used for defining the background signal. Striatal lysate from GdnfF/KO + Nestin-Cre adult mice was included in every analysis to define the background signal. The uncorrected ELISA raw signal in GdnfF/KO + Nestin-Cre striatal lysates was significantly lower [t(4) = 9.059, P = 0.001]; after background correction with the GdnfF/KO + Nestin-Cre signal the values were not applicable (n.a.) for statistical analysis (Fig.1e).All tissue lysates were analyzed with at least three protein concentrations.
Surgery
GdnfF and wt mice were injected unilaterally at University of Lund, Sweden with a replication deficient, recombinant adeno-associated viral (AAV) vector serotype 5 expressing Cre recombinase under the synapsin (syn) promoter (virus titre 5.8E13 gc/ml). Viral vectors were produced in Lund as previously described14. For all surgical procedures, mice were induced and maintained under anaesthesia using 2 % isoflurane delivered in a mix of air and N2O at a ratio of 4:1. When complete anaesthesia was achieved the mice were placed into a stereotaxic frame fitted with a mouse adaptor (Stoelting). A 5 µl Hamilton syringe with a 22-gauge needle fitted with a pulled glass capillary (tip with external diameter 60–80 µm) was used for injection, in order for more precise targeting and injection, and also to minimise tissue damage. For intrastriatal injection, a total volume of 2 µl of AAV5-Syn-Cre was injected at two depths within the striatum at the following coordinates relative to bregma: A/P: +1.0; M/L: −2.3; D/V: −3.2 and −2.2 from the surface of the dura mater. Animals woke up under close supervision and kept warm until they fully recovered from anaesthesia before being returned to a clean home cage.
Tamoxifen application
Tamoxifen was prepared and applied in doses as in7 as described in15.
Behavioural analyses
All animal experiments were conducted in accordance with the Council of Europe (Directive 2010/63/EU), the National Institute of Health Guidelines for the Care and Use of Laboratory Animals and Finnish and Swedish guidelines. Efforts were made to minimize animal suffering and to use only the number of animals necessary to produce reliable scientific data. Testing apparatuses were thoroughly cleaned after each animal using 70 % ethanol.
Rotational asymmetry in response to amphetamine (i.p., 5 mg/kg) and apomorphine (s.c., 0.1 mg/kg) was used to monitor motor deficits affecting the nigrostriatal dopamine system. Data are presented as net turns per minute, as recorded over 40 minute sessions. Lateralised sensorimotor neglect was monitored using the corridor task, performed as previously described10. Data are presented as percentage of the total number of contralateral retrievals divided by the total retrievals made on both sides as described in10. Behavioural analysis of AAV5-Syn-Cre injected mice was done on independent cohorts of animals in parallel in Lund University, Sweden and University of Helsinki, Finland.
All the other behavioral experiments were conducted in the order presented below, and the interval between subsequent tests was 2–3 days during the light phase. The experimenter was blind to the animal genotype and/or treatment. At the beginning of behavioral testing, the animals were 2–3 months or 12 months old.
Elevated Plus Maze
Elevated Plus Maze (EPM) asses anxiety-like behavior in rodents. The test was performed in a maze consistent of two open arms (30 × 5 cm), two enclosed arms (30 × 5 cm with 15 cm high transparent side- and end walls) and a connecting central platform (5 × 5 cm). The maze was raised 40 cm from above the floor. The mouse was placed on the central platform facing one of the enclosed arms and observed for 5 minutes. A video camera positioned above the maze recorded the experiments. Time spent on open arms (s) was calculated. Testing took place in reduced light condition.
Light–dark test
Light-dark test (LD) addresses anxiety-like behavior in rodents. Testing was performed for 10 min in an acrylic cage (28.5 × 28.5 × 20 cm; MED Associates, St. Albans, VT, USA) divided into two equal size compartments: one part was with transparent walls, open topped and brightly illuminated (~ 530 lx by a 40 W light bulb fixed 55 cm above the floor), the other part was made from black plastic (passing infrared light) and covered by a lid. The two compartments were separated by a partition containing an opening (7 × 5 cm) in its centre at floor level. The animal was placed in the center of the light compartment facing away from the opening, and time spent in the compartments and the number of entries between compartments were measured over 10 min.
