Abstract
Background
A 2% to 5% background rate of rare SCN5A nonsynonymous single nucleotide variants (nsSNVs) among healthy individuals confounds clinical genetic testing. Therefore, the purpose of this study was to enhance interpretation of SCN5A nsSNVs for clinical genetic testing using estimated predictive values derived from protein-topology and 7 in silico tools.
Methods and Results
Seven in silico tools were used to assign pathogenic/benign status to nsSNVs from 2888 long-QT syndrome cases, 2111 Brugada syndrome cases, and 8975 controls. Estimated predictive values were determined for each tool across the entire SCN5A-encoded Nav1.5 channel as well as for specific topographical regions. In addition, the in silico tools were assessed for their ability to correlate with cellular electrophysiology studies. In long-QT syndrome, transmembrane segments S3−S5+S6 and the DIII/DIV linker region were associated with high probability of pathogenicity. For Brugada syndrome, only the transmembrane spanning domains had a high probability of pathogenicity. Although individual tools distinguished case- and control-derived SCN5A nsSNVs, the composite use of multiple tools resulted in the greatest enhancement of interpretation. The use of the composite score allowed for enhanced interpretation for nsSNVs outside of the topological regions that intrinsically had a high probability of pathogenicity, as well as within the transmembrane spanning domains for Brugada syndrome nsSNVs.
Conclusions
We have used a large case/control study to identify regions of Nav1.5 associated with a high probability of pathogenicity. Although topology alone would leave the variants outside these identified regions in genetic purgatory, the synergistic use of multiple in silico tools may help promote or demote a variant's pathogenic status.
Keywords: Brugada syndrome, genetic testing, long-QT syndrome
After the discovery of the first inherited cardiac arrhythmia-associated mutation (ΔKPQ or p. KPQdel) in the SCN5A-encoded Nav1.5 α-subunit of the cardiac sodium channel,1 hundreds of additional SCN5A genetic variants have been linked to a heterogeneous group of heritable sudden cardiac death predisposing cardiac diseases, including type 3 long-QT syndrome (LQT3), type 1 Brugada syndrome (BrS1), cardiac conduction disease, sick sinus syndrome, atrial fibrillation, dilated cardiomyopathy, and overlap syndromes, whereby a single SCN5A variant results in clinical manifestations of >1 disease in an individual patient or pedigree.2
Although the overall contribution of SCN5A mutations to the pathogenesis of cardiac conduction disease, sick sinus syndrome, atrial fibrillation, and dilated cardiomyopathy is limited or not well defined, previous studies indicate that loss-of-function mutations in SCN5A underlie 20% to 30% of BrS (BrS1, prevalence: ≈1:10 000–25 000),3–5 whereas gain-of-function mutations account for 10% to 15% of LQTS (LQT3, prevalence: ≈1:13 000–20 000), respectively.6,7 Furthermore, the elucidation of the molecular and electrophysiological basis of SCN5A-mediated LQTS and BrS has led to the enumeration of clinically meaningful genotype–phenotype correlations that now aid in the risk stratification and clinical management of individuals with SCN5A-mediated disease.5,8,9
Given that SCN5A is associated with a relatively high background rate of rare, and most likely innocuous, non-synonymous single-nucleotide variants (nsSNVs; ≈2% for whites and ≈5% for nonwhites), the task of assigning the probability of pathogenicity to those rare SCN5A nsSNVs identified in patients suspected of possibly having either BrS or LQTS is an even greater challenge.10–13 Although attempts to enhance the classification of nsSNVs using protein topology–driven estimated predictive values (EPVs) or the use of in silico phenotype prediction algorithms has proven successful for other major cardiac channelopathy genes (namely KCNQ1/LQT1 and KCNH2/LQT2),12,14 previous attempts to use similar approaches to enhance the classification of nsSNVs in SCN5A-mediated disease have been either limited to a single in silico algorithm15 or severely underpowered.16,17
In an effort to improve the diagnostic interpretation of SCN5A nsSNVs in BrS and LQTS genetic testing, a large case–control study, using genotypic data from previously published BrS and LQTS compendia and recently released public data sets from population-based exome/genome sequencing initiatives, was used to: (1) establish and refine the protein topology–specific EPVs for major regions of the SCN5A-encoded Nav1.5 channel in BrS and LQTS, respectively, (2) objectively assess the ability of a set of 7 commonly used in silico tools to differentiate between case- and control-derived nsSNVs in SCN5A, (3) determine if the individual or synergistic use of the aforementioned in silico tools can enhance the diagnostic interpretation of BrS and LQTS genetic testing by enhancing the classification of nsSNVs localizing to problematic topological regions of the Nav1.5 channel, and finally (4) to determine the degree to which in silico phenotype prediction tools and in vitro heterologous expression system/cellular electrophysiology studies agree on the phenotypic manifestations of putative BrS- and LQTS-associated SCN5A nsSNVs.
Methods
Study Design
For this study, large compendia of previously published BrS and LQTS cases were assembled. The LQTS cases were derived from 388 clinically definite cases and 2500 cases referred for clinical LQTS genetic testing. In addition, 2111 previously genotyped BrS cases were assembled. The 388 clinically definite LQTS cases were defined as having a Schwartz score ≥3.5 or a QTc≥480 ms on ECG. For the 2500 cases referred for clinical LQTS genetic testing, limited phenotypic and demographic information was available, and the samples were accepted for genotyping based on the referring cardiologist's request after a presumed LQTS diagnosis. Similarly, the 2111 BrS cases had limited phenotypic and demographic information and again were genotyped based on a referring physician's clinical impression of BrS.
For comparison, a compendium of 8975 controls was assembled from 1380 in-house controls, 1092 exomes from the 1000 Genomes project (1kG), and 6503 exomes from the National Heart, Lung, and Blood Institute Exome Sequencing Project (ESP). For the purpose of this study, the term controls is meant to imply a group of anonymous volunteers not enriched or selected for the presence of either a 1:3000 to 5000 disorder (BrS, 1:10 000–25 000 for BrS1) or a 1:2000 disorder (LQTS, 1:13 000–20 000 for LQT3). Accordingly, a normal ECG was not a prerequisite for inclusion as a control.
BrS/LQTS Genetic Testing
Genomic DNA from all cases and the 1380 in-house controls were analyzed for genetic variants in the translated exons and splice-site junctions of the SCN5A gene using polymerase chain reaction and either denaturing high-performance liquid chromatography, followed by direct DNA sequencing or direct high-throughput DNA sequencing as described previously.12 The SCN5A variants from the 1kG and ESP samples were obtained from the corresponding online databases. As the 1kG and ESP samples were genotyped using whole-exome sequencing, the yields among these cohorts were compared with that of the 1380 in-house controls genotyped via Sanger sequencing and were equivalent statistically.
nsSNV Interpretation and Sequence Analysis
For the purposes of this study, only those nsSNVs (by definition codon- and amino acid–altering genetic variants) identified in at least 1 LQTS or BrS case that were also absent in the entire control population were considered to be rare, case-derived nsSNVs. Importantly, the term rare control–derived nsSNV in this study is not intended to imply pathogenicity or even functional abnormality; but rather it merely indicates that a particular nsSNV is rare (ie, seen in only a single control, 1 of 8975), amino acid altering, and if encountered during the course of clinical LQTS/BrS, genetic testing would be considered a potentially pathogenic variant.
The identified nsSNVs were overlaid on the linear protein topology of Nav1.5. The linear protein topology was annotated to define functionally important domains and structural regions within the cardiac sodium channel according to the UniProtKB/Swiss-Prot data-bank (http://ca.expasy.org/uniprot)18 and studies of the genomic and protein organization of Nav1.5.19
In Silico Phenotype Prediction Analyses
Each of the 7 commonly used in silico phenotype prediction algorithms have been described in detail elsewhere,14 and the specific versions and parameters used to assess the pathogenicity of the case- and control-derived SCN5A nsSNVs identified in this study are described in detail in Methods in the Data Supplement. In addition, we evaluated the recently reported APPRAISE algorithm.20 As this algorithm was designed and optimized on the variants derived from the cases and controls within our own study, the use of this tool was limited to a subset analysis of the functionally characterized variants.
Variants for Comparison of In Silico Phenotype Prediction and Cellular Electrophysiological Phenotype
A literature search was performed to identify all functionally characterized SCN5A variants. All variants identified with cellular electrophysiological data were included regardless of the frequency in the controls, and variants were classified as having abnormal or wild-type functional characteristic based on the electrophysiological findings. Those variants where there was conflicting data about the observed electrophysiological phenotype were noted but were not included for the comparison with the in silico predictions. In addition, 33 distinct (29 previously uncharacterized) SCN5A variants were characterized functionally in either the Mayo Clinic Windland Smith Rice Sudden Death Genomics Laboratory or the Makielski laboratory at the University of Wisconsin-Madison for the purpose of this case/control study. The methods for site-directed mutagenesis, heterologous expression, electrophysiological measurements, and data analysis can be found in the Methods in the Data Supplement. In short, variants were introduced into the SCN5A cDNA within the pcDNA3 expression vector via site-directed mutagenesis. These vectors were then used to heterologously express mutant or wild-type channels in HEK-293 cells and were characterized using standard whole-cell patch clamp technique.
