Summary
Copper (Cu) is an essential micronutrient that functions as a cofactor in several important enzymes, like respiratory heme-copper oxygen reductases. Yet, Cu is also toxic and therefore cells engage a highly coordinated Cu uptake and delivery system to prevent the accumulation of toxic Cu concentrations. In the current work we analyzed Cu delivery to the cbb3-type cytochrome c oxidase (cbb3-Cox) of Rhodobacter capsulatus. We identified the PCuAC-like periplasmic chaperone PccA and analyzed its contribution to cbb3-Cox assembly. Our data demonstrate that PccA is a Cu-binding protein with a preference for Cu(I), which is required for efficient cbb3-Cox assembly, in particular at low Cu concentrations. By using in vivo and in vitro crosslinking we show that PccA forms a complex with the Sco1-homologue SenC. This complex is stabilized in the absence of the cbb3-Cox specific assembly factors CcoGHIS. In cells lacking SenC, the cytoplasmic Cu content is significantly increased, but the simultaneous absence of PccA prevents this Cu accumulation. These data demonstrate that the interplay between PccA and SenC is not only required for Cu delivery during cbb3-Cox assembly, but that it also regulates Cu homeostasis in R. capsulatus.
Keywords: cbb3-type cytochrome c oxidase biogenesis, periplasmic copper chaperones, copper homeostasis, respiration, Rhodobacter capsulatus
Graphical abstract
Introduction
Cytochrome c oxidases (Cox) are ubiquitous multi-subunit enzyme complexes that terminate aerobic respiratory chains in prokaryotic and eukaryotic cells. They are members of the heme copper oxidase (HCO) superfamily and are characterized by the presence of a membrane integral catalytic subunit I, which contains a low-spin heme and a high-spin heme-CuB binuclear center (Ekici et al., 2012a, Richter & Ludwig, 2003). Cox enzymes are generally classified into three groups: type A includes the mitochondrial-like aa3-Cox, type B the ba3-Cox and type C, the cbb3-Cox, which is the most distant member of the HCO superfamily and present only in bacteria (Brzezinski & Gennis, 2008). Significant diversity in terms of subunit composition, co-factor content and proton transfer pathways is observed between these enzymes, yet their subunit I shows substantial amino acid sequence conservation.
The differences in subunit composition and co-factor content among the different Cox are also reflected in their assembly processes and their requirements for distinct assembly factors and chaperones. This is particularly evident for the process of copper delivery and insertion into Cox. The aa3-Cox and some ba3-Cox contain two redox-active copper centers: in addition to the membrane-embedded CuB center in the catalytic subunit I, they also have a binuclear CuA center in the surface-exposed and hydrophilic domain of subunit II (Brzezinski & Gennis, 2008). This CuA center is required for accepting electrons from electron donors, such as cytochromes, located in the periplasm of bacteria or in the inter-membrane space of mitochondria (Witt et al., 1995). The delivery and insertion of Cu into both the CuA and CuB centers have been investigated in both bacteria and mitochondria, and seem to follow distinct pathways. The formation of the CuB center in aa3-Cox and ba3-Cox relies on the Cox11 protein, which is also termed CoxG or CtaG in some bacteria (Carr et al., 2002, Carr et al., 2005, Buhler et al., 2010, Gurumoorthy & Ludwig, 2015). Cox11 has a periplasmically exposed Cu binding domain, and binds two Cu(I) atoms at the dimer interface of two Cox11 subunits (Carr et al., 2002, Banci et al., 2004). A direct interaction between the Paracoccus (P.) denitrificans Cox11 homologue CtaG and the aa3-Cox subunits I and II was recently shown (Gurumoorthy & Ludwig, 2015). In mitochondria, Cox11 receives Cu from the small soluble copper chaperone Cox17 (Horng et al., 2004), but it is currently unknown whether copper loading onto Cox11 in bacteria requires additional proteins. The formation of the CuA center on the other hand requires the coordinated function of the Sco1 and PCuAC proteins (Banci et al., 2005). Sco1, also called SenC (Buggy & Bauer, 1995) or PrrC (Eraso & Kaplan, 2000, McEwan et al., 2002) in some bacteria, is a single-spanning membrane protein, which contains a thioredoxin-like domain that protrudes into the periplasm (Balatri et al., 2003). Sco1 binds either Cu(I) or Cu(II) via two cysteine residues, and Sco1 mutants are impaired in Cox assembly (Balatri et al., 2003). The PCuAC copper chaperones are located in the bacterial periplasm, but are absent in mitochondria (Banci et al., 2010a). PCuAC adopts a cupredoxin-like fold, and probably coordinates Cu(I) via two methionine and two histidine residues (Banci et al., 2005). In vitro data show that PCuAC inserts copper into the CuA site of subunit II of the Thermus thermophilus ba3-Cox (Abriata et al., 2008).
In the case of aa3-Cox and ba3-Cox, available data suggest Cox11 dependent formation of the CuB center and Sco1/PCuAC-dependent formation of the CuA center. However, the formation of the CuB center appears to follow a different pathway in the cbb3-Cox, which lacks the CuA center (Gray et al., 1994, Ekici et al., 2012a). The cbb3-Cox consists of three to four subunits: CcoN is the catalytic subunit, and is homologous to subunit I of other members of the HCO superfamily. It contains a CuB-heme b binuclear center and a low-spin heme b (Gray et al., 1994). Importantly, instead of a CuA containing subunit, cbb3-Cox contains two membrane-bound c-type cytochrome subunits, CcoO and CcoP (Gray et al., 1994). Electrons are probably transferred from the di-heme cytochrome CcoP via the mono-heme cytochrome CcoO to CcoN (Buschmann et al., 2010). The fourth subunit, CcoQ, is a small membrane protein without any known cofactor and is required for the stability of cbb3-Cox (Peters et al., 2008). In addition, at least in Rhodobacter (R.) capsulatus, the assembly factor CcoH remains stably associated with cbb3-Cox (Pawlik et al., 2010).
Copper delivery to cbb3-Cox was mainly studied in Bradyrhizobium (B.) japonicum and in Rhodobacter species. In particular, R. capsulatus is an attractive model organism for studying cbb3-Cox assembly, because in this organism cbb3-Cox is the only Cox present (Marrs & Gest, 1973, Gray et al., 1994), while many other species (e.g. B. japonicum or R. sphaeroides) contain several Cox enzymes (Bott et al., 1992, Hosler, 2012). Unlike in mitochondria, Cox11 seems not always required for CuB assembly of cbb3-Cox, as R. capsulatus lacks a Cox11 homologue, and cbb3-Cox assembly studies in B. japonicum and R. sphaeroides support Cox11-independent CuB insertion into cbb3-Cox (Buhler et al., 2010, Thompson et al., 2010). Copper trafficking to R. capsulatus cbb3-Cox is a highly complex and coordinated process. Copper uptake is mediated by CcoA, a member of the major-facilitator superfamily of transporters (Ekici et al., 2012b). The intracellular copper homeostasis is controlled by the antagonistic activities of CcoA and the P1B-type ATPase CopA, which exports excess Cu (Ekici et al., 2014). A second P1B-type-type ATPase, CcoI, is absolutely required for cbb3-Cox assembly and appears to export Cu from the cytoplasm to the periplasm for insertion into the CuB center of this enzyme (Koch et al., 2000, Preisig et al., 1996). Moreover, under low Cu concentrations, the Sco1 homologue SenC is required for cbb3-Cox assembly (Lohmeyer et al., 2012), and it was suggested that CcoI and SenC could cooperate in the assembly of the CuB center of cbb3-Cox (Thompson et al., 2012). Sco1 homologues were furthermore suggested to cooperate with PCuAC-type copper chaperones during CuB center assembly of cbb3-Cox (Serventi et al., 2012, Thompson et al., 2012). However, the individual effects of SenC/ScoI and PCuAC on cbb3-Cox assembly and on cellular copper distribution/availability are largely unexplored.
In the current study, the R. capsulatus PCuAC homologue PccA was identified. The data showed that it is indeed a copper binding protein and required for cbb3-Cox assembly and steady-state activity. The phenotype of a ΔpccA strain on cbb3-Cox assembly was less pronounced than that of a ΔsenC strain, even though both single mutants and a ΔpccA/ΔsenC double mutant were rescued by copper supplementation. This indicated that both proteins are required for cbb3-Cox assembly under conditions of low copper availability. Analyses of the cellular copper content and distribution showed an increased intracellular Cu content in the ΔsenC strain, while the Cu content in the ΔpccA strain remained similar to that of a wild type. Finally, by using chemical crosslinking, we provide evidence for the formation of a SenC-PccA complex during cbb3-Cox assembly.
Results
The periplasmic chaperone PccA is required for cytochrome cbb3-Cox activity
Recent data have demonstrated that the Sco1 homologue SenC is required for cbb3-Cox assembly in R. capsulatus (Lohmeyer et al., 2012). As Sco1 proteins have been shown to cooperate with PCuAC-like copper chaperones during Cox assembly (Thompson et al., 2012), the genome of R. capsulatus was searched for PCuAC-homologues and a putative ORF (Rcc0246), encoding for a hypothetical protein with a predicted signal-sequence (amino acids (aa) 1–23) and a potential signal sequence cleavage site between positions 23–24 (Fig. 1A). The candidate protein showed strong homology to PCuAC proteins from other species (Fig. 1A) and contained the ‘HX10MX21HXM’ motif, which is typical of PCuAC-like chaperones (Abriata et al., 2008). As R. capsulatus contains only cytochrome cbb3-Cox, which lacks a CuA center, we termed the protein PccA (for periplasmic copper chaperone A) rather than PCuAC. In the R. capsulatus genome, PccA is located in a cluster of several genes associated with amino acid metabolism (Fig. 1B), while in B. japonicum, the gene for the PCuAC-like chaperone PcuC is located in the pcuABCDE operon, which is transcribed in response to copper limitation (Serventi et al., 2012). Yet another genetic organization of PCuAC is observed in R. sphaeroides, where the PCuAC-gene (RSP_2017) is sandwiched between the gene encoding the 50S ribosomal protein L28 (RSP_2016) and a gene encoding a 102 amino acids long hypothetical protein (RSP_2018) of no significant homology to known proteins. It therefore appears that there is no synteny of PCuAC-encoding genes and those surrounding it among different bacterial genomes.
Figure 1. Sequence alignment and genetic organization of the periplasmic copper chaperone PccA in R. capsulatus.