Open field test
We used Open field –test (OF) to measure basic locomotor activity of mice in open, novel environment. Testing was performed for 30 min in a well illuminated (~ 300 lx) transparent acrylic cage (28.5 × 8.5 × 20 cm) (Med Associates, St. Albans, VT). Interruptions of infrared photo beams were used to measure locomotor activity, resting time and rearings.
Accelerating Rotarod
The motor coordination and learning of mice was measured on the accelerating rotating rod. The mice were trained on an accelerating rotarod (RR) (Ugo Basile, Comerio, Italy) equipped with automatic fall detector in six trials on 2 succeeding days (three trials a day). The speed of rotation increased at a constant rate (from 4 to 40 r.p.m.) over 5 min and cutoff time was 6 min. For analysis, average of each day (3 trials) was calculated.
Pre-Pulse Inhibition
Lack of pre-pulse inhibition (PPI) is usually present in patients with schizophrenia. We used acoustic startle response to measure PPI. The experiment was performed in MED Associates (St. Albans, VT) PPI chambers. Data acquisition was performed using Med Associates software.
Mice were placed in the startle chamber with a background white noise of 65 dB and left undisturbed for 5 minutes. Testing was performed in 12 blocks of 5 trials and five trial types were applied. One trial type was a 40-ms, 120-dB white noise acoustic startle stimulus (SS) presented alone. In the remaining four trial types the startle stimulus was preceded by the acoustic pre-pulse. The 20-ms pre-pulse stimuli (PPS) were white noise bursts of 68, 72, 76 and 80 dB. The delay between onsets of PSS and SS was 100 ms. The 1st and 12th block consisted of SS-alone trials. In remaining blocks the SS and PPS+SS trials were presented in pseudo-randomized order such that each trial type was presented once within a block of 5 trials. The inter-trial interval ranged between 10 and 20 seconds. The startle response was recorded for 65 ms starting with the onset of the startle stimulus. The maximum startle amplitude recorded during the 65-ms sampling window was used as the dependent variable. The startle response was averaged over 10 trials from blocks 2–11 for each trial type. The pre-pulse inhibition for each PPS was calculated by using the following formula: 100–[(startle response on PPS+SS trials / startle response on SS trials) × 100].
Immunohistochemistry (IHC) for light microscopy
The mice were anesthetized with sodium pentobarbital (100 mg/kg, i.p.) and intracardially perfused with PBS followed by 4 % paraformaldehyde (PFA) in 0.1 M phosphate buffer, pH 7.4. The brains were postfixed in PFA for 4 h, and stored in phosphate buffer containing 20 % sucrose at 4 °C. Coronal striatal and nigral sections were cut and saved in serial order at –20 °C until immunostained.
Tyrosine hydroxylase (TH) immunohistochemistry
Free floating sections from the striatum (30 µm), ventral midbrain (40 µm) and locus coeruleus (40 µm) were stained using standard immunohistochemical procedures. Briefly, after quenching with 3 % hydrogen peroxide (H2O2) and 10 % methanol for 5 min sections were preincubated in 2 % normal goat serum (NGS; Vector Laboratories, Burlingame, CA) and 0.3 % Triton X-100 for 60 min followed by incubation with rabbit anti-TH polyclonal antibody (AB 152, 1:2000, Millipore, Bedford, MA) overnight, followed by incubation for 2 h with the biotinylated goat anti-rabbit antibody (BA1000, 1:200, Vector Laboratories) the next day. Vectastain Elite ABC peroxidase kit (Vector Laboratories) was used for visualization using 0.06 % diaminobenzidine and H2O2. The sections were mounted on gelatin/chrome coated slides, air-dried, dehydrated, cleared and mounted using Pertex mounting medium (Cellpath, Hemel Hempstead, UK). See 16 for further details. Histological images for the figures were generated using 3DHISTECH Pannoramic 250 FLASH II digital slide scanner at Genome Biology Unit (Institute of Biotechnology, Biocenter Finland, University of Helsinki).
Immunostaining for Cre recombinase was performed on free-floating sections as previously described10 using rabbit anti-Cre polyclonal antibody (PRB-106C, 1:5 000, Nordic BioSite, Sweden).