Statistical Analysis
To identify regions tolerant to variation and regions critical to the pathogenesis of disease, frequencies of affected amino acids were calculated using the number of amino acids affected by case or control rare nsSNVs for the overall protein, as well as described topological regions. The frequency of affected amino acids for each region was compared between cases and controls using the Fisher exact test with a threshold of significance set at P<0.05.
For the purpose of calculating the frequency of nsSNVs predicted to be pathogenic by each in silico tool, rare control–derived nsSNVs (ie, identified in only 1 of the 8975 controls) were compared with rare case–derived nsSNVs (identified in a LQTS or BrS case but not in the control population). A two-tailed Fisher exact test with a threshold of significance set at P<0.05 was then used to compare raw counts of rare case nsSNVs and rare control nsSNVs predicted to be pathogenic by each algorithm on the basis of their phylogenetic or physicochemical properties.
To estimate the likelihood of disease causation, a modified EPV (defined as the probability of pathogenicity for a mutation identified in a case) was used as described previously12,20,21 and within the Methods in the Data Supplement. Importantly, variants seen in multiple controls were considered polymorphisms and excluded from EPV analysis, whereas those seen only once constituted the rare, control-derived nsSNVs. As such, common polymorphisms, such as SCN5AH558R, did not influence the calculation of EPVs. The sensitivity and specificity were calculated based on the number of case and control nsSNVs predicted as pathogenic or benign by each in silico prediction algorithm. The 95% confidence interval was calculated using the Wilson score method. The Matthews correlation coefficient was calculated similarly based on the in silico predictions.
Results
Mutation Rates for BrS- and LQTS-Associated SCN5A nsSNVs
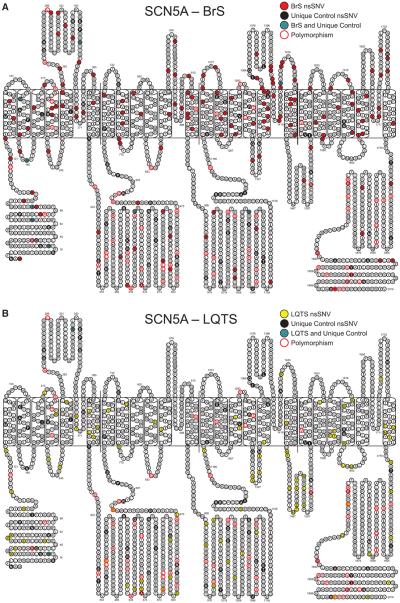
Overall, rare nsSNVs were identified in 273/2111 (13.0%) of the BrS cases, 20/388 (5.2%) of the clinically definite LQTS cases, and 109/2888 (4.4%) referral LQTS cases. As the yield was statistically similar between the 2 LQTS cohorts, they were combined into 1 LQTS cohort with a yield of 4.5% (129/2888). Collectively, 255 rare case–derived SCN5A nsSNVs were identified in the LQTS and BrS cohorts. In contrast, 101 rare control–derived SCN5A nsSNVs were identified in 101/8975 (1.1%) controls (18/1380 [1.3%] ostensibly healthy in-house and 83/7595 (1.1%) public exomes/genomes, P=0.49). As expected, rare SCN5A nsSNVs were significantly more common in cases than controls (LQTS: 129/2888 versus 101/8975; BrS: 273/2111 versus 101/8975; all cases: 402/4999 versus 101/8975; all comparisons P<1.0×10−16). All case and control nsSNVs are localized in Figure 1A and 1B and are listed in Table I in the Data Supplement.
Figure 1.
Location of variants in the SCN5A-encoded Nav1.5 pore-forming α subunit. A, Location of variants identified in Brugada syndrome (BrS) cases compared with the location of variants derived from controls. B, Location of variants identified in long-QT syndrome (LQTS) cases compared with the location of variants derived from controls. Filled circles represent residues harboring a nonsynonymous single-nucleotide variant (nsSNV). Red filled circles denote type 1 BrS (BrS1) case–derived nsSNVs; yellow filled circles denote type 3 LQTS (LQT3) case–derived nsSNVs; grey filled circles indicate amino acid residues hosting both case- and control-derived variants; black filled circles represent rare nsSNVs identified in only 1 of the 8975 controls; and circles with red trim indicate positions hosting a nsSNV identified in ≥1 control (polymorphism).
Despite the over-representation of nsSNVs in disease, the low yield in disease (LQTS: 4.5%, BrS: 13.0%) compared with ≈1% background rate for nsSNVs results in a fair signal-to-noise ratio (SNR) for SCN5A genetic testing in BrS (11.8:1), but a poor SNR in LQTS (4.1:1). In an effort to improve the overall interpretability of BrS and LQTS genetic testing, the relative incidence of rare SCN5A nsSNVs in cases versus controls was first used to generate region-specific mutation frequencies and EPVs/probability of pathology for SCN5A nsSNVs in BrS and refine the previously published region-specific mutations rates and EPVs for SCN5A nsSNVs in LQTS.
Distribution of nsSNVs in Nav1.5
In an effort to identify regions critical to the pathogenesis of disease as well as identify regions with high tolerance for genetic variation, we examined the frequency of amino acids hosting a rare case– or a rare control–derived nsSNV. Overall, the 101 rare control nsSNVs affected 4.8 per 100 amino acids (Table 1) and were distributed evenly (P>0.05) between the major topology regions (N-terminal, transmembrane, interdomain linkers [IDL], and C-terminal). The frequency in controls was lower than the overall frequency of 6.8 per 100 amino acids affected by BrS nsSNVs (P=0.003), but statistically indistinguishable from the 4.3 per 100 amino acids for LQTS nsSNVs (P=0.50).
Table 1.
Comparison of the Distribution of Control- and Case-Derived Nonsynonymous Single-Nucleotide Variants
| Control |
BrS Case |
LQTS Case |
|||||
|---|---|---|---|---|---|---|---|
| Region | Exon Length (aa) | No. | Per 100 aa | No. | Per 100 aa | No. | Per 100 aa |
| N-terminal | 126 | 10 | 7.9 | 10 | 7.9 | 10 | 7.9 |
| Transmembrane | 1036 | 35 | 3.4 | 105* | 10.1* | 37 | 3.6 |
| S1−S2/3+S5/6 | 580 | 25 | 4.3 | 60* | 10.3* | 9 | 1.6 |
| S3−S5+S6 | 456 | 10 | 2.2 | 45* | 9.9* | 28* | 6.1* |
| Interdomain linker | 610 | 39 | 6.4 | 18 | 3 | 32 | 5.2 |
| DI/DII | 296 | 22 | 7.4 | 10 | 3.4 | 10 | 3.4 |
| DII/DIII | 261 | 15 | 5.7 | 6 | 2.3 | 11 | 4.2 |
| DIII/DIV | 53 | 2 | 3.8 | 2 | 3.8 | 11* | 20.8* |
| C-terminal | 244 | 13 | 5.3 | 5 | 2 | 8 | 3.3 |
| All coding | 2016 | 97 | 4.8 | 138* | 6.8* | 87 | 4.3 |
aa indicates amino acids; BrS, Brugada syndrome; DI, transmembrane domain I; DII, transmembrane domain II; DIII, transmembrane domain III; DIV, transmembrane domain IV; and LQTS, long-QT syndrome.
Regions where there is an over-representation of case variants.
Examining the major regions for BrS, only the transmembrane region carried a higher frequency of affected amino acids with 10.1 per 100 amino acids being affected by BrS nsSNVs compared with only 3.4 per 100 amino acids that were affected by rare control nsSNVs (P=7.2×10−10). This highlights the critical nature of the transmembrane region for the pathogenesis of BrS. For the LQTS nsSNVs, no major region hosted a higher rate of affected amino acids for LQTS than controls.
As subregions within the transmembrane and IDLs may impart different biogenic and biophysical properties thereby making specific subregions more or less likely to harbor pathogenic LQTS-associated gain-of-function nsSNVs, the subregions were analyzed separately. Frequencies were calculated separately for the 4 transmembrane domains (DI through DIV), the 6 segments of the transmembrane domains (S1 through S6), and 3 IDLs (DI/DII, DII/DIII, and DIII/DIV). Consistent with the functional significance of the DIII/DIV IDL, this region showed a frequency of 20.8 per 100 amino acids for LQTS nsSNVs, whereas only 3.8 per 100 amino acids were affected by control nsSNVs (P=0.015). Although there were not enough LQTS-associated nsSNVs to independently calculate accurate frequencies for each collective segment of the transmembrane domains, lumping the S3 through S5 segments and linkers plus the S6 segment (S3−S5+S6) of all transmembrane domains resulted in a higher frequency for LQTS nsSNVs (6.1 per 100 amino acids) compared with control nsSNVs (2.2 per 100 amino acids; P=0.001). This suggests that specific subregions of the Nav1.5 transmembrane region may serve distinct functional roles that are perturbed by LQT3-associated SCN5A nsSNVs. The frequency of affected amino acids for all nsSNVs is summarized in Table 1.
Protein Topology–Derived EPV Analysis for BrS-and LQTS-Associated SCN5A nsSNVs
In an effort to guide genetic test interpretation, EPVs were calculated based on the regions identified as hosting a higher frequency of case nsSNVs, as well as the remaining regions. As the EPV calculation uses the frequency of cases hosting nsSNVs, a single nsSNV identified in a large number of cases could skew the EPV. Therefore, in an effort to determine the impact of the next new nsSNV throughout SCN5A, over-represented case-derived nsSNVs were removed from subsequent analyses comparing case and control sample counts.