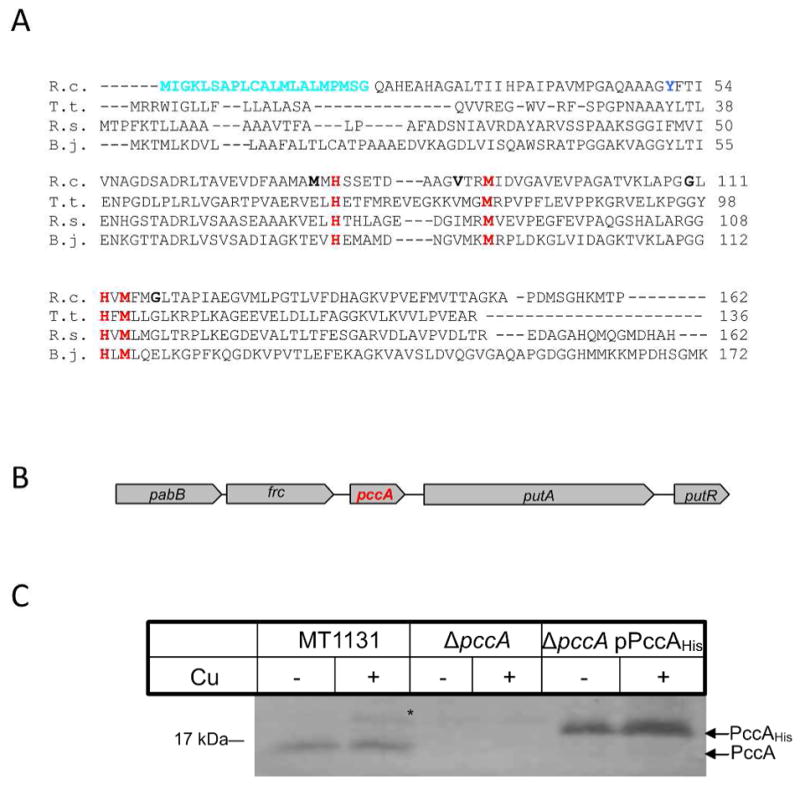
(A) The predicted amino acid sequence of PccA of R. capsulatus (R.c.) was aligned to other members of the PCuAC copper chaperone family using the Clustal sequence alignment algorithm: T. thermophilus PCuAC (T.t.); R. sphaeroides RSP_2017 (R.s.); and B. japonicum PcuC (B.j.). Indicated are the predicted cleavable signal sequence of R. capsulatus PccA (cyan) and the conserved residues of the putative Cu-binding site (red). The single tyrosine residue used for measuring the intrincisc fluorescence of PccA is indicated in blue and the positions used for cysteine substitutions in PccA are indicated in bold. (B) Genetic organization of pccA in R. capsulatus. The nucleotide sequence of R. capsulatus was retrived from NCBI and analyzed using the NCBI tool box (www.ncbi.nlm.nih.gov). Displayed is the R. capsulatus genomic region encompassing pabB, p-amino-benzoat synthase (Rcc02644); frc, CoA transferase (Rcc02645); pccA (Rcc02646) putA, proline dehydrogenase (Rcc02647) and putR, proline dehydrogenase regulator (Rcc02648). (C) Wild type R. capsulatus cells (MT1131), cells of the ΔpccA strain and of the ΔpccA strain carrying pccA on a low copy plasmid (ΔpccA/pPccAHis) were grown on MPYE medium or on MPYE medium supplemented with 10 μM CuSO4. Cells were harvested, and after washing, TCA precipitated and separated on SDS-PAGE. After western blotting, the samples were decorated with α-PccA antibodies. *indicates a band that cross-reacted with the α-PccA antibody in wild type cells grown in the presence of Cu.
For analyzing the function of PccA in R. capsulatus, a strain carrying a chromosomal deletion of PccA was constructed. Immune detection using polyclonal α-PccA antibodies detected a band of approx. 17 kDa in MT1131 wild type cells, but not in the ΔpccA strain (Fig. 1C). When a His-tagged PccA derivative was expressed in the ΔpccA strain, the PccA band was again detected but it migrated slightly slower due to the presence of the His-tag. Since the PccA homologues presumably function as Cu chaperones, it was analyzed whether pccA expression was affected by addition of Cu. Cells were grown on MPYE medium, which contains approx. 250 nM Cu (Koch et al., 2000) and on MPYE medium supplemented with an additional 10 μM CuSO4. However, the level of PccA was not significantly affected by the addition of Cu (Fig. 1C), indicating that pccA expression does not respond to Cu-supplementation.
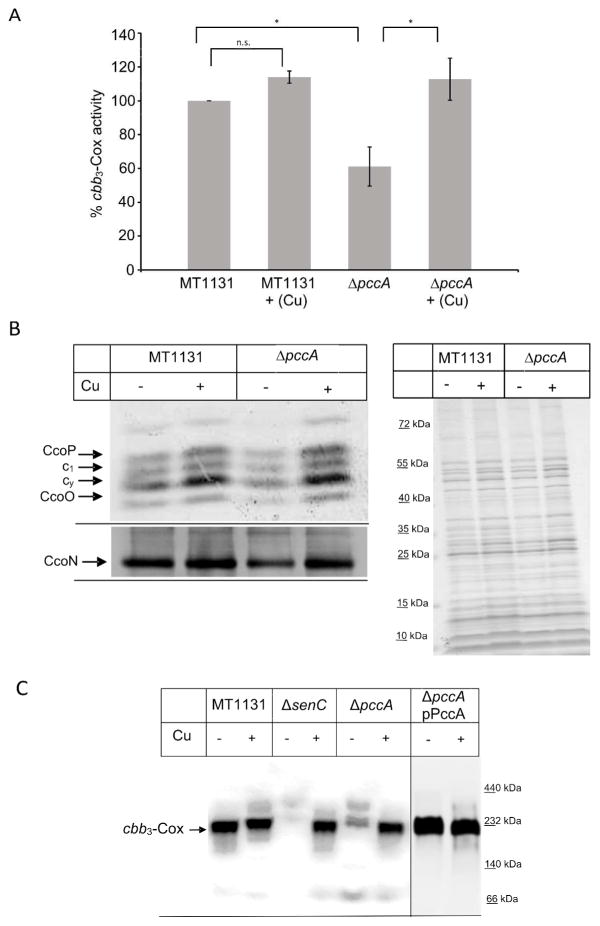
Next, cbb3-Cox activity and stability was analyzed in the ΔpccA strain. Oxygen-uptake measurements with intracytoplasmic membranes of the deletion strain revealed an approx. 40% reduction of cbb3-Cox activity (Fig. 2A) compared to the wild type. Intriguingly, when the ΔpccA strain was grown on Cu-supplemented media (+10 μM CuSO4), its intracytoplasmic membranes displayed only a slightly reduced cbb3-Cox activity (Fig. 2A). Cu supplementation of wild type cells did not significantly influence their cbb3-Cox activity in membranes.
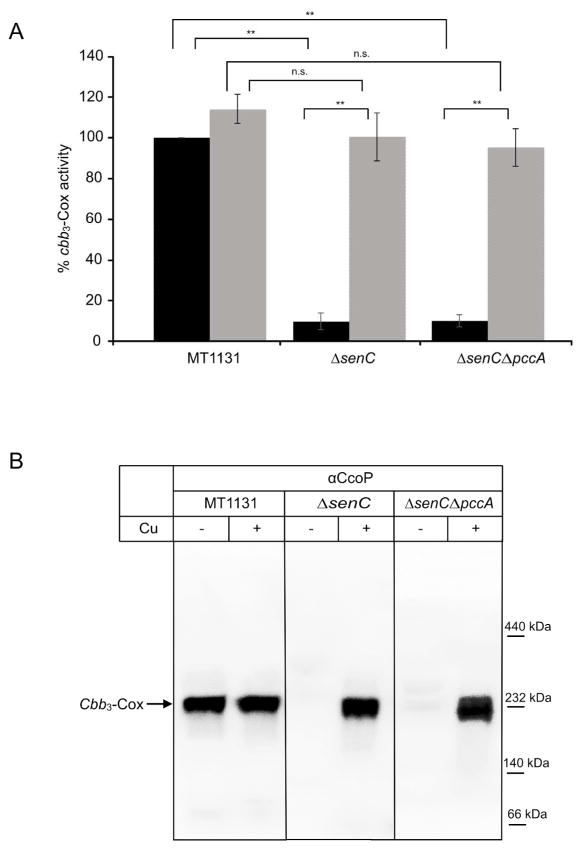
Figure 2. PccA is required for cbb3 Cox assembly in R. capsulatus.
(A) Oxygen uptake activity of intracytoplasmic membranes of R. capsulatus MT1131 (wild type) and ΔpccA strains was measured using a fiber optic oxygen meter. Cells for membrane preparation were grown either on regular MPYE medium or on MPYE medium supplemented with 10 μM CuSO4 (+Cu). Oxygen uptake activity in MT1131 corresponds to 20 μmol O2/min per mg protein and was set to 100%. Values shown are mean values of at least three independent experiments and the standard deviations are as indicated by error bars. * p<0.01; n.s. not significant. (B) The presence of cbb3-Cox subunits was determined by either heme staining, which visualizes the membrane-bound c-type cytochrome profile of R. capsulatus membranes (upper left panel) or by western blotting using antibodies against the catalytic subunit CcoN (lower left panel). CcoP and CcoO correspond to the membrane-bound c-type cytochrome subunit of cbb3-Cox, while c1 is subunit of the cytochrome bc1 complex and cy a membrane bound electron carrier. Approximately 50 μg of protein were loaded per lane and separated on a 16.5% Tris-Tricine SDS-PAGE. When indicated, cells were grown in the presence of 10 μM CuSO4 for membrane preparation. Right panel: Coomassie-brilliant blue stained samples after separation on SDS-PAGE (50 μg membranes). (C) Membranes of the indicated strains containing approximately 100 μg of protein, each, were solubilized with dodecyl-maltoside, separated on a Blue-Native PAGE (BN-PAGE), transferred to a polyvinylidene difluoride (PVDF) membrane, and decorated with antibodies against the CcoN subunit. The fully assembled cbb3-Cox migrates at about 230 kDa on BN-PAGE. ΔpccA pPccAHis corresponds to the ΔpccA strain carrying a plasmid borne copy of pccA with a C-terminal His-tag.
For correlating the reduced activity of cbb3-Cox in the ΔpccA strain with the steady state amounts of cbb3-Cox, heme staining and western blotting was performed. Heme staining, which detects the two membrane bound c-type cytochrome subunits of cbb3-Cox, CcoP and CcoO, showed decreased amounts of both subunits (Fig. 2B, upper left panel), but also a slight decrease in the cytochrome c1 subunit of the cytochrome bc1 complex. These phenotypes were restored when cells were grown in the presence of 10 μM CuSO4 (Fig. 2B, upper left panel). The steady state presence of the catalytic subunit CcoN was also reduced in the ΔpccA strain, and restored when cells were grown on Cu-supplemented media (Fig. 2B, lower left panel). A Coomassie-stained SDS-PAGE of the samples served as loading control (Fig. 2B, right panel). Finally, cbb3-Cox assembly was analyzed by BN-PAGE and western blot analyses. In R. capsulatus, cbb3-Cox assembles into an active 230 kDa complex, and in wild type membranes, formation of this complex was independent of Cu supplementation (Fig. 2C). A Cu-dependent cbb3-Cox assembly has been observed previously in a ΔsenC deletion strain. The Sco1-homologue SenC is a single spanning membrane protein with a periplasmically exposed Cu binding motif, which is required for cbb3-Cox activity (Lohmeyer et al., 2012). In agreement with previous data, no 230 kDa cbb3-Cox complex was detectable in ΔsenC membranes, unless the cells were grown in presence of supplementary Cu (Fig. 2C). In ΔpccA membranes, the 230 kDa complex was significantly reduced but still weakly detectable (Fig. 2C). In membranes of the ΔpccA strain grown on Cu supplemented media, the amount of the 230 kDa cbb3-Cox complex almost reached wild type level (Fig. 2C). Finally, when the ΔpccA strain carried a copy of pccA on a low-copy number plasmid (pRK415), the 230 kDa complex formation was restored independently of Cu supplementation during growth (Fig. 2C).
