Summary
Nitrification is a two-step process where ammonia is considered to first be oxidized to nitrite by ammonia-oxidizing bacteria (AOB) and/or archaea (AOA), and subsequently to nitrate by nitrite-oxidizing bacteria (NOB). Described by Winogradsky already in 18901, this division of labour between the two functional groups is a generally accepted characteristic of the biogeochemical nitrogen cycle2. Complete oxidation of ammonia to nitrate in one organism (complete ammonia oxidation; comammox) is energetically feasible and it was postulated that this process could occur under conditions selecting for species with lower growth-rates but higher growth-yields than canonical ammonia-oxidizing microorganisms3. Still, organisms catalysing this process have not yet been discovered. Here, we report the enrichment and initial characterization of two Nitrospira species that encode all enzymes necessary for ammonia oxidation via nitrite to nitrate in their genomes, and indeed completely oxidize ammonium to nitrate to conserve energy. Their ammonia monooxygenase (AMO) enzymes are phylogenetically distinct from currently identified AMOs, rendering recent acquisition by horizontal gene transfer from known ammonia-oxidizing microorganisms unlikely. We also found highly similar amoA sequences (encoding the AMO subunit A) in public sequence databases, which were apparently misclassified as methane monooxygenases. This recognition of a novel amoA sequence group will lead to an improved understanding on the environmental abundance and distribution of ammonia-oxidizing microorganisms. Furthermore, the discovery of the long-sought-after comammox process will change our perception of the nitrogen cycle.
Nitrification, the aerobic oxidation of ammonium to nitrate is divided into two subsequent reactions: ammonium oxidation to nitrite (equation (1)) and nitrite oxidation to nitrate (equation (2)). These two reactions are catalysed by physiologically distinct clades of microorganisms.
| (1) |
| (2) |
| (3) |
Even though the existence of a single microorganism capable of oxidizing ammonium to nitrate (equation (3)) was not previously reported, it was proposed that such a microorganism could have a competitive advantage in biofilms and other microbial aggregates with low substrate concentrations3.