Among the case-derived nsSNVs, 21/173 (12.1%) of the BrS-derived nsSNVs and 5/94 (5.3%) of the LQTS-derived ns SNVs were not only absent from controls but were also statistically over-represented (P<0.05; Table 2). E1784K was over-represented in both LQTS and BrS cases. Although accounting for a small subset of the nsSNVs identified, these over-represented nsSNVs accounted for a relatively large portion of the genotype-positive cases [BrS: 100/273 (36.6%); LQTS: 31/129 (24.0%)]. This over-representation suggests these nsSNVs have a high likelihood of being pathogenic mutations; however, the disproportion between the number of cases and the number of amino acids altered heavily skews any case/control analysis.
Table 2.
Twenty-Five Nonsynonymous Single-Nucleotide Variants Absent in Controls and Statistically Over-Represented in BrS or LQTS
| Nucleotide Change | Amino Acid Change | Region | BrS (2111) | LQTS (2888) |
|---|---|---|---|---|
| 311 G>A | R104Q | N-terminal | 3 | 0 |
| 1066 G>A | D356N | DI-S5/S6 | 8 | 0 |
| 1100 G>A | R367H | DI-S5/S6 | 6 | 0 |
| 1231 G>A | V411M | DI-S6 | 0 | 3 |
| 2204 C>T | A735V | DII-S1 | 4 | 0 |
| 2254 G>A | G752R | DII-S2 | 5 | 0 |
| 2633 G>A | R878H | DII-S5/S6 | 5 | 0 |
| 2678 G>A | R893H | DII-S5/S6 | 3 | 0 |
| 2701 G>A | E901K | DII-S5/S6 | 3 | 0 |
| 2780 A>G | N927S | DII-S6 | 3 | 0 |
| 2893 C>T | R965C | DII/DIII | 3 | 0 |
| 3157 G>A | E1053K | DII/DIII | 3 | 1 |
| 3673 G>A | E1225K | DIII-S1/S2 | 4 | 0 |
| 3694 C>T | R1232W | DIII-S1/S2 | 3 | 0 |
| 3823 G>A | D1275N | DIII-S3 | 3 | 0 |
| 3974 A>G | N1325S | DIII-S4/S5 | 0 | 4 |
| 4222 G>A | G1408R | DIII-S5/S6 | 7 | 0 |
| 4642 G>A | E1548K | DIV-S1/S2 | 3 | 0 |
| 4720 G>A | E1574K | DIV-S2 | 4 | 0 |
| 4868 G>A | R1623Q | DIV-S4 | 1 | 3 |
| 4978 A>G | I1660V | DIV-S5 | 5 | 0 |
| 5228 G>A | G1743E | DIV-S5/S6 | 6 | 0 |
| 5227 G>A | G1743R | DIV-S5/S6 | 5 | 0 |
| 5302 A>G | I1768V | DIV-S6 | 0 | 3 |
| 5350 G>A | E1784K | C-Terminal | 14 | 18 |
BrS indicates Brugada syndrome; DI, transmembrane domain I; DII, transmembrane domain II; DIII, transmembrane domain III; DIV, transmembrane domain IV; and LQTS, long-QT syndrome.
With the removal of these over-represented nsSNVs, the yield fell to 8.2% (173/2111) in BrS and 3.4% (98/2888) in LQTS resulting in reduced SNRs of 7.3:1 for BrS and 3.0:1 for LQTS for those rare variants that are not statistically over-represented in cases. This highlights the necessity for enhancements in genetic interpretation for SCN5A nsSNVs that have not achieved statistical over-representation.
SCN5A-Associated BrS (BrS1)
Having identified regions hosting a higher frequency of case nsSNVs, EPVs were calculated to help guide the interpretation of BrS nsSNVs. As the only region identified to host an over-representation of case-derived nsSNVs, the transmembrane region yielded a high probability of pathogenicity (EPV=94%; 95% CI, 91–96). The remaining regions (N-terminal, IDLs, and C-terminal), which were identified as hosting a similar frequency of alterations between BrS- and control-derived nsSNVs, were associated with a low probability of pathogenicity (60%; 95% CI, 0–78) of the Nav1.5 channel. The relative incidence of rare nsSNVs identified in BrS cases and controls are displayed in Table 3.
Table 3.
Region-Specific Mutation Rates and Topology-Derived EPVs for SCN5A variants in Control-, BrS-, and LQTS-Associated Individuals
| Control (8975) |
BrS (2111) |
LQTS (2888) |
|||
|---|---|---|---|---|---|
| Region | Count (%) | Count (%) | EPV* (95% CI) | Count (%) | EPV* (95% CI) |
| N-terminal | 10 (0.11) | 14 (0.66) | 83 (62–93) | 10 (0.35) | 68 (23–87) |
| Transmembrane | 36 (0.40) | 135 (6.40) | 94 (91–96) | 41 (1.42) | 72 (56–82) |
| S1−S2/3+S5/6† | 26 (0.29) | 79 (3.74) | 92 (88–95) | 10 (0.35) | 16 (0–60) |
| S3−S5+S6# | 10 (0.11) | 56 (2.65) | 96 (92–98) | 31 (1.07) | 90 (79–95) |
| Interdomain linker | 42 (0.47) | 19 (0.90) | 48 (11–70) | 36 (1.25) | 62 (42–76) |
| DI/DII | 23 (0.26) | 11 (0.52) | 51 (0–76) | 11 (0.38) | 33 (0–67) |
| DII/DIII | 17 (0.19) | 6 (0.28) | 33 (0–74) | 12 (0.42) | 54 (5–78) |
| DIII/DIV | 2 (0.02) | 2 (0.09) | 76 (0–97) | 13 (0.45) | 95 (78–99) |
| C-terminal | 13 (0.14) | 5 (0.24) | 39 (0–78) | 11 (0.38) | 62 (15–83) |
| All regions | 101 (1.13) | 173 (8.20) | 86 (83–89) | 98 (3.39) | 67 (56–75) |
CI indicates confidence interval; BrS, Brugada syndrome; DI, transmembrane domain I; DII, transmembrane domain II; DIII, transmembrane domain III; DIV, transmembrane domain IV; EPV, estimated predictive values; and LQTS, long-QT syndrome.
EPV=(case rate−control rate)/case rate.
S1−S2/3+S5/6 represents the transmembrane segments and linkers which had a low EPV in LQTS.
S3−S5+S6 represents the transmembrane segments and linkers which had high EPVs.
SCN5A-Associated LQTS (LQT3)
Similar to BrS, the regions hosting an over-representation of case-derived nsSNVs were the only regions associated with a high probability of pathogenicity. The separate calculation for the IDL3 (DIII/DIV) resulted in an EPV of 95% (95% CI, 78–99), pushing this specific region into the high probability of pathogenicity category. The S3−S5+S6 segment of all transmembrane domains resulted in an EPV of 90% (95% CI, 79–95), whereas the remaining regions generated an EPV of 47% (95% CI, 26–62). Rates and EPVs in LQTS for these regions are depicted in Table 3.
Through the use of the regions most affected by the case-derived nsSNVs, the SNRs were improved to 15.9 to 1 for BrS and 11.4 to 1 for LQTS. In contrast, for the remaining regions, the SNRs dropped substantially to 2.5 to 1 for BrS and 1.9 to 1 for LQTS. Although the identification of these critical regions resulted in 2-fold and 5-fold improvements in the SNR for BrS and LQTS, respectively, the pathogenic likelihood of a rare nsSNV localizing to any of the remaining regions is ambiguous.
Comparison of 7 Distinct In Silico Bioinformatic Tools
Although the topology-derived EPVs provide regions with high probability of pathogenicity, nsSNVs scattered throughout large regions of the protein remain uninterpretable. Given that recent studies have demonstrated the ability of phylogenetic- and or physicochemical-based in silico phenotype prediction algorithms to effectively distinguish between case- and control-derived nsSNVs,14 and that when used synergistically, these tools enhance the EPVs associated with regions of low and moderate probability of pathogenicity in other cardiac channelopathy–associated genes, we next looked to assess whether a set of 7 separate in silico tools could differentiate between BrS1-associated, LQT3-associated, and control-derived nsSNVs in the SCN5A-encoded Nav1.5 channel. Given the previously identified nsSNVs over-represented in cases already carry a high probability of pathogenicity, these variants were removed to assess the ability of the in silico tools to distinguish the difficult to interpret nsSNVs.