In summary, these data indicate that PccA is required in particular for cbb3-Cox assembly or stability under low Cu concentrations, which is in agreement with data in R. sphaeroides (Thompson et al., 2012) or B. japonicum (Serventi et al., 2012, Buhler et al., 2010). However, the lack of PccA appears to cause a less severe cbb3-Cox defect than the lack of SenC, which suggests that the function of PccA can be partially bypassed even in the absence of supplementary Cu.
PccA is a copper binding protein
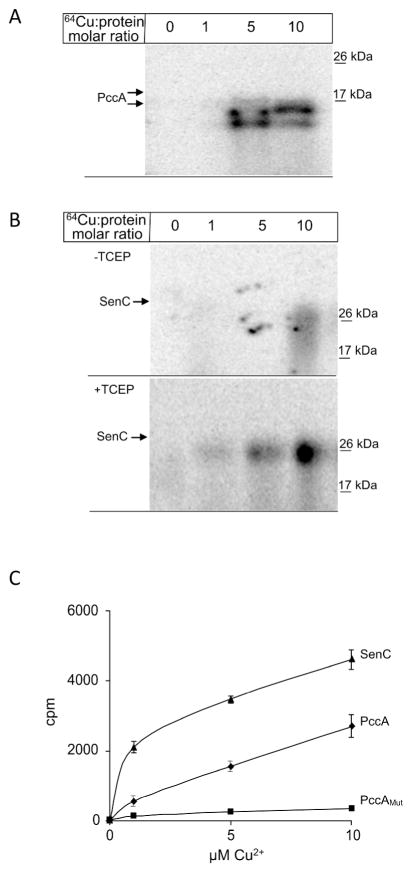
PccA contains the signature motif of Cu-binding PCuAC chaperones and Cu binding to PccA was analyzed by using radioactive 64Cu. A signal sequence-less variant of PccA containing a C-terminal Strep-tag was expressed in E. coli and purified. Purified PccA was incubated with 64Cu at different molar ratios, separated on native PAGE and analyzed by phospho-imaging. At a molar protein:Cu ratio of 1:5 and 1:10, two radioactive bands were observed, indicating Cu binding to PccA (Fig. 3A). The band with the lower molecular mass corresponds to PccA lacking the Strep-tag, as it was not recognized by the α-Strep antibody (data not shown). As a control, 64Cu binding to the R. capsulatus Sco1 homologue SenC was also analyzed, for which copper binding has previously been shown (Lohmeyer et al., 2012) (Fig. 3B). Similar to PccA, a SenC variant lacking its N-terminal transmembrane domain was expressed in E. coli and purified. The Cu binding motif in SenC includes two conserved cysteine residues, which are required for Cu binding (Lohmeyer et al., 2012). We therefore reasoned that for in vitro Cu binding, SenC probably had to be in its reduced form. Purified SenC was incubated with 64Cu with and without prior reduction using Tris(2-carboxyethyl)phosphine (TCEP). Without prior reduction, no significant 64Cu binding to SenC was observed. In contrast, after reduction, a Cu concentration dependent binding of 64Cu to SenC was detected (Fig. 3B).
Figure 3. PccA is a copper binding protein.
(A) PccA was purified via a C-terminal Strep-tag and 10 μM of purified proteins were incubated with increasing concentrations of 64CuCl2. Samples were subsequently separated on native PAGE and analyzed by phosphor-imaging. (B) Purified SenC (10μM) was incubated with increasing concentrations of 64CuCl2 with and without prior reduction of the cysteine-containing copper binding motif by TCEP. Samples were then analyzed as in (A). (C) Quantification of the 64Cu binding to SenC (▲), or PccA (◆), or PccAMut, a mutant lacking the two conserved methionine residues of the Cu-binding motif of PCuAC chaperones (■). After 64Cu binding for 10 min at room temperature, samples were purified via Strep-tag and Cu binding was analyzed using a scintillation counter. The data displayed are the mean value of four independent experiments and the error bars represent the standard deviation.
64Cu binding to PccA and SenC was further quantified using a scintillation counter. Purified proteins were incubated with 64Cu, re-purified via their Strep-tag, and the radioactivity in the samples was measured. For both PccA and SenC, a Cu concentration-dependent increase of the radioactive signal was observed (Fig. 3C), whereas 64Cu-binding to a mutant variant of PccA, lacking the two methionine residues of its ‘HX10MX21HXM’ motif (PccAMut), was insignificant (Fig. 3C). These data showed that PccA is a Cu-binding protein, which binds Cu via the canonical ‘HX10MX21HXM’ motif of PCuAC-type chaperones. These data also support a specific binding of 64Cu to SenC and PccA, and exclude significant non-specific Cu binding to these proteins.
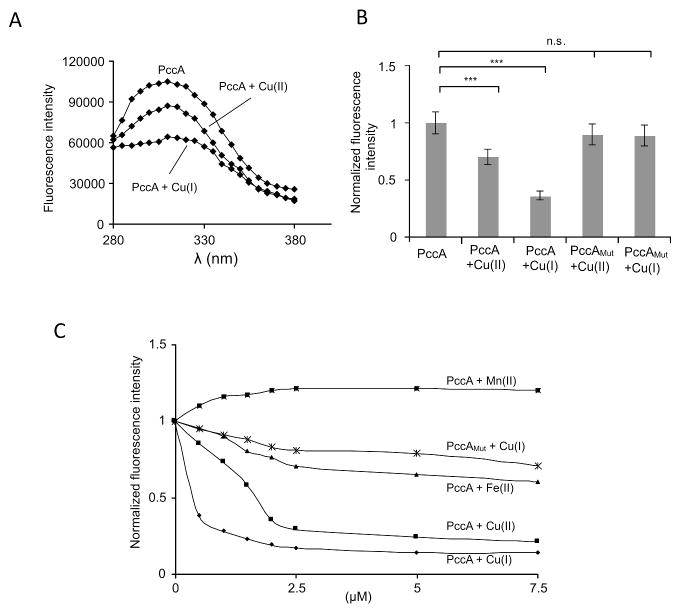
Cu displays two oxidation states, and the reduced Cu(I) form appears to be the preferred substrate for P1B-type Cu-exporting ATPases (Gonzalez-Guerrero & Arguello, 2008) and for PCuAC-type chaperones (Banci et al., 2005). We determined binding of Cu(I) or Cu(II) to PccA by following the intrinsic protein fluorescence of PccA. The aromatic amino acids tryptophan, tyrosine and phenylalanine display intrinsic fluorescence, and changes in their fluorescence emission have been used to monitor conformational changes or ligand binding to proteins. PccA lacks tryptophan but contains a single tyrosine residue at position 51 (Fig. 1A), which in the NMR structure of D. radiodurans PCuAC is located close to the Cu binding ‘HxM’ motif (Banci et al., 2005), and is expected to respond to Cu binding. When excited at 276 nm, purified PccA displayed an emission maximum at approx. 310 nm (Fig. 4A). For analyzing Cu binding to PccA, purified protein was incubated in an anaerobic chamber with either Cu(I) or Cu(II). The emission was readily quenched by the addition of Cu(I) (Fig. 4A), indicative for changes in the local environment of the tyrosine residue. Quenching was also observed when Cu(II) was added, but the effect was weaker than in the presence of Cu(I) (Fig. 4A). Quantification of several independent experiments revealed that Cu(II) reduced emission by approx. 30%, while Cu(I) reduced it by approx. 60% (Fig. 4B). As control, Cu binding to PccAMut was also analyzed, but neither Cu(I) nor Cu(II) significantly reduced fluorescence emission of the mutant protein. The specificity of Cu(I) binding to PccA was further verified by analyzing tyrosine fluorescence in the presence of different metals and at different metal concentrations. When purified PccA was incubated under anaerobic conditions with increasing concentrations of Cu(I), we observed a rapid decrease in fluorescence (Fig. 4C). Cu(II) also quenched fluorescence, however only at higher concentrations as compared to Cu(I). No significant quenching was observed for PccMut. Fe2+ had also only a weak quenching effect and Mn2+ did not quench fluorescence of PccA (Fig. 4B). In summary, our data demonstrate that the R. capsulatus PCuAC homologue PccA is a Cu binding protein, possibly with a preference for Cu(I).
Figure 4. PccA preferentially binds Cu+.
(A) Intrinsic tyrosine fluorescence of PccA (10 μM protein) was determined under anaerobic conditions after excitation at 276 nm and emission was monitored between 280 and 380 nm. When indicated Cu(I) or Cu(II) was added at a final concentration of 0.5 μM. (B) Intrinsic fluorescence emission of PccA at 310 nm in the presence of either Cu(I) or Cu(II) is plotted relative to the initial signal in the absence of Cu, which was set to 1.0. In addition, the intrinsic fluorescence emission of the PccAMut variant was determined in the presence of either Cu(I) or Cu(II). The error bars indicate the SD (n=4). *** p<0.001; n.s. not significant. (C) The intrinsic fluorescence emission of PccA or PccAMut was plotted against different metal concentrations. Plotted are the mean values of three experiments. The SDs were ≤ 10% and are not displayed for clarity.
PccA interacts with SenC in vivo and in vitro
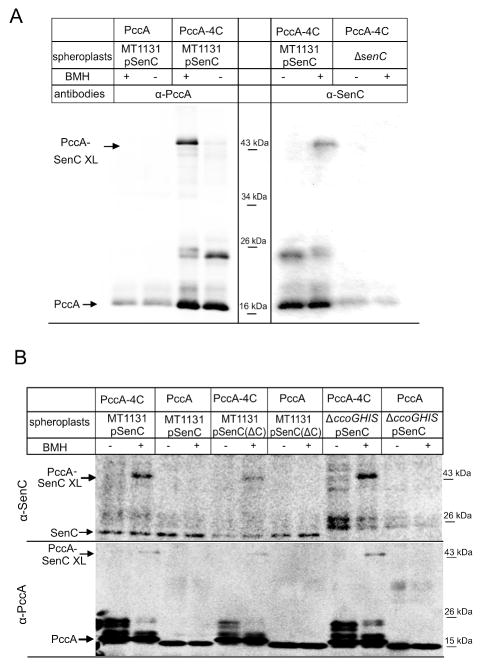
PCuAC-like chaperones were suggested to interact with the SenC-homologue ScoI during the assembly of the CuA center of aa3-Cox (Banci et al., 2010b, Banci et al., 2005, Thompson et al., 2012, Serventi et al., 2012). Thus, both in vitro and in vivo chemical crosslinking approaches were used to probe whether PccA and SenC interact during Cu transfer to cbb3-Cox. In the first approach, the cysteine-specific cross-linker BMH was used. SenC contains two cysteine residues within its copper binding motif, but PccA has only one cysteine residue at its position 10, which is within the predicted cleaved signal sequence. Therefore, a PccA derivative (PccA-4C) containing four cysteine residues at positions 76, 93, 109 and 117, respectively was constructed (Fig. 1A). These residues are very close to the putative Cu-binding motif of PccA. PccA-4C was expressed as a Strep-tagged protein in E. coli and purified. Spheroplasts derived from wild type R. capsulatus cells (MT1131), expressing senC from a low copy plasmid, were incubated with purified PccA-4C in the presence or absence of BMH. PccA-4C and potential crosslinked proteins were subsequently purified and separated on SDS-PAGE followed by western transfer using α-Pcc1 antibodies. As a control, native PccA lacking these cysteine residues was also incubated with MT1131 spheroplasts. In the presence of PccA-4C and the crosslinker BMH, a strong band at approx. 43 kDa, was observed, which was only weakly detectable in the absence of BMH (Fig. 5A). Furthermore, this band was not detected when native PccA lacking cysteine residues was incubated with MT1131 spheroplasts in the presence of BMH (Fig. 5A). Unlike the native PccA, PccA-4C migrated on SDS-PAGE as multiple bands at approx. 25 kDa and 16 kDa (Fig. 5A), which could reflect intramolecular disulfide-bridges or disulfide-bridge linked dimers, but this was not further analyzed.