Stereological analysis of TH-positive cells
The number of TH-positive neurons in the substantia nigra pars compacta (SNpc) and ventral tegmental area (VTA) was assessed by a person blinded to the identity of the samples. Briefly, TH-positive cell counts were assessed at medial levels of the SNpc, around the medial terminal nucleus (MTN). From each animal, every third section between levels −3.08 and −3.28 mm from the bregma was selected (3 sections per animal). StereoInvestigator (MBF Bioscience, Williston, VT) was used to outline the SNpc, and positively stained cells were counted within the outlines according to optical dissector rules17. Cells were counted at regular predetermined intervals (x = 100 μm; y = 80 μm) within the counting frame (60 μm × 60 μm) superimposed on the image using a 60× oil objective [Olympus BX51 (Olympus Optical, Tokyo, Japan) equipped with an Optronics camera]. Positions of the counting frame within the SNpc were randomized by the software, which created a systematic random sample of the area. The coefficient of error was used as an estimate of precision and values <0.1 were accepted. Failure in staining or perfusion resulting in spoiled sections was an exclusion criterion. Please see16 for further details.
Striatal densitometry measurements
The optical density (OD) analysis was performed blinded for the identity of the coded slides. Striatal TH-positive fiber immunostaining OD measurements were performed using an Optronics (Goleta, CA) digital camera and Image-Pro Plus software (Version 3.0.1; Media Cybernetics, Silver Spring, MD) from four to five striatal sections from each animal and the final reading was calculated as an average. The nonspecific background correction in each section was done by subtracting the OD value of the corpus callosum from the striatal OD value of the same section. Failure in staining or perfusion resulting in spoiled sections was an exclusion criterion. Please see16 for further details.
Estimation of monoamines and their metabolites
Dopamine and its metabolites were analyzed as previously reported in18 using high performance liquid chromatography (HPLC) with electrochemical detection.
Statistical analysis
Due to non-homogeneity of variances, Gdnf mRNA and protein levels were analyzed either with non-parametric Mann-Whitney U test or Kruskal-Wallis test with stepwise-stepdown multiple comparison option of the SPSS software. The data from experiments where tamoxifen was used as treatment was additionally analyzed with two-way ANOVA in order to reveal possible genotype-treatment interaction. All the other comparisons were performed using either one-way ANOVA or Student’s two-tailed t-test with the unequal variance option to control possible violations in normal distribution. In case of technical repeats (qPCR, ELISA) mean values were used in calculations. A priori power analyses were not performed, the sample sizes were similar to those generally employed in the field. All numerical results are reported as mean ± standard error of mean. SPSS version 20 for Windows software (IBM Corp., Armonk NY, USA) or STATISTICA 11 software (StatSoft Inc., Tulsa, OK, USA) was used for analysis.
Supplementary Material
A supplementary methods checklist is available.
Acknowledgements
J.O.A. was supported by the Academy of Finland grants 136591, 140983 and 263700, NIH (NS 070825) and by the Institute of Biotechnology. M.S. was supported by grants from Sigrid Jusélius Foundation, H. Lundbeck Foundation, M. J. Fox Foundation for Parkinson’s Research, Academy of Finland (grant 11186236), NIH (NS 0708259) and the University of Helsinki. T.P.P. was supported by Päivikki and Sakari Sohlberg Foundation. A.B. was supported by grants from the Swedish Research Council. Mouse Behavioural Phenotyping Facility (V.V.) is supported by Biocenter Finland. E.C. is supported by the ERC iPlasticity project (322742), Sigrid Jusélius foundation and the Academy of Finland. S.G. was supported by a postdoctoral stipend from the Swedish Brain Foundation (Hjärnfonden). K.V. is supported by a grant from BnM. J.K. was supported by Research Foundation of the University of Helsinki and The Finnish Cultural Foundation. We thank Susanna Wiss and Kati Rautio for expert technical assistance.