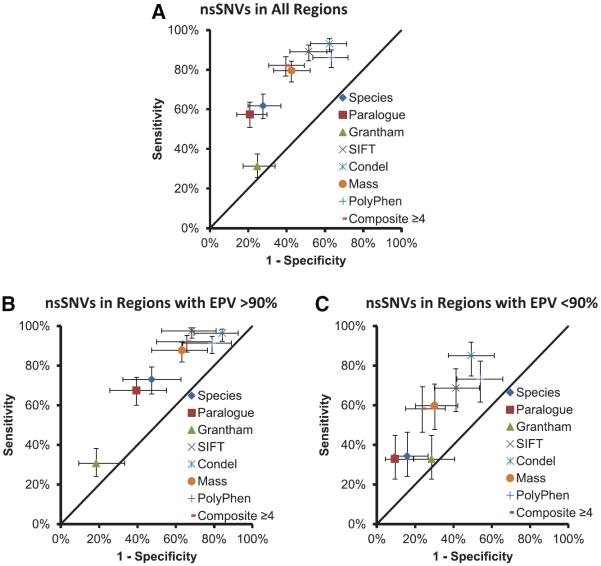
Collectively, 6 of the 7 in silico algorithms displayed a statistically significant ability (P<1.0×10−5) to differentiate between rare case– and control–derived nsSNVs over the entire protein. Only Grantham matrix values failed to distinguish between case- and control-derived nsSNVs. For each algorithm, the percent predicted pathogenic/damaging for case- and control-derived rare nsSNVs are displayed in Figure 2A. Despite the ability of the tools to distinguish case from control nsSNVs, there was a poor level of concordance between tools with all tools agreeing on the prediction for only 45.9% (152/331) of the nsSNVs. In addition, the sensitivity and specificity of the tools was variable.
Figure 2.
Classification of case- and control- derived nonsynonymous single-nucleotide variants (nsSNVs) by individual algorithms and when multiple algorithms are in agreement presented as sensitivity and 1-specificity for each tool. A, Classification of all nsSNVs across entire protein. B, Classification of nsSNVs residing within the identified critical regions. C, Classification of nsSNVs residing outside the identified critical regions. EPV indicates estimated predictive value.
In an attempt to maximize the sensitivity and specificity by leveraging the more conservative predictions (ie, more potential false-negatives) rendered by ortholog conservation and paralog conservation algorithms and the more liberal predictions (ie, more potential false-positives) rendered by the SIFT, Condel, MAss, and PolyPhen2 algorithms, we created a composite score intended to reflect the number of individual algorithms in agreement about the potential pathogenic nature of a given nsSNV. As Grantham values failed to distinguish case from control nsSNVs, this tool was excluded from the composite score. Based on an receiver operating characteristic curve (Figure I in the Data Supplement) and the Matthews correlation coefficient, detection of both benign and pathogenic SCN5A nsSNVs in BrS and LQTS was optimized by the use of composite scores of ≥4 tools in agreement. Using this threshold, 189 of 230 (82.2%) case-derived nsSNVs were predicted to be pathogenic by ≥4 tools compared with 40 of 101 (39.6%; P=5.3×10−14) control-derived nsSNVs (Figure 2A). Comparing the sensitivity and specificity of each in silico tool (including the composite score) via the Matthews correlation coefficient, the composite tool provided the best performance (Table 4). Of note, except for a slightly lower specificity, the SIFT algorithm alone performs nearly as well as the composite score.
Table 4.
In Silico Predictions for Case- and Control-Derived Nonsynonymous Single-Nucleotide Variants
| Tool | Sensitivity (95% CI) | Specificity (95% CI) | MCC | P Value (FET) |
|---|---|---|---|---|
| Grantham | 25.84% (17.88–35.80) | 75.25% (66.01–82.64) | 1.3% | 0.868868 |
| PolyPhen | 78.65% (69.05–85.89) | 36.63% (27.89–46.36) | 16.7% | 0.025639 |
| Ortholog conservation | 50.56% (40.37–60.71) | 72.28% (62.85–80.07) | 23.4% | 0.001647 |
| Mass | 73.03% (63.00–81.16) | 57.43% (47.68–66.62) | 30.7% | 3.46E–05 |
| SIFT | 82.02% (72.77–88.62) | 48.51% (39.00–58.14) | 32.1% | 8.59E–06 |
| Paralog conservation | 53.93% (43.63–63.91) | 79.21% (70.30–85.98) | 34.4% | 2.41E–06 |
| Condel | 93.26% (86.06–96.87) | 37.62% (28.79–47.36) | 36.5% | 2.69E–07 |
| Composite (≥4 tools) | 76.40% (66.61–84.02) | 60.40% (50.65–69.38) | 37.1% | 4.42E–07 |
| Region | 46.07% (36.09–56.37) | 88.12% (80.37–93.07) | 38.0% | 1.43E–07 |
Sensitivity and specificity are indicated for each in silico algorithm, as well as the use of the identified critical region (inside region denoted as pathogenic for sensitivity and specificity). The P value column indicates the calculated Fisher exact test (FET) P value comparing the predictions for case and control variants. CI indicates confidence interval; and MCC, Matthews correlation coefficient.
As we have shown that channel topology (ie, variant location) enhances the interpretation of rare nsSNVs, we next assessed whether the tools could distinguish between case- and control-derived nsSNVs inside and outside of those identified critical regions (transmembrane region and the DIII/DIV-IDL). Again, 6 of the 7 tools and the composite tool displayed a statistical ability to collectively distinguish between case and control nsSNVs that localized either inside (Figure 2B) or outside (Figure 2C) of the critical regions.
Interestingly, the in silico predictions were higher statistically for those case- and control-derived nsSNVs that resided within these critical topologic regions when compared with those localizing outside the critical regions for all tools except for Grantham values. For example, the composite tool predicted that 92.0% (150/163) of case-derived nsSNVs that resided within 1 of the critical regions as being pathogenic, but only predicted 58.2% (39/67) of case-derived nsSNVs as being pathogenic when residing outside the critical regions (P=8.2×10−9).
For control-derived nsSNVs, the composite tool predicted 65.8% (25/38) as pathogenic within the critical regions and 23.8% (15/63) as pathogenic outside the critical regions (P=5.0×10−5). This provides evidence that protein topology may supersede the properties assessed by each in silico tool. However, given the evidence that these tools can still distinguish case from control nsSNVs when analyzed inside and outside of the critical regions, this would suggest that their use may enhance interpretation within the regions of the channel.
Correlation Between Predicted In Silico and Observed In Vitro Electrophysiological Phenotypes
Given that the use of in silico phenotype prediction tools and electrophysiological studies represent 2 of the primary means to further explore the possible pathogenicity of rare nsSNVs, we examined the level of agreement between these 2 approaches using a catalog of previously characterized SCN5A nsSNVs in the literature as well as novel, unpublished SCN5A nsSNVs functionally characterized by our collective research groups in an effort to further evaluate the ability of in silico tools to aid in nsSNV classification.
Overall, 145 SCN5A variants with heterologous expression/cellular electrophysiological data were identified. Importantly, all variants with cellular electrophysiological data were included regardless of frequency in the controls. Of the 146 functionally characterized nsSNVs, 117 were reported previously and 29 had novel electrophysiological data generated for this study. The electrophysiological parameters for the novel functional variants are summarized in Table 5 (all electrophysiological data in Tables II–IV in the Data Supplement). Conflicting electrophysiological data was found for 6 variants and these were removed from the subsequent comparative analyses with the in silico tools. Among the remaining 139 functionally characterized nsSNVs, 106 displayed an abnormal functional phenotype and 33 were wild type.
Table 5.
Summary of Cellular Electrophysiological Parameters for Novel Heterologously Expressed and Functionally Characterized SCN5A Variants
| SCN5A Variants | Current Density % Change | Activation V1/2 Shift, mV | Inactivation V1/2 Shift, mV | Late Current % Change | Functional Status | Resource |
|---|---|---|---|---|---|---|
| R18W | −19 | 1 | +11* | +129* | Pathogenic | Madison |
| E30G-Q1077del | 10 | 0 | +3* | 83 | Benign | Mayo |
| E48K-Q1077del | −37 | +4* | −3 | 0 | Benign† | Mayo |
| E48K-Q1077 | −22 | 4 | 0 | 38 | Benign† | Mayo |
| Y87C-Q1077del | −31 | 1 | −2 | 15 | Benign | Mayo |
| V125L | −29 | 4 | 4 | 57 | Benign | Madison |
| A185T | −44 | 3 | +7* | 143 | Pathogenic | Madison |
| R190Q-Q1077del | −8 | 0 | 1 | 21 | Benign | Mayo |
| Q245K | 30 | −1 | 2 | 29 | Benign | Madison |
| Q245K-Q1077del | −16 | −4 | 1 | 0 | Benign | Mayo |
| I397F-Q1077del | −37* | −7* | +5* | +382* | Pathogenic | Mayo |
| I397F-Q1077 | −46* | 0 | +9* | +575* | Pathogenic | Mayo |
| E462A-Q1077del | −9 | −3 | 1 | −5 | Benign | Mayo |
| E462A-Q1077 | −2 | −2 | 1 | 33 | Benign | Mayo |
| E462K-Q1077del | −9 | −2 | −3 | −15 | Benign | Mayo |
| E462K-Q1077del/R558 | 28 | −3 | −1 | 19 | Benign | Mayo |
| R569G-Q1077del | −7 | −7* | 1 | −10 | Pathogenic | Mayo |
| R569W-Q1077del | 10 | −1 | 1 | −33 | Benign | Mayo |
| R620C-Q1077del | −12 | 1 | +2* | −40 | Benign† | Mayo |
| P627L-Q1077del | −12 | −1 | 1 | −18 | Benign | Mayo |
| Q692K-Q1077del | −6 | −2 | −4* | −19 | Benign† | Mayo |
| Q692K-Q1077 | 40 | −1 | 2 | 23 | Benign | Mayo |
| Q750R-Q1077del | −52* | 3 | −2 | 14 | Pathogenic | Mayo |
| Q750R-Q1077 | −6 | 0 | −3* | 0 | Benign† | Mayo |
| R800L-Q1077del | −37* | −1 | 2 | 26 | Pathogenic | Mayo |
| R878H | No current | … | … | … | Pathogenic | Madison |
| R893H in H/R558 | No current | … | … | … | Pathogenic | Madison |
| G897E-Q1077del | No current | … | … | … | Pathogenic | Mayo |
| E1208K-Q1077del/R558 | −18 | −4* | −8* | −29 | Pathogenic | Mayo |
| M1320V-Q1077del | 8 | 1 | 2 | 63 | Benign | Mayo |
| I1485V-Q1077del | 1 | – | +7* | 21 | Pathogenic | Mayo |
| I1593M-Q1077del/R558 | 11 | −4* | 2 | −10 | Benign† | Mayo |
| T1708N-Q1077del/H558 | −25 | 6 | 3 | +133* | Pathogenic | Madison |
| T1708N-Q1077del/R558 | −63* | 4 | 1 | +133* | Pathogenic | Madison |
| T1779M-Q1077del | −17 | 2 | 2 | 44 | Benign | Mayo |
| T1779M-Q1077del/R558 | 10 | −3 | −1 | −23 | Benign | Mayo |
| D1819N-Q1077del | −24 | −2 | −1 | −12 | Benign | Mayo |
| D1819N-Q1077del/R558 | 31 | −2 | −5* | 13 | Benign† | Mayo |
| Q1909R | −42 | 4 | 7 | +271* | Pathogenic | Madison |
Mayo indicates Mayo Clinic Windland Smith Rice Sudden Death Genomics Laboratory; Madison, Makielski laboratory at the University of Wisconsin–Madison; and WT, wild-type.