Figure 5. PccA interacts with SenC in vitro.
(A) Spheroplasts of MT1131 cells expressing SenC or from a ΔsenC strain were incubated with Strep-tag purified wild type PccA or PccA-4C, which contained four engineered cysteine residues close to the metal binding site. When indicated, the cysteine-specific crosslinker BMH was added. Samples were subsequently analyzed under non-reducing PAGE, transferred to PVDF membranes and decorated with antibodies against PccA or SenC. PccA-SenC XL indicates the crosslinking product. (B) Spheroplasts were prepared from MT1131 cells expressing either SenC or SenCΔC, which corresponds to a SenC derivative, in which the two cysteine residues of the Cu-binding motif were replaced by alanine. In addition, spheroplasts of the ΔccoGHIS strain expressing SenC were analyzed. In the ΔccoGHIS strain, the cbb3-Cox assembly factors CcoGHIS are absent. The upper panel was decorated with antibodies against SenC and the lower panel with antibodies against PccA.
The crosslinked product of approx. 43 KDa would be consistent with a crosslink between PccA (16 kDa) and SenC (25 kDa). In support of this, the crosslink product was also detected by using α-SenC antibodies. However, the α-SenC antibodies also cross-reacted with purified PccA (Fig. 5A). To validate that the 43 kDa band represents a SenC-PccA complex, PccA-4C was incubated with spheroplasts derived from the ΔsenC strain. Here, α-SenC antibodies did not recognize the 43 kDa band, indicating that the 43 kDa crosslinked product corresponded to a PccA-SenC complex (Fig. 5A, Pcc1-SenC XL (crosslink)).
The BMH crosslinking was further controlled by incubating either PccA-4C or native PccA with spheroplasts from cells expressing a SenC variant with a mutated Cu-binding motif (SenCΔC), in which cysteine residues 83 and 89 were replaced by serine). Although the crosslinking product was still weakly detectable, probably due to the presence of additional cysteine residues within SenC, it was significantly weaker than with spheroplasts expressing wild type SenC (Fig. 5B). Incubating either SenC or SenCΔC spheroplasts with wild type PccA did not result in any specific crosslinked products.
A periplasmic PccA-SenC complex could potentially link Cu-export via CcoI with the subsequent Cu insertion into the catalytic CcoN subunit of cbb3-Cox. Thus, the PccA-SenC interaction was analyzed in strain CW1 (ΔccoGHIS), which lacks the CcoGHIS proteins required for cbb3-Cox assembly. The PccA-SenC crosslink product was also detected in the absence of the CcoGHIS proteins and present even in slightly higher amounts than in wild type cells (Fig. 5B).
As the BMH crosslinking experiments used a modified derivative of PccA (PccA-4C), it could be argued that the observed interaction between PccA and SenC might be due to the non-native cysteine residues added to PccA. To address this possibility, the crosslinking experiment was repeated in living cells using para-formaldehyde (PFA). Wild type cells and wild type cells expressing PccA with a C-terminal His-tag were incubated with PFA, and PccA was subsequently purified via metal affinity chromatography. Purified PccA and potential crosslinked products were then separated on SDS-PAGE and analyzed using α-PccA and α-SenC antibodies. Both antibodies recognized again a band at approx. 43 kDa in the PFA treated sample, which was not observed in the absence of PFA or in cells not expressing PccAHis (Fig. 6). This provides in vivo verification for the existence of a PccA-SenC complex.
Figure 6. PccA interacts with SenC in vivo.
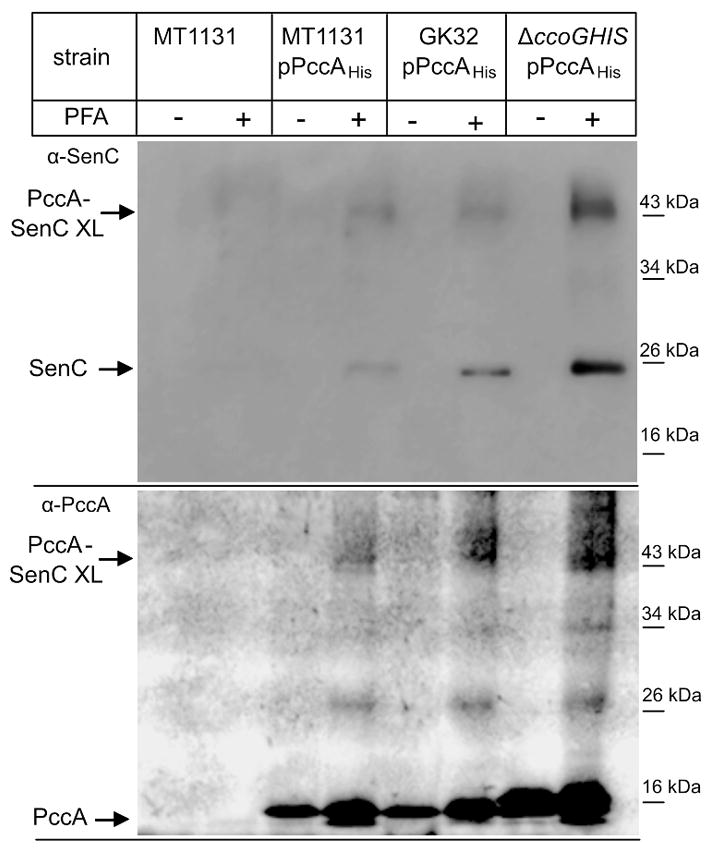
The indicated strains were incubated with para-formaldehyde (PFA) (+) or the reaction buffer only (−). After membrane preparation, cells were solubilized with DDM, and PccAHis was purified via metal affinity chromatography. Purified protein was then separated on SDS-PAGE, transferred to a PVDF membrane and decorated with antibodies against SenC (upper panel) or PccA (lower panel). GK32 carries a ccoNO deletion and lacks cbb3-Cox. In the ΔccoGHIS strain, the cbb3-Cox assembly factors CcoGHIS are absent.
The increased crosslinking between PccA and SenC in the ΔccoGHIS strain was also observed with PFA in vivo (Fig. 6). In the absence of ccoGHIS, cbb3-Cox is not assembled, raising the possibility that the increased interaction between SenC and PccA could result from the absence of cbb3-Cox. This was analyzed by testing the PccA-SenC interaction in the cbb3-Cox deletion strain GK32 (ΔccoNO). The PccA-SenC crosslinking product was observed in GK32 cells, but the crosslink was not stronger than in wild type cells (Fig. 6). In summary, the crosslink data show that PccA and SenC form a complex of 43 kDa. The data also suggested an increased interaction between SenC and PccA in the absence of the CcoGHIS proteins. However, considering that the senC mRNA and protein levels also increase in a ΔccoI strain (Lohmeyer et al., 2012) (c.f. Fig. 6), we cannot completely exclude that this increased crosslinking results from increased levels of SenC.
A SenC-PccA double knock-out lacks cbb3-Cox
The cbb3-Cox defect seen in both the ΔpccA and the ΔsenC strain is rescued by increasing copper concentrations in the growth medium. This finding suggested that either the delivery of Cu to cbb3-Cox via the SenC-PccA pathway is completely bypassed at high Cu concentrations, or that either SenC or PccA is sufficient to deliver Cu to cbb3-Cox at high Cu concentrations. For differentiating between these two possibilities, a senC-pccA double knock-out strain was constructed. cbb3-Cox activity assays and BN-PAGE analyses revealed that the phenotype of the ΔsenC-ΔpccA strain was identical to that of the ΔsenC single deletion mutant (Fig. 7A & 7B); i.e. basically no cbb3-Cox activity was detectable and the 230 kDa cbb3-Cox complex was absent, unless the cells were grown in Cu-supplemented media. Thus, the SenC-PccA copper delivery pathway to cbb3-Cox can be bypassed by a currently unknown SenC/PccA-independent pathway when cells are grown in the presence of high Cu concentrations. These data furthermore indicate that SenC and PccA cannot substitute for each other at low Cu concentrations, rationalizing why over-expression of either PccA or SenC is unable to rescue the cbb3-Cox deficiency of the ΔsenC or ΔpccA strains, respectively (Lohmeyer et al., 2012).
Figure 7. The phenotype of a senC-PccA double knock-out mimics that of the ΔsenC strain.
(A) cbb3-Cox activity was determined in membranes of the indicated strains grown on MPYE medium (■) or on MPYE medium supplemented with 10 μM CuSO4 (□). The activity observed in membranes isolated from cells grown on MPYE medium was set to 100%. Displayed are the mean values of three independent experiments and the error bars represent the SD. ** p< 0.001; n.s. not significant. (B) BN-PAGE of the membranes described in (B). After western transfer, the blot was decorated with α-CcoN antibodies.
Differential effects of PccA and SenC on copper homeostasis in Rhodobacter capsulatus
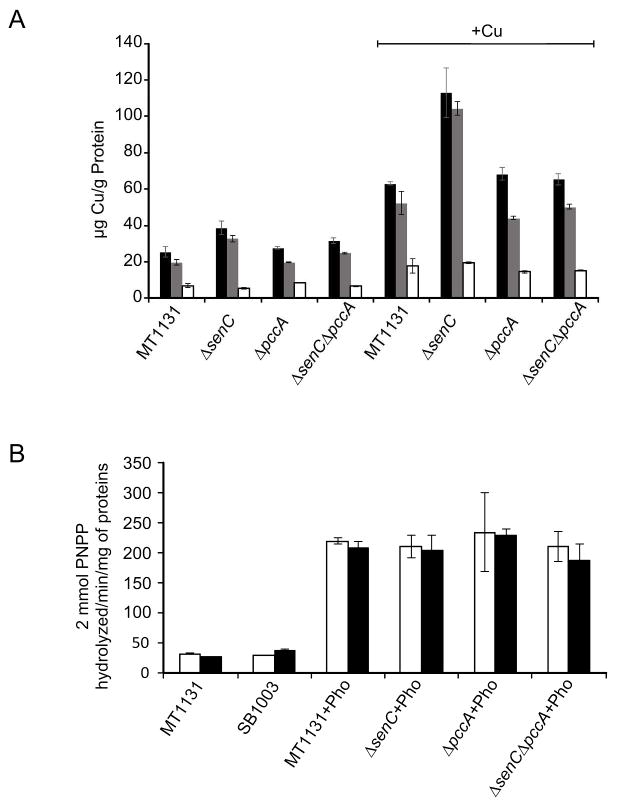
As the addition of Cu rescued the cbb3-Cox defect seen in the ΔpccA, the ΔsenC and the ΔsenC-ΔpccA strains, the total Cu content and its distribution between the cytoplasm and the periplasm was examined in these mutants. The Cu concentrations were determinend by atomic absorption spectroscopy using the manganese and the zink concentration as internal references.