Footnotes
Author contributions
J.O.A. designed and generated the GDNF conditional knock-out mice, K.V., C.V., J.O.A. and M.A.H. performed qPCR measurements, J.O.A. and M.A.H. performed GDNF ELISA, J.K. and C.V. performed immunohistochemistry and stereology, J.K., C.V. and S.G. dissected the brains, J.K. measured dopamine levels and optical density, S.G. did AAV-Cre injections, C.V. did tamoxifen injections, J.L., C.V. and S.G. performed all the behavioral assays, J.O.A., M.S., T.P.P., A.B., V.V. and E.C. planned experiments, M.S., J.O.A., T.P.P. and A.B. provided funding, J.K. and J.O.A. prepared the figures and wrote the paper.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Lin LF, Doherty DH, Lile JD, Bektesh S, Collins F. GDNF: a glial cell line-derived neurotrophic factor for midbrain dopaminergic neurons. Science. 1993;260:1130–1132. doi: 10.1126/science.8493557. [DOI] [PubMed] [Google Scholar]
- 2.Arenas E, Trupp M, Akerud P, Ibanez CF. GDNF prevents degeneration and promotes the phenotype of brain noradrenergic neurons in vivo. Neuron. 1995;15:1465–1473. doi: 10.1016/0896-6273(95)90024-1. [DOI] [PubMed] [Google Scholar]
- 3.Meissner WG, et al. Priorities in Parkinson 's disease research. Nat Rev Drug Discov. 2011;10:377–393. doi: 10.1038/nrd3430. [DOI] [PubMed] [Google Scholar]
- 4.Airaksinen MS, Saarma M. The GDNF family: signalling, biological functions and therapeutic value. Nat Rev Neurosci. 2002;3:383–394. doi: 10.1038/nrn812. [DOI] [PubMed] [Google Scholar]
- 5.Jain S, et al. RET is dispensable for maintenance of midbrain dopaminergic neurons in adult mice. J Neurosci. 2006;26:11230–11238. doi: 10.1523/JNEUROSCI.1876-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kramer ER, et al. Absence of Ret signaling in mice causes progressive and late degeneration of the nigrostriatal system. PLoS Biol. 2007;5:e39. doi: 10.1371/journal.pbio.0050039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pascual A, et al. Absolute requirement of GDNF for adult catecholaminergic neuron survival. Nat Neurosci. 2008 doi: 10.1038/nn.2136. [DOI] [PubMed] [Google Scholar]
- 8.Tronche F, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 9.Heuer A, Smith GA, Lelos MJ, Lane EL, Dunnett SB. Unilateral nigrostriatal 6-hydroxydopamine lesions in mice I: motor impairments identify extent of dopamine depletion at three different lesion sites. Behavioural brain research. 2012;228:30–43. doi: 10.1016/j.bbr.2011.11.027. [DOI] [PubMed] [Google Scholar]
- 10.Grealish S, Mattsson B, Draxler P, Bjorklund A. Characterisation of behavioural and neurodegenerative changes induced by intranigral 6-hydroxydopamine lesions in a mouse model of Parkinson's disease. Eur J Neurosci. 2010;31:2266–2278. doi: 10.1111/j.1460-9568.2010.07265.x. [DOI] [PubMed] [Google Scholar]
- 11.Hudson JL, et al. Correlation of apomorphine- and amphetamine-induced turning with nigrostriatal dopamine content in unilateral 6-hydroxydopamine lesioned rats. Brain Res. 1993;626:167–174. doi: 10.1016/0006-8993(93)90576-9. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, et al. Tamoxifen-independent recombination in the RIP-CreER mouse. PLoS One. 2010;5:e13533. doi: 10.1371/journal.pone.0013533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taraviras S, et al. Signalling by the RET receptor tyrosine kinase and its role in the development of the mammalian enteric nervous system. Development. 1999;126:2785–2797. doi: 10.1242/dev.126.12.2785. [DOI] [PubMed] [Google Scholar]
- 14.Ulusoy A, Bjorklund T, Buck K, Kirik D. Dysregulated dopamine storage increases the vulnerability to alpha-synuclein in nigral neurons. Neurobiol Dis. 2012;47:367–377. doi: 10.1016/j.nbd.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 15.Guo C, Yang W, Lobe CG. A Cre recombinase transgene with mosaic, widespread tamoxifen-inducible action. Genesis. 2002;32:8–18. doi: 10.1002/gene.10021. [DOI] [PubMed] [Google Scholar]
- 16.Mijatovic J, et al. Constitutive Ret activity in knock-in multiple endocrine neoplasia type B mice induces profound elevation of brain dopamine concentration via enhanced synthesis and increases the number of TH-positive cells in the substantia nigra. J Neurosci. 2007;27:4799–4809. doi: 10.1523/JNEUROSCI.5647-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gundersen HJ, et al. The new stereological tools: disector, fractionator, nucleator and point sampled intercepts and their use in pathological research and diagnosis. APMIS. 1988;96:857–881. doi: 10.1111/j.1699-0463.1988.tb00954.x. [DOI] [PubMed] [Google Scholar]
- 18.Airavaara M, et al. In heterozygous GDNF knockout mice the response of striatal dopaminergic system to acute morphine is altered. Synapse. 2006;59:321–329. doi: 10.1002/syn.20245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.