P<0.05 vs WT.
≤5 mV shift of V1/2 even though P<0.05 vs WT.
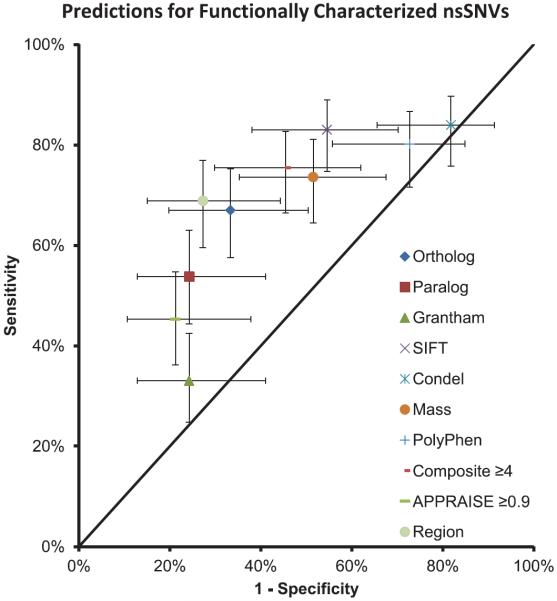
Overall, 75% of those SCN5A nsSNVs that imparted either an LQT3-like gain-of-function or BrS1-like loss-of-function phenotype were predicted to be pathogenic by ≥4 in silico phenotype prediction tools (Figure 3). Furthermore, 45% of the SCN5A nsSNVs with a wild-type electrophysiological phenotype were predicted nevertheless to be pathogenic by ≥4 in silico phenotype prediction tools (P=0.002; Figure 3). As seen with the comparison of case- and control- nsSNVs, there is a significant false-positive issue for SIFT, Condel, MAss, and PolyPhen2 and a potential false-negative issue with Grantham values and paralog conservation. Interestingly, ortholog conservation and SIFT alone seem to be comparable with the composite scores. In addition, we assessed the performance of the recently reported APPRAISE algorithm, which was designed using a Bayesian model and optimized for cardiac channelopathy/cardiomyopathy genes.20 Although this optimized tool was able to statistically distinguish variants with abnormal cellular electrophysiological phenotypes from those with wild-type properties, this tool's sensitivity was lower than the use of composite score. Interestingly, the simple use of the identified critical topological regions actually had the highest Matthews correlation coefficient value, again suggesting that the protein topology may supersede the properties assessed by each in silico tool. However, this collectively provides evidence that in silico phenotype prediction tools are concordant overall with the observed in vitro electrophysiological phenotype of SCN5A nsSNVs but still with an alarming number of mismatches.
Figure 3.
Correlation of in silico pathogenicity predictions with cellular electrophysiological studies of SCN5A variants. The points on the graph represent the sensitivity and 1-specificity for each in silico tool when assessing only the functionally characterized variants. The error bars represent the 95% confidence interval for both the sensitivity and specificity. nsSNVs indicates nonsynonymous single-nucleotide variants.
Enhancement of Topology-Derived EPVs for SCN5A nsSNVs in BrS and LQTS by the Synergistic Use of These In Silico Tools
Given the ability of the in silico tools to distinguish rare case– from control–derived nsSNVs, even when removing the over-represented nsSNVs, as well as provide statistically concordant predictions with in vitro electrophysiological phenotype, we next sought to determine if the in silico tools could enhance the topology-derived EPVs.
We applied a synergistic approach to the BrS and LQTS cases to generate topology plus in silico–derived EPVs. For BrS, region-specific EPVs for rare nsSNVs localizing to the suboptimal regions outside the transmembrane region (EPV=60%; 95% CI, 40–73) were enhanced to an EPV of 83% (95% CI, 69–91) when ≥4 tools were in agreement and dropped the EPV to 19 (95% CI, 0–55) when <4 tools were in agreement. Surprisingly, the composite score also separated the EPVs within the transmembrane region with the EPV maintained at >90% when ≥4 tools were in agreement (EPV=96%; 95% CI, 93–97), but fell to 66% (95% CI, 21–85) when <4 tools were in agreement (Tables 6 and 7).
Table 6.
Topology-Speicific EPVs When ≥4 of 6 Tools Are in Agreement
| Control (8975) |
BrS (2111) |
|||
|---|---|---|---|---|
| Region | Tools | Count (%) | Count (%) | EPV (95% CI) |
| Transmembrane | ≥4 | 23 (0.26) | 126 (5.97) | 96 (93–97)* |
| <4 | 13 (0.14) | 9 (0.43) | 66 (21–85)* | |
| Outside transmembrane | ≥4 | 17 (0.19) | 24 (1.14) | 83 (69–91)* |
| <4 | 48 (0.53) | 14 (0.66) | 19 (0–55)* | |
CI indicates confidence interval; BrS, Brugada syndrome; and EPV, estimated predictive values.
Regions where the synergistic use of the in silico tools statistically augmented the topology-based predictions.
Table 7.
Topology-Specific EPVs When ≥4 of 6 Tools Are in Agreement
| Control (8975) |
LQTS (2888) |
|||
|---|---|---|---|---|
| Region | Tools | Count (%) | Count (%) | EPV (95% CI) |
| S3−S5+S6 or DIII/DIV IDL | ≥4 | 10 (0.11) | 44 (1.52) | 93 (85–96) |
| <4 | 2 (0.02) | 0 (0.00) | 0 (NA) | |
| Outside S3−S5+S6 or DIII/DIV IDL | ≥4 | 30 (0.33) | 32 (1.11) | 70 (50–82)* |
| <4 | 59 (0.66) | 22 (0.76) | 14 (0–47)* | |
CI indicates confidence interval; DIII, transmembrane domain III; DIV, transmembrane domain IV; EPV, estimated predictive values; IDL, interdomain linker; and LQTS, long-QT syndrome.
Regions where the synergistic use of the in silico tools statistically augmented the topology-based predictions.
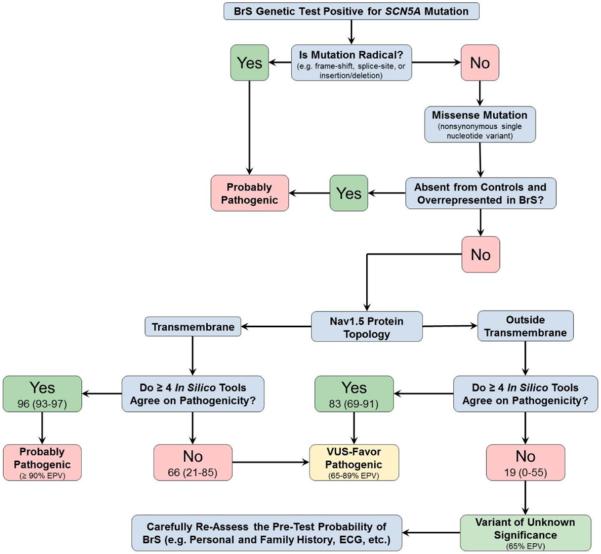
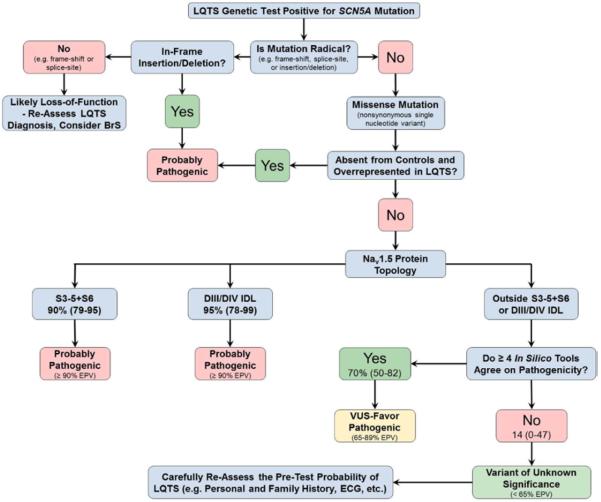
Similar to BrS, when ≥4 tools were in agreement, the LQTS-associated EPVs for rare nsSNVs localizing to the suboptimal regions outside of the S3−S5+S6 transmembrane regions and the DIII/DIV IDL (EPV=47%; 95% CI, 26–68) rose to 70% (95% CI, 50–82), whereas the EPV dropped to 14% (95% CI, 0–47; Table 6) when <4 tools suggested pathogenicity. Although the composite score was unable to raise the EPVs outside the critical regions to a high probability of pathogenicity, they were able to separate the EPVs for those nsSNVs with ≥4 from those with <4 providing the physician with an additional piece of evidence in the interpretation of nsSNVs. The EPVs for all examined regions are shown in Table V in the Data Supplement. An evidence-based decision tree was generated for interpretation of an SCN5A variant in the context of either a BrS (Figure 4) or LQTS (Figure 5) evaluation using the empirical data provided in this study.