In whole cells grown on MPYE medium without Cu supplementation, the total Cu content of all three mutants was comparable to the Cu content of wild type cells (Fig. 8A, black bars). For testing whether the Cu distribution between the cytoplasm and the periplasm was changed, the Cu content in spheroplasts, which reflects the cytoplasmic Cu pool (i.e. the Cu content in the cytosol and in the cytoplasmic membrane), and the Cu content in the periplasm were determined. In wild type cells and in all three mutants, most of the copper was present in the spheroplast fraction (Fig. 8A, grey bars) and only a small amount was found in the periplasmic fraction (Fig. 8A, white bars). Thus, the cbb3-Cox defect observed in the ΔpccA, the ΔsenC and the ΔsenC-ΔpccA strains cannot be attributed to a decrease of the total cellular Cu concentration or to a different Cu distribution between cytoplasm and periplasm.
Figure 8. Copper distribution in R. capsulatus cells is influenced by the deletion of SenC.
(A) Total copper concentration (black bars) of MT1131 (wild type), ΔsenC, ΔpccA and ΔsenC-ΔpccA cells was monitored by atomic absorption spectroscopy after growth on MPYE medium or MPYE medium supplemented with 10μM CuSO4 (+) as described in Material and Methods. The manganese and zink concentrations were determined as internal reference. In addition, the copper content of spheroplasts (grey bars) and the supernatants of the spheroplast preparations (periplasmic fraction) (white bars) were also analyzed. The values are the mean of at least three independent experiments and the error bars indicate the SD. Significant p-values (p<0.01) were only observed for the Cu content in the ΔsenC strain (+Cu) compared to the wild type (+Cu). (B) Alkaline phosphatase (PhoA) activity of a ccoA-phoA translational fusion was determined in cell extracts of the indicated strains grown either in the absence of presence of supplementary Cu. The values are the mean of at least two experiments and the error bars indicate the SD.
The Cu content and the Cu distribution of cells grown in Cu-supplemented media were also analyzed. Surprisingly, the total and the cytoplasmic Cu content of the ΔsenC cells was signficantly increased, as compared to wild type cells or the ΔpccA cells. This enhanced Cu accumulation suggested that either Cu import is increased in the absence of SenC, or Cu export is diminished. An increased Cu content was not observed in ΔsenC-ΔpccA cells (Fig. 8A), which indicates that the increased Cu content seen in the ΔsenC cells requires the presence of PccA. These data point to a so far unrecognized role of both proteins in regulating Cu homeostasis in response to high Cu concentrations.
So far, Cu uptake via the MFS-type transporter CcoA is the only known Cu uptake system in R. capsulatus (Ekici et al., 2012b). Hence, it was analyzed whether the increased cellular Cu content in the ΔsenC strain was caused by an increased expression of ccoA. CcoA was fused to the alkaline phosphatase (PhoA)-moiety and used as translational fusion protein to determine the expression of CcoA by alkaline phosphatase activity. As a control, the intrinsic PhoA activity in two wild type strains, MT1131 and SB1003, was determined, which revealed no significant background activity. PhoA activities in MT1131 and in the ΔsenC, ΔpccA and ΔsenC-ΔpccA strains expressing the CcoA-PhoA-fusion were comparable, and largely independent of Cu supplementation to the growth media (Fig. 8B). This demonstrated that the increased Cu content seen in the ΔsenC strain is not caused by enhanced expression of the Cu-importer CcoA, although an enhanced import activity of CcoA in the absence of SenC or a CcoA-independent Cu import at high Cu availability cannot be excluded. In any event, our data demonstrate that periplasmic Cu-chaperones are not only required for providing Cu for cbb3-Cox assembly, but they are also crucial determinants for maintaining cellular Cu homeostasis.
Discussion
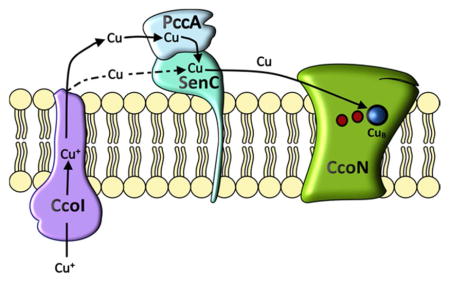
The acquisition of Cu is a crucial step during the biogenesis of heme copper oxidases and both eukaryotic and prokaryotic cells have developed sophisticated pathways for uptake and delivery of this potentially toxic metal (Arguello et al., 2012, Arguello et al., 2013). In bacteria, periplasmic Cu chaperones of the PCuAC family and the Sco1-family have been suggested to cooperate during the assembly of the CuA centers of aa3-Cox and ba3-Cox (Banci et al., 2007, Khalimonchuk et al., 2010, Rigby et al., 2008, Abriata et al., 2008, Banci et al., 2006). This cooperation also appears to be required for the assembly of the CuB center of cbb3-Cox (Thompson et al., 2012), even though the latter is deeply buried within the membrane, while the CuA center is exposed to the periplasm. Cu transfer between PCuAC and Sco1 is likely to require a physical interaction between both proteins, but so far a direct interaction between PCuAC-like and ScoI-like proteins has not been demonstrated in any organism. This was achieved in the present study by using in vitro and in vivo cross-linking. Complex formation between the PCuAC homologue PccA and the Sco1 homologue SenC was observed in wild type R. capsulatus cells. Importantly, the detected amounts of this complex increased in the absence of the assembly proteins CcoGHIS (Koch et al., 2000, Kulajta et al., 2006). Within the ccoGHIS gene cluster, ccoI codes for a putative Cu-transporting P-type ATPase that is absolutely essential for cbb3-Cox assembly (Koch et al., 2000). CcoI is thought to export Cu(I) from the cytoplasm to the periplasm (Gonzalez-Guerrero et al., 2010, Raimunda et al., 2011) and its deletion cannot be bypassed by increasing the Cu concentrations in the growth medium (Koch et al., 2000). A functional link between CcoI and SenC is suggested by the observation that SenC is up-regulated in the absence of CcoI (Lohmeyer et al., 2012). This is also reflected by increased co-purification of SenC with PccA when PFA-crosslinking is performed in the absence of ccoGHIS (Fig. 6). Up-regulation of periplasmic Cu-binding proteins when Cu-transport to the periplasm is impaired has also been observed in P. aeruginosa (Raimunda et al., 2013) and might be a general strategy for coping with perturbed Cu transport. The increased SenC levels could explain why more of the SenC-PccA complex is observed in the absence of ccoGHIS. However, SenC appears to be present in R. capsulatus cells at a higher concentration than PccA (data not shown), suggesting that SenC is unlikely to be limiting for complex formation. It rather seems that the stability of the SenC-PccA complex is increased when CcoI-dependent Cu delivery to the periplasm is impaired. In any event, the available data likely position the SenC-PccA complex between CcoI and cbb3-Cox during Cu delivery to CcoN (Fig. 9). It is conceivable that PccA might also convey Cu to other periplasmic proteins; however, this is currently unknown.
Figure 9. A putative Cu delivery pathway for cbb3-Cox assembly in R. capsulatus.
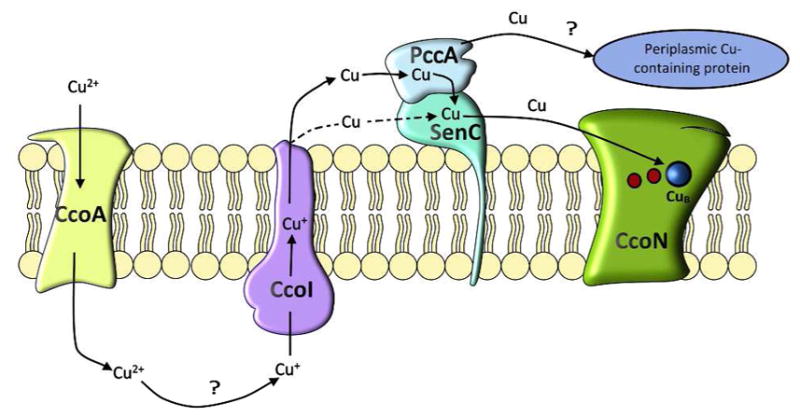
At low Cu concentrations, the MFS-type Cu transporter CcoA imports Cu2+, which is reduced to Cu+ in the cytoplasm by an unknown mechanism. The P1B-type ATPase CcoI transports Cu+ into the periplasm, where it is bound to PccA, which transfers it to the Sco1-homologue SenC and possibly to other periplasmic Cu-dependent proteins. SenC then delivers Cu+ to the CcoN subunit of cbb3-Cox (red dots indicate the two heme groups in CcoN). In the absence of PccA, Cu+ loading onto SenC and subsequent cbb3-Cox assembly is significantly reduced but still detectable (dashed arrows). At high Cu concentrations, cbb3-Cox assembly proceeds independently of CcoA, SenC and PccA, but still requires CcoI. Although SenC and PccA are dispensable for cbb3-Cox assembly at high Cu concentration, they are very important for regulating Cu homeostasis under these conditions. Absence of SenC leads to increased cytoplasmic Cu concentrations, which return to normal by the simultaneous elimination of PccA. A plausible explanation is the accumulation of Cu-loaded PccA in the absence of SenC, repressing Cu-export via CcoI and leading to increased cytoplasmic Cu pool.
Interestingly, in contrast to the ΔsenC strain, which does not assemble cbb3-Cox, partial assembly occurs in the absence of PccA, indicating that the PccA function can be partially bypassed at low Cu concentrations. This observation is not related to differences in the total cellular Cu pool, or to differences in the Cu distribution between cytoplasm and periplasm, because we did not observe significant changes in both mutants compared to wild type cells. It rather argues that SenC can deliver Cu to cbb3-Cox even in the absence of PccA. In R. sphaeroides and B. japonicum it was suggested that the PccA-homologues work upstream of the SenC-homologues during CuB center assembly (Serventi et al., 2012, Thompson et al., 2012). This would be in line with the hypothesis that PCuAC-like chaperones in bacteria are functionally related to mitochondrial Cox17 and donate Cu to either Sco1 or to Cox11 (Banci et al., 2010a). PccA might prevent re-oxidation of Cu(I) to Cu(II) and accelerate Cu(I) loading onto SenC, which then delivers it to cbb3-Cox. This assumption is supported by the observation that both proteins have a preference for Cu(I) and the observation of a direct interaction between SenC and cbb3-Cox (Lohmeyer et al., 2012) (Fig. 9).
The cbb3-Cox defect of the ΔsenC and the ΔpccA strains is rescued by Cu supplementation to the medium, which was previously also observed in B. japonicum and R. sphaeroides (Serventi et al., 2012, Thompson et al., 2012). The dispensability of PccA and SenC at high Cu concentrations could indicate that under these conditions Cu insertion into CcoN occurs unassisted directly from the periplasmic Cu pool. However, this appears unlikely for two reasons: (1) Cu supplementation primarily increases the cytoplasmic Cu pool, while the Cu concentrations in the periplasm of the ΔsenC and the ΔpccA strains remain rather low and are not higher than in wild type cells. (2) The cbb3-Cox defect in the ΔccoI mutant is not rescued by supplementary Cu (Koch et al., 2000, Ekici et al., 2014), suggesting that Cu transport from the cytoplasm to the periplasm is an essential step during Cu delivery to cbb3-Cox.
Unexpectedly, Cu supplementation increases the cytoplasmic Cu pool in the ΔsenC strain more than in the wild type, the ΔpccA or the ΔsenC-ΔpccA double knock-out strains. This indicates that SenC is not only required for cbb3-Cox, but also regulates Cu homeostasis. A regulatory role of Sco1 and Sco2 on Cu homeostasis has already been proposed for humans (Leary, 2010, Leary et al., 2007). Here, Sco1 transfers Cu(I) to the CuA site of CoxII, while Sco2 works as Cu-dependent thiol reductase for the cysteine ligands of CoxII (Morgada et al., 2015). Conceptually, the absence of SenC could either stimulate Cu import or reduce Cu export. R. capsulatus imports Cu primarily via the MFS-type Cu-importer CcoA (Ekici et al., 2014, Ekici et al., 2012b), but we did not find indications that CcoA expression is increased in the absence of SenC. A very slow CcoA-independent Cu uptake by an unknown protein is also detectable in R. capsulatus cells (Ekici et al., 2014), and we can currently not exclude that this CcoA-independent uptake system is stimulated by the absence of SenC. Cu export in R. capsulatus is mediated by the P1B-type ATPases CopA and CcoI (Ekici et al., 2014, Koch et al., 2000) and their deletion leads to a weak (ccoI, (Lohmeyer et al., 2012)) or strong (copA (Ekici et al., 2014)) increase in the cellular Cu concentration, which reflects their different Cu export kinetics (Raimunda et al., 2011, Gonzalez-Guerrero et al., 2010). Thus, Cu accumulation is observed in the absence of either CopA/CcoI or SenC. In humans, Sco1/Sco2 appear to regulate Cu efflux from mitochondria (Briere & Tzagoloff, 2007), and our data are also consistent with a role of SenC in Cu export regulation. Whether this involves CopA or CcoI or both, is currently unknown. CcoI appears to be more likely, because it was functionally linked to SenC (Lohmeyer et al., 2012) and is essential for cbb3-Cox assembly (Kulajta et al., 2006), while CopA does not seem to influence cbb3-Cox assembly (Ekici et al., 2014).
The ΔsenC-ΔpccA double knock-out strain accumulates much less cytoplasmic Cu than the ΔsenC strain, suggesting that the absence of PccA somehow counterbalances the Cu accumulation seen in the ΔsenC strain. The underlying molecular mechanisms are currently unknown and need to be further explored. A possibility is that the Cu occupancies of PccA or SenC are important for regulating Cu homeostasis. If PccA works upstream of SenC, deletion of PccA would probably result in the accumulation of Cu-free SenC, while the deletion of SenC would result in the accumulation of Cu-loaded PccA. Cu-loaded PccA could repress Cu-export via the CcoI or CopA pathways, explaining the accumulation of intracellular Cu. In the absence of both proteins, this repression would fail, explaining why in the double knock-out the Cu content is normal. It has been shown that P1B-type ATPases are activated by their cognate cytosolic chaperones (Gonzalez-Guerrero & Arguello, 2008) and it is conceivable that periplasmic chaperones also influence the activity of these Cu-exporting proteins.
In summary, our data illustrate that an intricate interplay between two periplasmic Cu-binding chaperones is required for both the assembly of the Cu-containing cbb3-Cox and maintenance of cellular Cu homeostasis.
Experimental Procedures
Bacterial strains and growth conditions
The strains and plasmids used are listed in Table S1. The E. coli strains harboring plasmids were grown in LB medium suplemented with appropriate antibiotics (ampiciline [Amp] 100 μg/ml, kanamycine [Kan] 50 μg/ml and tetracycline [Tet] 10 μg/ml) at 37° C in liquid medium or on LB agar plates. R. capsulatus strains were grown in Sistrom’s minimal medium A, or in enriched MPYE medium at 35° C with the apropriate antibiotics (10 μg/ml of Kan and 2,5 μg/ml of Tet) (Daldal, 1988, Jenney & Daldal, 1993). For semi-aerobic growth, 500 ml of liquid culture were grown in 1000 ml flask at 35° C and 110 rpm shaking in the dark. Metal-free MPYE medium and metal-free water was obtained by mixing 100 ml MPYE medium/water with 5 g of Chelex-100 resin and stirring for 1 hour at room temperature. The Chelex-100 resin was then removed by filtration.
Molecular genetic techniques
Standard molecular genetic techniques were performed as described previously (Sambrook & Russel, 2001, Koch et al., 1998b). To construct chromosomal knockout alleles of desired genes, the gene transfer agent (GTA) was used as described earlier (Koch et al., 1998b). Genomic DNA of R. capsulatus was extracted from 10 ml cultures of wild type (MT1131) or ΔPccA strains, which were grown in MPYE medium by using the Dnasy Blood & Tissue kit (Qiagen Inc.). The isolated DNA was then analysed qualitatively and quantitatively by Nanodrop spectrophotometer and by agarose gel electrophoresis.
To construct a deletion-insertion allele of PccA (R. capsulatus RCC02646), a DNA fragment including ~500 bp 5′ upstream and 3′ downstream of PccA was PCR amplified using wild-type (MT1131) chromosomal DNA as a template and the primers PccA_SacI_for and PccA_KpnI_rev containing 5′-SacI and 3′-KpnI restriction sites (Table S2). This DNA fragment was digested with appropriate restriction enzymes and cloned into pBlueScriptIIKS+ and into pRK415 to yield the plasmids pBS-PccA and pRK415-PccA, respectively (Table S1). PccA was deleted from pBS-PccA using the primers pB_circ_upll (forward primer) and pB_circ_downl (reverse primer) containing a 5′-NdeI and a 3′-XhoI restriction site, which amplified the remaining flanking regions of PccA and the vector. The Kan cassette of pKD4 vector was amplified using the primers pKD_Kan_for and pKD_Kan_Rev containing NdeI and XhoI restriction sites, and appropriate digestions ligated between the two flanking regions of PccA to yield pBS-ΔPccA carrying the Δ(PccA::kan) allele. The fragment between SacI and KpnI of pBS_ΔPccA was cloned into the respective cloning sites of pRK415 to yield pRK415_ΔPccA. This plasmid was used to generate the ΔPccA knockout by GTA crosses with the wild-type (MT1131) and a ΔsenC (Swem et al., 2005) mutant, to yield the strains IT1 and IT10, respectively.
Plasmid pBS-PccA was used as a template with the PccA_Strep_For and PccA_Strep_Rev primers (Table 2), and the PCR product thus obtained was cloned into pET22b plasmid to yield pET22b-PccAStrep carrying a Strep-tagged version of PccA (Table 1). This latter plasmid was also used as a template for constructing variants of PccA in which the amino acid residues at positions 76, 93, 109 and 117 were replaced by cysteines (PccA-4C; Fig. 1). This plasmid was also used to replace the two methionine residues of the Cu-binding motif by leucine residues (PccAMut).
Construction of ccoA::phoA fusions
For the ccoA::phoA fusions, the CcoA-XhoIFor primer located 400bp upstream the ATG of CcoA was used as a forward primer (Table S2). This forward primer was used in conjunction with BamHI249Rev and BamHI281Rev reverse primers to amplify the DNA fragments of ccoA encoding its N-termini up to His249 and Arg281 residue of CcoA, respectively. The PCR products obtained were digested with XhoI and BamHI restriction enzymes and cloned into the similarly digested pTFD342 to yield pBK86 (ccoA249::phoA) and pBK88 (ccoA281::phoA).
Protein purification, fluorescence measurements and 64Cu binding assays
For fluorescence measurements and Cu binding assays, PccA-Strep and PccA-4CStrep variants of the PccA protein were expressed in E.coli BL21 (DE3) carrying the plasmids pET22b-PccAStrep and pET22b-PccA-4Cstrep, respectively, and were purified via their C-terminal Strep-Tag. E.coli cells were grown aerobically in LB medium with apropriate antibiotics, and when they reached an OD600 of 0.6, freshly prepared IPTG was added to the culture at a final concentration of 1mM. After 2 hours incubation at 30° with 170 rpm shaking, cells were harvested and washed once with 40 ml washing buffer (100 mM Tris/HCl pH 7.5, 150 mM NaCl and 1 mM EDTA). Following centrifugation cells were resuspended in 15 ml washing buffer, and disrupted by using a French pressure cell at 8000 psi (pounds per square inch). Cell lysates were centrifuged at 28,720 g for 30 min using the SS-34 rotor (Evolution RC6 centrifuge, Thermo Scientific Inc.) to remove cell debris. The supernatant was ultracentrifuged for 1 hour at 174,000 g in the Ti50 rotor (Beckmann-Coulter, Krefeld, Germany), then loaded onto a polypropylene column containing Strep-Tactin matrix, equilibrated with the washing buffer. After five washing steps with 1 column volume of washing buffer each, elution was initiated using the elution buffer (100 mM TrisHCl pH 7.5, 150 mM NaCl, 1mM EDTA and 2.5 mM desthiobiotin), and fractions were collected.
For measuring intrinsic tyrosine fluorescence of PccA, purified protein (10 μM final concentration) was incubated with different concentrations of Cu(II) or Cu(I) for 10 min at room temperature (25°C) under anaerobic conditions (3.8% H2 and 96.2% N2) in an anaerobic chamber (Coy Laboratory products). Cu(II) was added as CuSO4, which was dissolved in Cu-free washing buffer lacking EDTA. Cu(I) was added as Tetrakis(acetontrile)Cu(I) hexafluorophosphate ([Cu(CH3CN)4]PF6; Sigma Aldrich, Munich, Germany) and was dissolved under anaerobic conditions in acetonitrile to a final concentration of 100 mM. Further dilutions were performed in Cu-free washing buffer lacking EDTA under anaerobic conditions. Samples were subsequently transferred under anaerobic conditions to a TECAN infinite M200 fluorescence microplate reader and were excited at 276 nm. Fluorescence emmision intensity was recorded between 280 and 380 nm. For quantification, the intrinsic fluorescence observed in the absence of metals at 310 nm was set to 1.0 and the values observed in the presence of different metals and different metal concentrations were normalized in respect to this value.
For measuring 64Cu binding to SenC, PccA or PccAMut, 1 μM purified protein was incubated for 10 min at room temperature with 1, 5 and 10 μM radioactive copper and then precipitated with TCA for 30 min on ice (final TCA concentration 5%). After 10 min centrifigation at 13000 rpm, the pellet was resuspended in scintillation liquid and its radioactivity was measured using a scintillation counter to determine copper binding to these proteins. Alternatively, the pellet was resuspended in 25 μl TCA loading buffer (150 mM TRIS, 3.3% (w/v) SDS, 12% (w/v) glycerol, 2 mM EDTA, and 0.1 M DTT) and run on a 15 % native PAGE. Subsequently the gel was dryed using a gel dryer (BioRad Inc.) and visualised by phosphor imaging. 64Cu was obtained as CuCl2 from the National Centre for Nuclear Research, Otwock, Poland and delivered by Hartmann Analytic GmbH (Braunschweig, Germany). It was dissolved in washing buffer lacking EDTA to the appropriate concentrations.
Preparation of intracytoplasmic membranes (ICMs) and Blue Native Page analyses
Inverted cytoplasmic membranes from wild-type R. capsulatus (MT1131) and its mutants were prepared as described previously (Pawlik et al., 2010, Koch et al., 1998a). For Blue Native (BN) PAGE analyses ICM’s (~100 μg of total protein) were incubated at 25° C for 15 minutes with continous shaking (550 rpm Eppendorf Thermomixer), in lysis buffer (50 mM NaCl, 50 mM imidazole/HCl, pH 7.0, 5 mM 6-aminohexanoic acid) containing 1% n-dodecylmaltoside (Thermo Scientific Inc.). After 15 min centrifugation at 70000 rpm in a TLA 100.3 rotor, 15 μl of the supernatant was mixed with 2 μl loading dye (5% Coomassie blue G250 in 0.5 M 6-aminohexanoic acid) and 50% glycerol and then analyzed using a 4–16% gradient BN-PAGE.