Figure 4.
Evidence-based decision tree for SCN5A nonsynonymous single-nucleotide variant classification in the setting of a suspected Brugada syndrome (BrS) case. EPV indicates estimated predictive value; and VUS, variant of uncertain/unknown significance.
Figure 5.
Evidence-based decision tree for SCN5A nonsynonymous single-nucleotide variant classification in the setting of a suspected long-QT syndrome (LQTS) case. BrS indicates Brugada syndrome; DIII, transmembrane domain III; DIV, transmembrane domain IV; EPV, estimated predictive values; IDL, interdomain linker; and VUS, variant of uncertain/unknown significance.
Discussion
Rare Variant Interpretation in the Era of Exome Sequencing
With the release of large public databases of genetic variation from next-generation sequencing projects such as the 1000 genomes (1kG), the National Heart Lung and Blood Institute ESP, and now most recently the 60 000+ exomes represented in the Exome Aggregation Consortium, our understanding of the normal range of background genetic variation within cardiac channelopathy–susceptibility genes has grown substantially in recent years. Although it is well established that the interpretation of rare nsSNVs in the major BrS- (SCN5A) and LQTS-susceptibility (KCNQ1, KCNH2, and SCN5A) genes is complicated by the existence of a background noise rate of 3% to 4% among whites and 6% to 8% among nonwhites, recent descriptive analyses revealed that ≈10% (38/355)22 of previously published BrS1-associated mutations and ≈5% (33/631)23 of previously published LQT3-associated mutations reside in the ESP population suggesting that many of these mutations may be annotated wrongly as pathogenic mutations.
These exome-based studies reveal a glaring discrepancy between the prevalence of a putative BrS1 or LQT3 genotype (≈1:84 for LQTS and ≈1:145 for BrS) and the highest reported prevalence of either disease phenotype (≈1:13 000 for LQT3 and ≈1:17 000 for Br1) in the general population.22,23 Thus, in the absence of any clinical index of suspicion, the identification of a rare SCN5A variant alone has only a 1 in 200 chance of being pathogenic. When discovered in a patient with clinical suspicion for either LQTS or BrS, that rare variant's possibility of pathogenicity climbs significantly but as shown here, its identification should not be elevated to pathogenic status without careful scrutiny.
Regardless of how one chooses to interpret these recent findings, the troubling discordance between genotype and phenotype prevalence in both BrS and LQTS serves to re-enforce the fact that the interpretation of genetic test results for these disorders should be viewed always as strictly probabilistic, rather than binary/deterministic, in nature.12 Of course, the clinical use of genetic testing is enhanced greatly when limited to those individuals with a high index of suspicion/pretest probability of disease based on personal/family history and objective electrocardiographic parameters as well as interpreted in conjunction with available in vitro functional data and previously established high-EPV criteria,12,14 such as mutation type and location. In reality, however, pertinent clinical/historical data are not always available before genetic testing is pursued and the vast majority of nsSNVs encountered clinically have not been characterized functionally.
As such, several nsSNVs still fall within gray areas that are difficult to interpret and the numbers of such nsSNVs are expected to grow as the use of exome/genome sequencing becomes more widespread in clinical practice.14 In addition, this issue is compounded by the understanding that only 25 specific nsSNVs (BrS, 12.1%; LQTS, 5.3%), which are over-represented statistically in disease, account for a disproportionately large percentage of SCN5A positive genetic tests (BrS, 36.6%; LQTS, 24.0%), making the interpretation of the next new nsSNV in SCN5A even more difficult. Despite these issues, the emergence of exome/genome sequencing also promises to allow researchers to systematically study background genetic variation like never before, providing an important opportunity to use population-scale genetic data to significantly improve the criteria used to distinguish rare putative pathogenic mutations from equally rare, yet innocuous genetic variants identified in heritable cardiac channelopathy–susceptibility genes.
Use of Mutation Compendia and Public Exome/Genome Data Sets to Define/Refine the Topology-Based Probability of Pathogenicity for BrS- and LQTS-Associated SCN5A nsSNVs
Given the myriad of issues currently associated with the use of in vitro heterologous expression studies and patient-specific induced pluripotent stem cells, most recent attempts to enhance the interpretability of cardiac channelopathy genetic testing results have focused on the development of gene, mutation type, and topology/region-specific probability of pathogenicity criteria derived from the systematic evaluation of all genetic variation present in the genes of interest within large case and control cohorts. Unfortunately, attempts to improve the interpretability of a SCN5A-positive LQTS genetic test through the identification of specific structure–function domains in the Nav1.5 channel associated with a high probability of pathogenicity (ie, EPV >90%) have languished in comparison with the other 2 major LQTS-susceptibility genes (KCNQ1 and KCNH2) as a result of the (1) the significant background noise rate in SCN5A (≈2% in whites and ≈5% in nonwhites), (2) the relative paucity of SCN5A mutations in clinically compelling cases, and (3) the even scattering of these case and control variants throughout the significantly larger coding region of the SCN5A gene (ie, more nucleotides/amino acids to harbor rare unique nsSNVs).14 Likely secondary to a similar set of issues, attempts to improve the interpretability of a SCN5A-positive BrS genetic test or use a comparative approach to determine if different structure–function domains are more/less likely to harbor BrS-associated, LQTS-associated, or functionally pleiotropic SCN5A mutations have not been undertaken.
To circumvent these issues, in this case–control study, we examined a large number of case- and control-derived SCN5A nsSNVs identified in 2888 previously published LQTS cases and 2111 previously published BrS cases to identify topological regions likely to be critical to the pathogenesis of disease via an over-representation of case nsSNVs and use this information to refine the previously published topology-based probability of pathogenicity for SCN5A nsSNVs identified in LQTS cases and for the first time generate topology-based probability of pathogenicity for SCN5A nsSNVs identified in BrS.
Through this analysis, BrS-derived SCN5A nsSNVs localizing to any of the transmembrane spanning domains had a high probability of pathogenicity (EPV>90%). In addition, unlike previous LQT3 studies, there was now adequate power to assess specific subdomains within the transmembrane and IDL regions of the Nav1.5 channel. In this subanalysis, LQTS-derived SCN5A nsSNVs localizing to certain transmembrane segments (S3−S5+S6) and DIII/DIV IDL had a high probability of pathogenicity (EPV>90%).
Although the overall SNR for nsSNVs after the removal of the 25 over-represented variants dropped to 7.3:1 for BrS and 3.0:1 for LQTS, the use of these identified critical regions raised the SNR to 15.9:1 for nsSNVs within the transmembrane for BrS and to 11.4:1 for nsSNVs within either the S3−S5+S6 domains or DIII/DIV's IDL for LQTS. In contrast, based on topology-alone, extreme caution must still be exercised when attempting to interpret the pathogenicity of similarly rare BrS or LQTS SCN5A nsSNVs that do not localize to these aforementioned regions of the Nav1.5 channel.
Synergistic Use of In Silico Tools to Enhance the Classification of BrS- and LQTS-Associated SCN5A nsSNVs Localizing to Suboptimal Regions of the Nav1.5 Channel
Although the use of larger case and control cohorts provided important insights in regards to the location of high-probability BrS- and LQTS-derived SCN5A nsSNVs, 22% of BrS-derived SCN5A nsSNVs and 52% of LQTS-derived SCN5A nsSNVs localize too difficult to interpret low probability of pathogenicity regions. As such, there is a pressing need to develop accurate, rapid, and cost-effective methods of identifying high- or low-probability of pathogenicity SCN5A nsSNVs that reside in these low EPV topologic regions of the Nav1.5 channel.