Oxygen uptake assays
Ascorbate-TMPD (N,N,N,N-tetramethyl-p-phenylendiamine) oxidase activity was measured with a fiber optic oxygen meter (Fibox 3; PreSens GmbH, Regensburg, Germany) at 28° C in a closed reaction chamber. R. capsulatus membranes were disolved in ICM buffer (50 mM triethanolamine acetate (TeaAc) pH 7.5, 1 mM EDTA, 1 mM DTT, and 0.5 mM PMSF) to a final concentration of about 0.1 μg/μl. The reaction was initiated by the addition of 10 μl of 1 M sodium ascorbate (10 mM final concentration) and 5 μl of 24 mM TMPD (N,N,N,N – tetramethyl-p-phenylendiamine, final concentration 0.2 mM). Oxygen consumption was recorded at 28° C using the OxyView 3.5.1 software (PreSens GmbH) and terminated after several minutes of recording by the addition of 2 μl NaCN (0.1 mM final concentration). cbb3-Cox activity was determined by substraction of the respiration rate remaining after addition of NaCN from that initiated by ascorbate-TMPD.
Alkaline phosphatase measurements
The alkaline phosphatase activities of strains carrying the ccoA::phoA fusions were measured in vitro using membranes (100 μg of total proteins) and p-nitrophenyl phosphate as a substrate, as described in (Brickman & Beckwith, 1975). The absorbance at 420 nm resulting from the pigments of R. capsulatus chromatophore membranes was recorded before adding the substrate, and subtracted from the final absorbance obtained at 420 nm after incubation with p-nitrophenyl phosphate. Alkaline phosphatase activity was calculated as follows: [A420 (after addition of substrate) – A420 (before addition of substrate)] x 1000/min/mg of total proteins.
In-gel heme staining
To visualize the c-type cytochromes, ICMs (100 μg of protein) were disolved in 20 μl Läemmli buffer for 5 minutes at 25° C with shaking a 1000 rpm, and denaturated at 37° C for 15 minutes. After separation, SDS-Tris-Tricine polyacrylamide gels were treated with 3,3,5,5-tetramethylbenzidine (TMBZ) to reveal the peroxidase activity associated with c-type cytochromes (Thomas et al., 1976) as performed earlier (Koch et al., 1998b).
Measurement of cellular copper accumulation
R. capsulatus cells were grown in MPYE media under aerobic conditions (100 ml of growth medium in 250 ml flask) at 35° C, 110 rpm to an OD685 of 1. Cells were harvested by centrifugation at 4° C, 5,000 rpm for 20 min, and the pellets were washed with 50 ml of metal-depleted MPYE and re-centrifuged. The pellet was then resuspended in 1 ml of metal-depleted H2O, and a small aliquot (about 10 to 20 μl) was taken for protein concentration determination by the Lowry assay. Exactly 1 ml of cell suspension was transferred to a new 15 ml tube to which 0.75 ml of 53% nitric acid was added and incubated at 80 °C for 1 hour followed by overnight incubation at 60 °C to yield a clear liquid. The reaction was terminated by the addition of 300 μl H2O2. Cu-free water was added up to a final volume of 10 ml, and samples were measured by using an atomic absorption spectrophotometer Perkin-Elmer 4110 ZL Zeeman. In case any insoluble impurities were observed within the sample, they were filtered through a 0.45 μm filter.
R. capsulatus spheroplasts were essentially prepared as described previously (Norell et al., 2014) with the following modifications: 10 ml of cells were grown at 35° C, 110 rpm up to OD685 of 0.8 – 1, and centrifuged at 5,000 rpm for 10 min at 4° C. The pellet was resuspended in 5 ml of 100 mM Tris/HCl pH 7.5 containing 0.5 M sucrose. Subsequentely, 0.1 mg/ml lysosyme dissolved in 8 mM EDTA, pH 8.0 was added, and the sample was incubated for 15 min on ice. Formation of spheroplasts was monitored microscopically, using a Olympus BX51 microscope at 100x magnification with a numerical aperture of 1.4. Spheroplasts were pelleted by centrifugation at 5,000 rpm for 20 min, and copper contents of the pellet and the supernatant were determined after acid digestions, as described above for intact cells.
Formaldehyde or cysteine cross-linking
In vivo formaldehyde cross-linking was performed as described previously (Lohmeyer et al., 2012). Cysteine crosslinking was performed with PccA and PccA-4C proteins that were purified using the Strep-Tag purification protocol of the supplier (IBA GmbH, Göttingen, Germany) from appropriate E. coli strains carrying the pET22b-PccAStrep and pET22b-PccA-4CStrep plasmids, respectively,. Spheroplasts of R. capsulatus cells carrying pRK415-SenCHis (Lohmeyer et al., 2012) were prepared as described above. 500 μl of spheroplasts resuspended in 100 mM Tris/HCl pH 7.5, 150 mM NaCl was incubated with ~ 0,3 mg/ml of purified PccA or PccA-4C protein, followed by incubation for 1 hour at room temperature with bis(maleimido) hexane (BMH) (Thermo scientific Inc.), added to a final concentration of 0.5 mM. The reaction was stopped by quenching BMH via the addition of 10 mM 1,4-dithiothreitol (DTT). Strep-tagged PccA and PccA-4C were purified from the incubation mixture via Strep-Tag, and the elution fractions were analyzed by SDS-PAGE.
Immune detection methods
For specific visualization of proteins on SDS- or BN-PAGE, proteins were electro-blotted onto different types of membranes as follows. Proteins from BN-PAGE were transferred a PVDF Immobilon-P membrane (GE Healthcare). Proteins from SDS-PAGE were transferred to a nitrocellulose membrane (GE Healthecare). Polyclonal antibodies against PccA, CcoN, CcoP were used with either peroxidase-conjugated goat anti-rabbit (Caltag Laboratories, Burlingame, CA) or alkaline phosphatase-conjugated goat anti-rabbit antibodies (Sigma, St. Louis, USA) antibodies as a secondary antibodies with ECL (GE Helthcare, Munich, Germany) or NBT/BCIP (Roche, Germany) as detection substrate, as approriate. Peptide antibodies against SenC were generated by GeneScript (New Jersey, USA) using the following peptide CLQTPGDTPAAGNGN. Antibodies against PccA were raised in rabbits (Gramsch Laboratories, Schwabhausen, Germany) against the mature protein purified from E. coli.
Supplementary Material
Acknowledgments
This work was supported by grants of the Deutsche Forschungsgemeinschaft (DFG ITRG 1478) to HGK, the German French PhD College on Membranes and Membrane Proteins to HGK, the Motivate MD College of the Else-Kröner Fresenius Stiftung and the University Freiburg Medical School to MU and HGK, the Deutsche Akademische Austauschdienst (DAAD) to PIT, by a grant from the National Institute of Health [NIH GM38237 to FD], and also partially (for construction of various strains used here) by a grant from the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of Department of Energy [DOE DE-FG02-91ER20052 to FD]. We gratefully acknowledge Anja Wüst and Oliver Einsle, Faculty of Chemistry and Pharmacy, University Freiburg, for their help with preparing Cu(I).
References
- Abriata LA, Banci L, Bertini I, Ciofi-Baffoni S, Gkazonis P, Spyroulias GA, Vila AJ, Wang S. Mechanism of Cu(A) assembly. Nature chemical biology. 2008;4:599–601. doi: 10.1038/nchembio.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello JM, Raimunda D, Gonzalez-Guerrero M. Metal transport across biomembranes: emerging models for a distinct chemistry. The Journal of biological chemistry. 2012;287:13510–13517. doi: 10.1074/jbc.R111.319343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello JM, Raimunda D, Padilla-Benavides T. Mechanisms of copper homeostasis in bacteria. Frontiers in cellular and infection microbiology. 2013;3:73. doi: 10.3389/fcimb.2013.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balatri E, Banci L, Bertini I, Cantini F, Ciofi-Baffoni S. Solution structure of Sco1: a thioredoxin-like protein Involved in cytochrome c oxidase assembly. Structure (London, England: 1993) 2003;11:1431–1443. doi: 10.1016/j.str.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, Calderone V, Ciofi-Baffoni S, Mangani S, Martinelli M, Palumaa P, Wang S. A hint for the function of human Sco1 from different structures. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8595–8600. doi: 10.1073/pnas.0601375103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci L, Bertini I, Cantini F, Ciofi-Baffoni S. Cellular copper distribution: a mechanistic systems biology approach. Cellular and molecular life sciences: CMLS. 2010a;67:2563–2589. doi: 10.1007/s00018-010-0330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci L, Bertini I, Cantini F, Ciofi-Baffoni S, Gonnelli L, Mangani S. Solution structure of Cox11, a novel type of beta-immunoglobulin-like fold involved in CuB site formation of cytochrome c oxidase. The Journal of biological chemistry. 2004;279:34833–34839. doi: 10.1074/jbc.M403655200. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, Cavallaro G, Rosato A. The functions of Sco proteins from genome-based analysis. Journal of proteome research. 2007;6:1568–1579. doi: 10.1021/pr060538p. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, Ciofi-Baffoni S, Katsari E, Katsaros N, Kubicek K, Mangani S. A copper(I) protein possibly involved in the assembly of CuA center of bacterial cytochrome c oxidase. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:3994–3999. doi: 10.1073/pnas.0406150102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci L, Bertini I, Ciofi-Baffoni S, Kozyreva T, Zovo K, Palumaa P. Affinity gradients drive copper to cellular destinations. Nature. 2010b;465:645–648. doi: 10.1038/nature09018. [DOI] [PubMed] [Google Scholar]
- Bott M, Preisig O, Hennecke H. Genes for a second terminal oxidase in Bradyrhizobium japonicum. Archives of microbiology. 1992;158:335–343. doi: 10.1007/BF00245362. [DOI] [PubMed] [Google Scholar]
- Brickman E, Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and phi80 transducing phages. J Mol Biol. 1975;96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- Briere JJ, Tzagoloff A. The scoop on Sco. Mol Cell. 2007;25:176–178. doi: 10.1016/j.molcel.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Brzezinski P, Gennis RB. Cytochrome c oxidase: exciting progress and remaining mysteries. Journal of bioenergetics and biomembranes. 2008;40:521–531. doi: 10.1007/s10863-008-9181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggy J, Bauer CE. Cloning and characterization of senC, a gene involved in both aerobic respiration and photosynthesis gene expression in Rhodobacter capsulatus. Journal of bacteriology. 1995;177:6958–6965. doi: 10.1128/jb.177.23.6958-6965.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler D, Rossmann R, Landolt S, Balsiger S, Fischer HM, Hennecke H. Disparate pathways for the biogenesis of cytochrome oxidases in Bradyrhizobium japonicum. The Journal of biological chemistry. 2010;285:15704–15713. doi: 10.1074/jbc.M109.085217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann S, Warkentin E, Xie H, Langer JD, Ermler U, Michel H. The structure of cbb3 cytochrome oxidase provides insights into proton pumping. Science (New York, NY) 2010;329:327–330. doi: 10.1126/science.1187303. [DOI] [PubMed] [Google Scholar]