Given the speed and low overhead associated with informatics-based analyses, the use of in silico phenotype prediction algorithms to enhance the classification of SCN5A nsSNVs localizing to problematic topological regions represents an attractive option. With the exception of the Grantham matrix score, the remaining 6 in silico tools all demonstrated a statistically significant ability to differentiate case- and control-derived SCN5A nsSNVs over the entire protein, as well as when nsSNVs inside and outside the identified critical regions were assessed separately. However, if the combined EPV for all BrS- and LQTS-derived SCN5A nsSNVs (79%; CI, 74% to 83%) reflects a relatively accurate estimate of the percentage of the case-derived nsSNVs that are truly pathogenic, it seems that certain in silico tools are overly conservative (ortholog and paralog conservation) classifying only 57% to 62% of case-derived nsSNVs as pathogenic, whereas other in silico tools are overly liberal (SIFT, Condel, Mass, PolyPhen2, and Composite) classifying 40% to 63% of control-derived nsSNVs as pathogenic. Thus, reliance solely on in silico tools may result in an unacceptably high false-negative rate or unacceptably high false-positive rate.
Although recent studies have highlighted issues with using electrophysiological studies to identify pathogenic nsSNVs, their use still represents the gold standard for the determination of the pathogenicity of a given variant.24 Therefore, we further assessed the in silico tools predictive ability using functionally characterized nsSNVs from the literature and 29 nsSNVs newly characterized for this study. Overall, the synergistic use of in silico tools may provide a reasonable albeit imperfect surrogate for in vitro functional characterization as ≥4 of the 6 in silico tools correctly predicted 75% of variants with abnormal electrophysiology as pathogenic and 66% of variants with wild-type electrophysiology as benign. Although electrophysiological studies are the current gold standard for assigning a pathogenic status, these studies are not without issue. This is highlighted by the identification of 6 functionally characterized variants with discordant electrophysiological phenotypes.
Despite the overall performance of in silico tools, the use of these tools is severely limited because of the major issues of potential false positives and false negatives derived from these tools. In addition, the finding that case-derived nsSNVs residing within the identified critical regions are more likely to be predicted pathogenic than case-derived nsSNVs localizing outside these identified regions may suggest that protein topology alone is the primary determinant of pathogenicity in many of the regions of the Nav1.5 channel. However, given that the in silico tools still maintained the ability to distinguish case- from control-derived nsSNVs when assessed inside and outside of the critical regions suggests that these tools may enhance the topology-derived prediction. The use of a ≥4 composite score separated the EPVs of SCN5A nsSNVs in the transmembrane (94% to 96% and 66% with ≥4 and <4 tools, respectively) and outside the transmembrane (60% to 83% and 19% with ≥4 and <4 tools, respectively) regions in BrS. In addition, the composite tool separated the EPV for nsSNVs falling outside of S3−S5+S6 or DIII/DIV IDL (47% to 70% and 14% with ≥4 and <4 tools, respectively). Although the in silico tools were unable to bring the EPV above 90% for BrS or LQTS, the use of the composite score can provide additional information elevating nsSNVs outside of the critical region from the purgatory of a variant of uncertain/unknown significance (VUS) classification to the VUS-favor pathogenic. The issues identified for the in silico tools should limit the clinical use of these tools, however, the identification that these tools can enhance the topology-derived EPVs may provide additional information in the setting of a difficult to interpret nsSNV.
Collectively, these findings suggest that BrS and LQTS genetic test results should be interpreted cautiously in the context of the topological location of the SCN5A nsSNV, particularly for those BrS-derived nsSNVs localizing outside of the transmembrane region and those LQTS-derived nsSNVs localizing outside the S3−S5+S6 subdomains or the DIII/DIV IDL. Unfortunately, those rare SCN5A nsSNVs that reside outside of these specified regions remain ambiguously classified as a VUS (ie, stuck in genetic purgatory) and require additional cosegregation or functional data to upgrade their probability of pathogenicity at this time. However, the use of a composite score that relies on 7 distinct in silico algorithms may enable rare SCN5A nsSNVs localizing outside of these high probability regions to be upgraded to possibly pathogenic when >4 in silico tools point toward pathogenicity or downgraded further when <4 of the tools do so (Figures 4 and 5).
Limitations
There are several limitations inherent to the nature of this case–control study. First and foremost, not every rare SCN5A nsSNV that was derived from 1 of the control populations is likely to be completely devoid of functional significance. Given that neither a 12-lead ECG nor comprehensive cardiac evaluation was a prerequisite for enrollment in either the Sanger-sequenced in-house controls or the public exome/genome databases, it stands to reason that a small number of control-derived nsSNVs could in fact be disease-causative mutations and therefore a small number of the 8975 controls may in fact have genetic susceptibility for either BrS1 or LQT3.
Thus, the catalog of genetic variants derived from next-generation sequencing projects, such as 1kG and ESP, is not implicitly nonpathogenic. Rather, as the number of individuals sequenced approaches or exceeds disease prevalence, the likelihood of encountering bona fide disease-causative mutations in the control population increases. However, given the highest estimates for the prevalence of SCN5A-mediated BrS (≈1:10 000) and SCN5A-mediated LQTS (≈1:13 000), only 5 of the 101 distinct control-derived nsSNVs from among the nearly 9000 controls have a statistical chance of actually being either a BrS1- or LQT3-causative SCN5A mutation. To that end, we have identified the most likely false negatives within the control population based on the low frequency in the controls, localization to either the BrS- or LQTS-associated critical regions, and a composite score of 6 (ie, all in silico tools predicting pathogenic). These variants are detailed in Table VI in the Data Supplement. Furthermore, even if larger number of control-derived SCN5A nsSNVs included in this study were truly pathogenic, the presented EPVs in this study would represent conservative underestimates.
Conversely, given the loosened clinical stringency associated with the use of published BrS and LQTS mutation compendia, it is possible that some cases may have been referred for genetic testing without a robust phenotype and as such, may in fact not have either BrS1 or LQT3. However, for the BrS cohort, the overall yield was in line with previous estimates in other BrS cohorts suggesting that the overall rate of misdiagnosis is equivalent to other cohorts studied.3–5 In addition for the LQTS cohort, the yield of nsSNVs in the clinically definite cases was statistically indistinguishable from the referral cases, suggesting again that the overall rate of misdiagnosis for LQT3 was similar between the cohorts. However, the rate of background genetic variation in SCN5A dictates that roughly 1:20 to 1:50 SCN5A mutations, identified in either BrS or LQTS cases represent potential false positives. Therefore, it is possible that 5 to 13 of the 255 distinct, case-derived SCN5A nsSNVs are false positives. However, their erroneous classification would again cause the derived EPVs to represent conservative underestimates. Here, based on their localization outside the critical regions, and composite score of zero (ie, all in silico tools predicting benign), we speculate on which case-derived variants are most likely the false positives (Table VI in the Data Supplement). Functional characterization of this false-positive and false-negative hit list of nsSNVs could provide a validation of these methods used within this study.
Second, because of an inability to determine allelic status with 100% certainty in ESP individuals, neither the case or control population was adjusted for the presence of nsSNV multiplicity. Third, because of the increased genetic diversity in nonwhites, the over-representation of nonwhite individuals in the control relative to the case population suggests the reported EPVs are overly conservative.
Finally, although we have used 7 commonly used in silico tools, there are many more which could be used. Rather than encompassing all known tools or providing a statistically optimized tool weighing all variables in the form of a new in silico algorithm, we hoped to provide a framework for the proper assessment of novel nsSNVs. Consistent with the recent American College of Medical Genetics and Genomics guidelines, the use of a single in silico prediction tool should be discouraged and should be replaced by the synergistic use of multiple tools.25 Although the addition of other tools, such as the APPRAISE algorithm, may further enhance the interpretation, the necessary time to assess an even greater number of in silico tools may reach a ceiling effect. However, despite these stated limitations, this study still gleaned several robust observations that will affect significantly how a SCN5A-positive BrS or LQTS genetic test result is interpreted in the future.
Conclusions
The significant background noise and relatively low yield of mutations in BrS1/LQT3 makes the interpretation of a rare SCN5A nsSNVs tremendously challenging for clinicians. Therefore, we have used a large case/control study to generate topology-based estimative predictive values to aid in the interpretation; identifying rare variants localizing to the transmembrane regions in BrS and the S3−S5+S6 subsegments of the transmembrane and the DIII/DIV interdomain linker in LQTS as high probability disease variants. Topology alone would leave the nsSNVs residing outside these identified regions in genetic purgatory. However, the synergistic use of in silico tools may enhance topology-derived predictions allowing for the elevation of nsSNVs within certain regions to moderate probability of pathogenicity. Importantly, extreme caution must be used when using in silico tools to enhance variant classification, especially when using a single tool. In addition, we have identified a correlation between the composite predictions of in silico tools and in vitro cellular functional studies, providing evidence for the potential use of multiple in silico tools by physicians in the time between identification of a novel SCN5A mutation and functional characterization.
Supplementary Material
CLINICAL PERSPECTIVE.
The significant background noise and relatively low yield of mutations in type 1 Brugada syndrome/type 3 long-QT syndrome makes the interpretation of a rare SCN5A nonsynonymous single-nucleotide variants tremendously challenging for clinicians. Therefore, we have used a large case/control study to generate topology-based estimative predictive values to aid in the interpretation; identifying rare variants localizing to the transmembrane regions in Brugada syndrome and the S3−S5+S6 subsegments of the transmembrane and the DIII/DIV interdomain linker in long-QT syndrome as high probability disease variants. Topology alone would leave the nonsynonymous single-nucleotide variants residing outside these identified regions in genetic purgatory. However, the synergistic use of in silico tools may enhance topology-derived predictions allowing for the elevation of nonsynonymous single-nucleotide variants within certain regions to moderate probability of pathogenicity. Importantly, extreme caution must be used when using in silico tools to enhance variant classification, especially when using a single tool. In addition, we have identified a correlation between the composite predictions of in silico tools and in vitro cellular functional studies, providing evidence for the potential use of multiple in silico tools by physicians in the time between identification of a novel SCN5A mutation and functional characterization.
Acknowledgments
Sources of Funding This work was supported by the Windland Smith Rice Sudden Comprehensive Sudden Cardiac Death Program (Dr Ackerman) and the Dutch Heart Foundation, Dutch Federation of University Medical Centres, the Netherlands Organisation for Health Research and Development and the Royal Netherlands Academy of Sciences (A. A. Wilde). J. D. Kapplinger is supported by the National Institutes of Health grant GM72474-08. J. D. Kapplinger and Dr Giudicessi thank the Mayo Clinic Medical Scientist Training Program, and Dr Giudicessi thanks the Mayo Clinic Clinician Investigator Training Program for fostering an outstanding environment for physician–scientist training.
Footnotes
The Data Supplement is available at http://circgenetics.ahajournals.org/lookup/suppl/doi:10.1161/CIRCGENETICS.114.000831/-/DC1.
Disclosures Dr Callis is an employee of Transgenomic, Inc. Dr Ackerman is a consultant for Boston Scientific, Gilead Sciences, Medtronic, and St. Jude Medical. Dr Ackerman, D. J. Tester, and Mayo Clinic receive royalties from Transgenomic's FAMILION-LQTS and FAMILION-CPVT genetic tests.
References
- 1.Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson JL, et al. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 2.Giudicessi JR, Ackerman MJ. Determinants of incomplete penetrance and variable expressivity in heritable cardiac arrhythmia syndromes. Transl Res. 2013;161:1–14. doi: 10.1016/j.trsl.2012.08.005. doi: 10.1016/j.trsl.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Priori SG, Napolitano C, Gasparini M, Pappone C, Della Bella P, Brignole M, et al. Clinical and genetic heterogeneity of right bundle branch block and ST-segment elevation syndrome: a prospective evaluation of 52 families. Circulation. 2000;102:2509–2515. doi: 10.1161/01.cir.102.20.2509. [DOI] [PubMed] [Google Scholar]
- 4.Priori SG, Napolitano C, Gasparini M, Pappone C, Della Bella P, Giordano U, et al. Natural history of Brugada syndrome: insights for risk stratification and management. Circulation. 2002;105:1342–1347. doi: 10.1161/hc1102.105288. [DOI] [PubMed] [Google Scholar]
- 5.Crotti L, Marcou CA, Tester DJ, Castelletti S, Giudicessi JR, Torchio M, et al. Spectrum and prevalence of mutations involving BrS1- through BrS12-susceptibility genes in a cohort of unrelated patients referred for Brugada syndrome genetic testing: implications for genetic testing. J Am Coll Cardiol. 2012;60:1410–1418. doi: 10.1016/j.jacc.2012.04.037. doi: 10.1016/j.jacc.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Splawski I, Shen J, Timothy KW, Lehmann MH, Priori S, Robinson JL, et al. Spectrum of mutations in long-QT syndrome genes. KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 2000;102:1178–1185. doi: 10.1161/01.cir.102.10.1178. [DOI] [PubMed] [Google Scholar]
- 7.Tester DJ, Will ML, Haglund CM, Ackerman MJ. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm. 2005;2:507–517. doi: 10.1016/j.hrthm.2005.01.020. doi: 10.1016/j.hrthm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 8.Zareba W, Moss AJ, Schwartz PJ, Vincent GM, Robinson JL, Priori SG, et al. Influence of genotype on the clinical course of the long-QT syndrome. International Long-QT Syndrome Registry Research Group. N Engl J Med. 1998;339:960–965. doi: 10.1056/NEJM199810013391404. doi: 10.1056/NEJM199810013391404. [DOI] [PubMed] [Google Scholar]
- 9.Meregalli PG, Tan HL, Probst V, Koopmann TT, Tanck MW, Bhuiyan ZA, et al. Type of SCN5A mutation determines clinical severity and degree of conduction slowing in loss-of-function sodium channelopathies. Heart Rhythm. 2009;6:341–348. doi: 10.1016/j.hrthm.2008.11.009. doi: 10.1016/j.hrthm.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 10.Ackerman MJ, Splawski I, Makielski JC, Tester DJ, Will ML, Timothy KW, et al. Spectrum and prevalence of cardiac sodium channel variants among black, white, Asian, and Hispanic individuals: implications for arrhythmogenic susceptibility and Brugada/long QT syndrome genetic testing. Heart Rhythm. 2004;1:600–607. doi: 10.1016/j.hrthm.2004.07.013. doi: 10.1016/j.hrthm.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 11.Kapplinger JD, Tester DJ, Salisbury BA, Carr JL, Harris-Kerr C, Pollevick GD, et al. Spectrum and prevalence of mutations from the first 2,500 consecutive unrelated patients referred for the FAMILION long QT syndrome genetic test. Heart Rhythm. 2009;6:1297–1303. doi: 10.1016/j.hrthm.2009.05.021. doi: 10.1016/j.hrthm.2009.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapa S, Tester DJ, Salisbury BA, Harris-Kerr C, Pungliya MS, Alders M, et al. Genetic testing for long-QT syndrome: distinguishing pathogenic mutations from benign variants. Circulation. 2009;120:1752–1760. doi: 10.1161/CIRCULATIONAHA.109.863076. doi: 10.1161/CIRCULATIONAHA.109.863076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapplinger JD, Tester DJ, Alders M, Benito B, Berthet M, Brugada J, et al. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm. 2010;7:33–46. doi: 10.1016/j.hrthm.2009.09.069. doi: 10.1016/j.hrthm.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giudicessi JR, Kapplinger JD, Tester DJ, Alders M, Salisbury BA, Wilde AA, et al. Phylogenetic and physicochemical analyses enhance the classification of rare nonsynonymous single nucleotide variants in type 1 and 2 long-QT syndrome. Circ Cardiovasc Genet. 2012;5:519–528. doi: 10.1161/CIRCGENETICS.112.963785. doi: 10.1161/CIRCGENETICS.112.963785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsh R, Peters NS, Cook SA, Ware JS. Paralogue annotation identifies novel pathogenic variants in patients with Brugada syndrome and catecholaminergic polymorphic ventricular tachycardia. J Med Genet. 2014;51:35–44. doi: 10.1136/jmedgenet-2013-101917. doi: 10.1136/jmedgenet-2013-101917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juang JM, Lu TP, Lai LC, Hsueh CH, Liu YB, Tsai CT, et al. Utilizing multiple in silico analyses to identify putative causal SCN5A variants in Brugada syndrome. Sci Rep. 2014;4:3850. doi: 10.1038/srep03850. doi: 10.1038/srep03850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riuró H, Campuzano O, Berne P, Arbelo E, Iglesias A, Pérez-Serra A, et al. Genetic analysis, in silico prediction, and family segregation in long QT syndrome. Eur J Hum Genet. 2015;23:79–85. doi: 10.1038/ejhg.2014.54. doi: 10.1038/ejhg.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Junker VL, Apweiler R, Bairoch A. Representation of functional information in the SWISS-PROT data bank. Bioinformatics. 1999;15:1066–1067. doi: 10.1093/bioinformatics/15.12.1066. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, Li Z, Shen J, Keating MT. Genomic organization of the human SCN5A gene encoding the cardiac sodium channel. Genomics. 1996;34:9–16. doi: 10.1006/geno.1996.0236. doi: 10.1006/geno.1996.0236. [DOI] [PubMed] [Google Scholar]
- 20.Ruklisa D, Ware JS, Walsh R, Balding DJ, Cook SA. Bayesian models for syndrome- and gene-specific probabilities of novel variant pathogenicity. Genome Med. 2015;7:5. doi: 10.1186/s13073-014-0120-4. doi: 10.1186/s13073-014-0120-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopes LR, Zekavati A, Syrris P, Hubank M, Giambartolomei C, Dalageorgou C, et al. Uk10k Consortium. Genetic complexity in hypertrophic cardiomyopathy revealed by high-throughput sequencing. J Med Genet. 2013;50:228–239. doi: 10.1136/jmedgenet-2012-101270. doi: 10.1136/jmedgenet-2012-101270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Risgaard B, Jabbari R, Refsgaard L, Holst AG, Haunsø S, Sadjadieh A, et al. High prevalence of genetic variants previously associated with Brugada syndrome in new exome data. Clin Genet. 2013;84:489–495. doi: 10.1111/cge.12126. doi: 10.1111/cge.12126. [DOI] [PubMed] [Google Scholar]
- 23.Refsgaard L, Holst AG, Sadjadieh G, Haunsø S, Nielsen JB, Olesen MS. High prevalence of genetic variants previously associated with LQT syndrome in new exome data. Eur J Hum Genet. 2012;20:905–908. doi: 10.1038/ejhg.2012.23. doi: 10.1038/ejhg.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe H, Yang T, Stroud DM, Lowe JS, Harris L, Atack TC, et al. Striking In vivo phenotype of a disease-associated human SCN5A mutation producing minimal changes in vitro. Circulation. 2011;124:1001–1011. doi: 10.1161/CIRCULATIONAHA.110.987248. doi: 10.1161/CIRCULATIONAHA.110.987248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–23. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.