- Carr HS, George GN, Winge DR. Yeast Cox11, a protein essential for cytochrome c oxidase assembly, is a Cu(I)-binding protein. The Journal of biological chemistry. 2002;277:31237–31242. doi: 10.1074/jbc.M204854200. [DOI] [PubMed] [Google Scholar]
- Carr HS, Maxfield AB, Horng YC, Winge DR. Functional analysis of the domains in Cox11. The Journal of biological chemistry. 2005;280:22664–22669. doi: 10.1074/jbc.M414077200. [DOI] [PubMed] [Google Scholar]
- Daldal F. Cytochrome c2-independent respiratory growth of Rhodobacter capsulatus. Journal of bacteriology. 1988;170:2388–2391. doi: 10.1128/jb.170.5.2388-2391.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekici S, Pawlik G, Lohmeyer E, Koch HG, Daldal F. Biogenesis of cbb(3)-type cytochrome c oxidase in Rhodobacter capsulatus. Biochimica et biophysica acta. 2012a;1817:898–910. doi: 10.1016/j.bbabio.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekici S, Turkarslan S, Pawlik G, Dancis A, Baliga NS, Koch HG, Daldal F. Intracytoplasmic Copper homeostasis controls Cytochrome c oxidase production. mBio. 2014;5:e01055–01013. doi: 10.1128/mBio.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekici S, Yang H, Koch HG, Daldal F. Novel transporter required for biogenesis of cbb3-type cytochrome c oxidase in Rhodobacter capsulatus. mBio. 2012b;3 doi: 10.1128/mBio.00293-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eraso JM, Kaplan S. From redox flow to gene regulation: role of the PrrC protein of Rhodobacter sphaeroides 2.4.1. Biochemistry. 2000;39:2052–2062. doi: 10.1021/bi9923858. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Guerrero M, Arguello JM. Mechanism of Cu+-transporting ATPases: soluble Cu+ chaperones directly transfer Cu+ to transmembrane transport sites. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5992–5997. doi: 10.1073/pnas.0711446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guerrero M, Raimunda D, Cheng X, Arguello JM. Distinct functional roles of homologous Cu+ efflux ATPases in Pseudomonas aeruginosa. Molecular microbiology. 2010;78:1246–1258. doi: 10.1111/j.1365-2958.2010.07402.x. [DOI] [PubMed] [Google Scholar]
- Gray KA, Grooms M, Myllykallio H, Moomaw C, Slaughter C, Daldal F. Rhodobacter capsulatus contains a novel cb-type cytochrome c oxidase without a CuA center. Biochemistry. 1994;33:3120–3127. doi: 10.1021/bi00176a047. [DOI] [PubMed] [Google Scholar]
- Gurumoorthy P, Ludwig B. Deciphering protein-protein interactions during the biogenesis of cytochrome c oxidase from Paracoccus denitrificans. The FEBS journal. 2015;282:537–549. doi: 10.1111/febs.13160. [DOI] [PubMed] [Google Scholar]
- Horng YC, Cobine PA, Maxfield AB, Carr HS, Winge DR. Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome C oxidase. The Journal of biological chemistry. 2004;279:35334–35340. doi: 10.1074/jbc.M404747200. [DOI] [PubMed] [Google Scholar]
- Hosler J. Biogenesis/assembly of respiratory enzyme complexes. Biochimica et biophysica acta. 2012;1817:849–850. doi: 10.1016/j.bbabio.2012.04.014. [DOI] [PubMed] [Google Scholar]
- Jenney FE, Jr, Daldal F. A novel membrane-associated c-type cytochrome, cyt cy, can mediate the photosynthetic growth of Rhodobacter capsulatus and Rhodobacter sphaeroides. The EMBO journal. 1993;12:1283–1292. doi: 10.1002/j.1460-2075.1993.tb05773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalimonchuk O, Bestwick M, Meunier B, Watts TC, Winge DR. Formation of the redox cofactor centers during Cox1 maturation in yeast cytochrome oxidase. Molecular and cellular biology. 2010;30:1004–1017. doi: 10.1128/MCB.00640-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch HG, Hwang O, Daldal F. Isolation and characterization of Rhodobacter capsulatus mutants affected in cytochrome cbb3 oxidase activity. Journal of bacteriology. 1998a;180:969–978. doi: 10.1128/jb.180.4.969-978.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch HG, Myllykallio H, Daldal F. Using Genetics to explore Cytochrome function and Structure in Rhodobacter. Methods in enzymology. 1998b;297:81–94. [Google Scholar]
- Koch HG, Winterstein C, Saribas AS, Alben JO, Daldal F. Roles of the ccoGHIS gene products in the biogenesis of the cbb(3)-type cytochrome c oxidase. Journal of molecular biology. 2000;297:49–65. doi: 10.1006/jmbi.2000.3555. [DOI] [PubMed] [Google Scholar]
- Kulajta C, Thumfart JO, Haid S, Daldal F, Koch HG. Multi-step assembly pathway of the cbb3-type cytochrome c oxidase complex. Journal of molecular biology. 2006;355:989–1004. doi: 10.1016/j.jmb.2005.11.039. [DOI] [PubMed] [Google Scholar]
- Leary SC. Redox regulation of SCO protein function: controlling copper at a mitochondrial crossroad. Antioxidants & redox signaling. 2010;13:1403–1416. doi: 10.1089/ars.2010.3116. [DOI] [PubMed] [Google Scholar]
- Leary SC, Cobine PA, Kaufman BA, Guercin GH, Mattman A, Palaty J, Lockitch G, Winge DR, Rustin P, Horvath R, Shoubridge EA. The human cytochrome c oxidase assembly factors SCO1 and SCO2 have regulatory roles in the maintenance of cellular copper homeostasis. Cell metabolism. 2007;5:9–20. doi: 10.1016/j.cmet.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Lohmeyer E, Schroder S, Pawlik G, Trasnea PI, Peters A, Daldal F, Koch HG. The ScoI homologue SenC is a copper binding protein that interacts directly with the cbb(3)-type cytochrome oxidase in Rhodobacter capsulatus. Biochimica et biophysica acta. 2012;1817:2005–2015. doi: 10.1016/j.bbabio.2012.06.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs B, Gest H. Genetic mutations affecting the respiratory electron-transport system of the phoosythetic bacterium Rhodopseudomonas capsulata. J Bacteriol. 1973;114:1045–1051. doi: 10.1128/jb.114.3.1045-1051.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan AG, Lewin A, Davy SL, Boetzel R, Leech A, Walker D, Wood T, Moore GR. PrrC from Rhodobacter sphaeroides, a homologue of eukaryotic Sco proteins, is a copper-binding protein and may have a thiol-disulfide oxidoreductase activity. FEBS letters. 2002;518:10–16. doi: 10.1016/s0014-5793(02)02532-2. [DOI] [PubMed] [Google Scholar]
- Morgada MN, Abriata LA, Cefaro C, Gajda K, Banci L, Vila AJ. Loop recognition and copper-mediated disulfide reduction underpin metal site assembly of CuA in human cytochrome oxidase. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:11771–11776. doi: 10.1073/pnas.1505056112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norell D, Heuck A, Tran-Thi TA, Götzke H, Jacob-Dubuisson F, Clausen T, Daley DO, Braun V, Müller M, Fan E. Versatile in vitro system to study translocation and functional integration of bacterial outer membrane proteins. Nat Comm. 2014;5:5396. doi: 10.1038/ncomms6396. [DOI] [PubMed] [Google Scholar]
- Pawlik G, Kulajta C, Sachelaru I, Schroder S, Waidner B, Hellwig P, Daldal F, Koch HG. The putative assembly factor CcoH is stably associated with the cbb3-type cytochrome oxidase. Journal of bacteriology. 2010;192:6378–6389. doi: 10.1128/JB.00988-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Kulajta C, Pawlik G, Daldal F, Koch HG. Stability of the cbb3-type cytochrome oxidase requires specific CcoQ-CcoP interactions. Journal of bacteriology. 2008;190:5576–5586. doi: 10.1128/JB.00534-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisig O, Zufferey R, Hennecke H. The Bradyrhizobium japonicum fixGHIS genes are required for the formation of the high-affinity cbb3-type cytochrome oxidase. Archives of microbiology. 1996;165:297–305. doi: 10.1007/s002030050330. [DOI] [PubMed] [Google Scholar]
- Raimunda D, Gonzalez-Guerrero M, Leeber BW, 3rd, Arguello JM. The transport mechanism of bacterial Cu+-ATPases: distinct efflux rates adapted to different function. Biometals: an international journal on the role of metal ions in biology, biochemistry, and medicine. 2011;24:467–475. doi: 10.1007/s10534-010-9404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimunda D, Padilla-Benavides T, Vogt S, Boutigny S, Tomkinson KN, Finney LA, Arguello JM. Periplasmic response upon disruption of transmembrane Cu transport in Pseudomonas aeruginosa. Metallomics: integrated biometal science. 2013;5:144–151. doi: 10.1039/c2mt20191g. [DOI] [PubMed] [Google Scholar]
- Richter OM, Ludwig B. Cytochrome c oxidase--structure, function, a nd physiology of a redox-driven molecular machine. Reviews of physiology, biochemistry and pharmacology. 2003;147:47–74. doi: 10.1007/s10254-003-0006-0. [DOI] [PubMed] [Google Scholar]
- Rigby K, Cobine PA, Khalimonchuk O, Winge DR. Mapping the functional interaction of Sco1 and Cox2 in cytochrome oxidase biogenesis. The Journal of biological chemistry. 2008;283:15015–15022. doi: 10.1074/jbc.M710072200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Molecular Cloning. Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- Serventi F, Youard ZA, Murset V, Huwiler S, Buhler D, Richter M, Luchsinger R, Fischer HM, Brogioli R, Niederer M, Hennecke H. Copper starvation-inducible protein for cytochrome oxidase biogenesis in Bradyrhizobium japonicum. The Journal of biological chemistry. 2012;287:38812–38823. doi: 10.1074/jbc.M112.406173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swem DL, Swem LR, Setterdahl A, Bauer CE. Involvement of SenC in assembly of cytochrome c oxidase in Rhodobacter capsulatus. Journal of bacteriology. 2005;187:8081–8087. doi: 10.1128/JB.187.23.8081-8087.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PE, Ryan D, Levin W. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfat polyacrylamide gels. Analytical biochemistry. 1976;75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- Thompson AK, Gray J, Liu A, Hosler JP. The roles of Rhodobacter sphaeroides copper chaperones PCu(A)C and Sco (PrrC) in the assembly of the copper centers of the aa(3)-type and the cbb(3)-type cytochrome c oxidases. Biochimica et biophysica acta. 2012;1817:955–964. doi: 10.1016/j.bbabio.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AK, Smith D, Gray J, Carr HS, Liu A, Winge DR, Hosler JP. Mutagenic analysis of Cox11 of Rhodobacter sphaeroides: insights into the assembly of Cu(B) of cytochrome c oxidase. Biochemistry. 2010;49:5651–5661. doi: 10.1021/bi1003876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt H, Zickermann V, Ludwig B. Site-directed mutagenesis of cytochrome c oxidase reveals two acidic residues involved in the binding of cytochrome c. Biochimica et biophysica acta. 1995;1230:74–76. doi: 10.1016/0005-2728(95)00050-s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.