In this study, to characterize the microorganisms responsible for nitrogen transformations in an ammonium-oxidizing biofilm, we sampled the anaerobic compartment of a trickling filter connected to a recirculation aquaculture system (RAS)4 with an ammonium effluent of less than 100 μM. To enrich for the N-cycling community, a bioreactor was inoculated and supplied with low concentrations of ammonium, nitrite and nitrate under hypoxic conditions (≤ 3.1 µM O2). Within 12 months, we obtained a stable enrichment culture, which efficiently removed ammonium and nitrite from the medium (Extended Data Fig. 1). The culture showed anaerobic ammonium-oxidizing (anammox) activity (Fig. 1a), and fluorescence in situ hybridization (FISH) revealed that anammox organisms of the genus Brocadia constituted ~45% of all FISH-detectable bacteria. Surprisingly, Nitrospira-like NOB accounted for ~15% of the community and co-occurred with the Brocadia species in flocs (Fig. 2a). This tight clustering with anammox bacteria was unexpected as both microorganisms require nitrite for growth. Together with the presence of Nitrospira at very low oxygen concentrations, this indicated that there could be a functional link between these organisms.
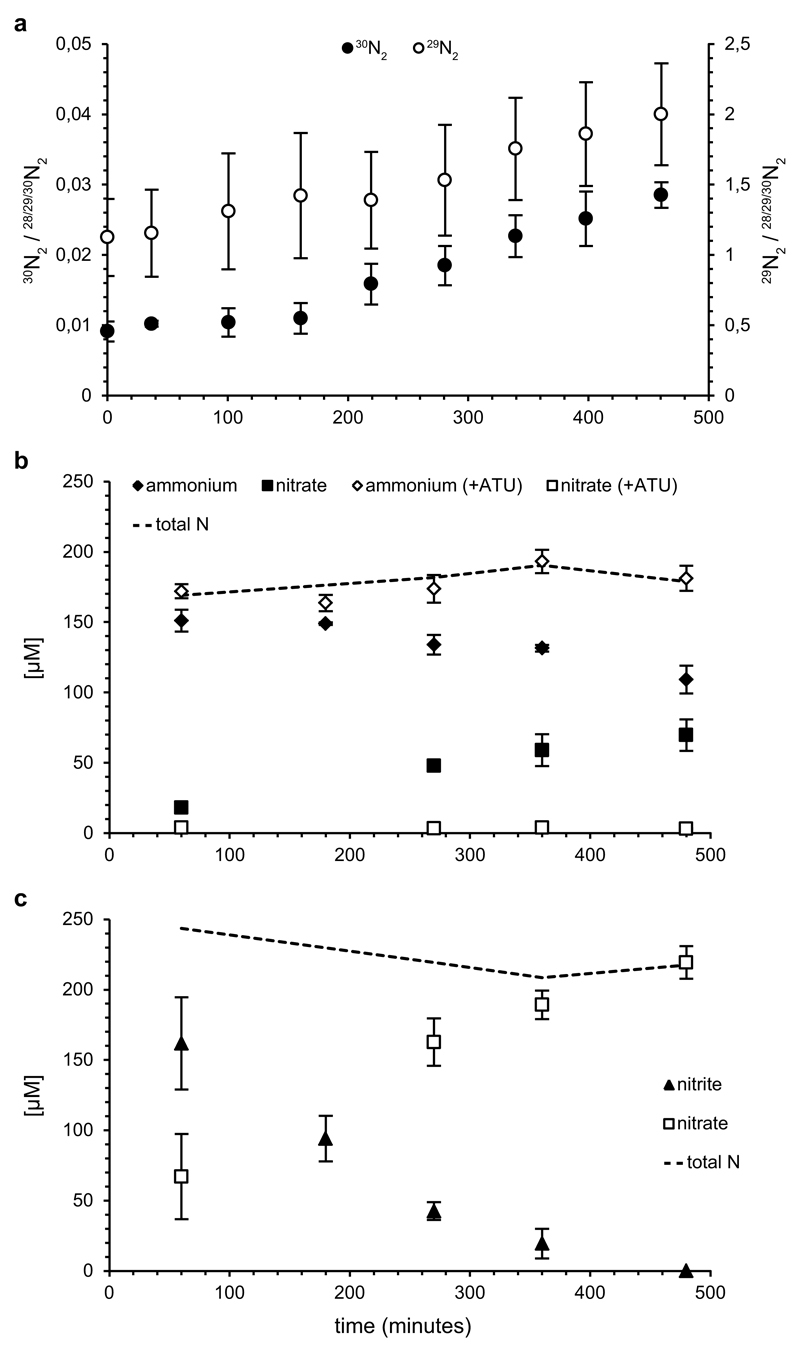
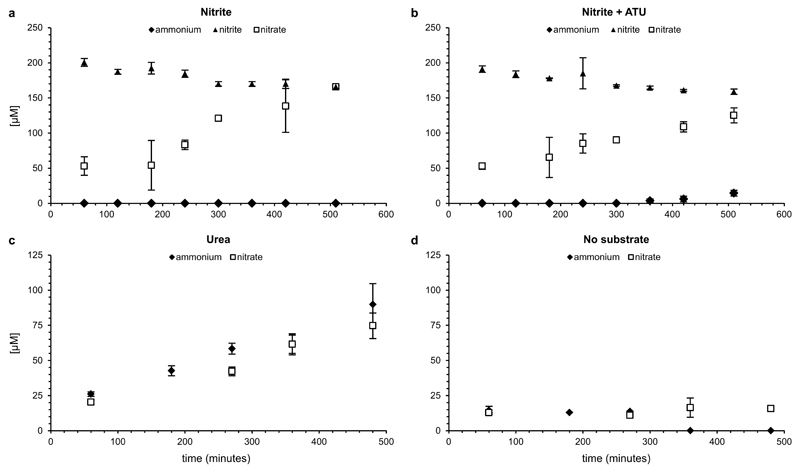
Figure 1. Ammonium oxidation by the enrichment culture.
a, 29N2 (open circles) and 30N2 (filled circles) production from 15NH4+ by the enrichment. b, Ammonium (diamonds) oxidation to nitrate (squares) in aerobic batch incubations in the absence (filled symbols) and presence (open symbols) of ATU. Nitrite concentrations were below the detection limit (<5 µM) at all time points. c, Nitrite (triangles) oxidation to nitrate (squares) in aerobic batch incubations. In b and c, total nitrogen balances are indicated (dashed lines). Symbols in all plots represent averages of three individual experiments. Ammonium concentrations were determined in single measurements, other compounds in triplicate. Error bars represent standard deviations of three biological replicates.
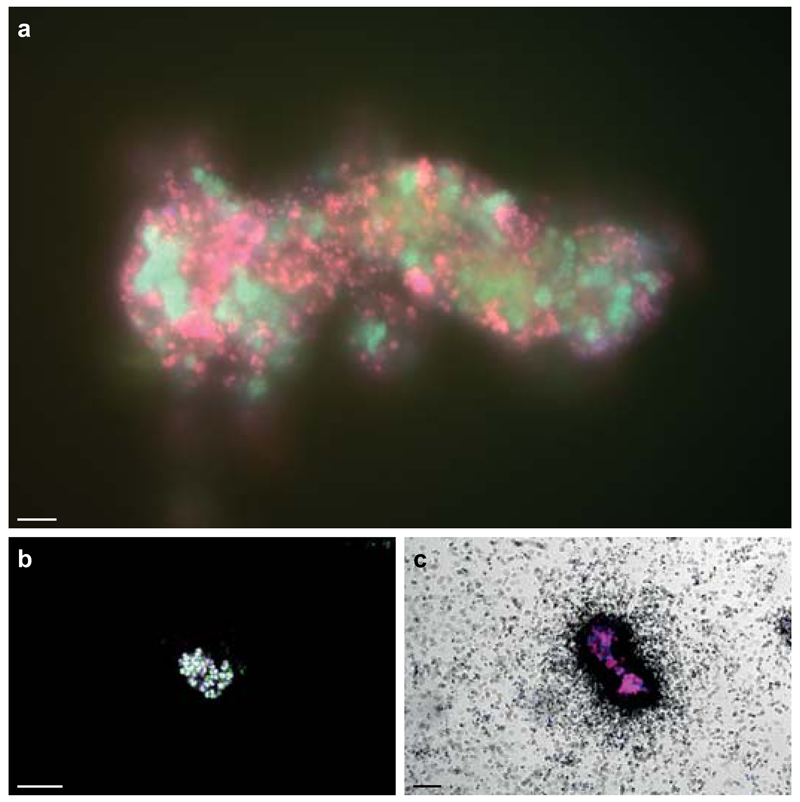
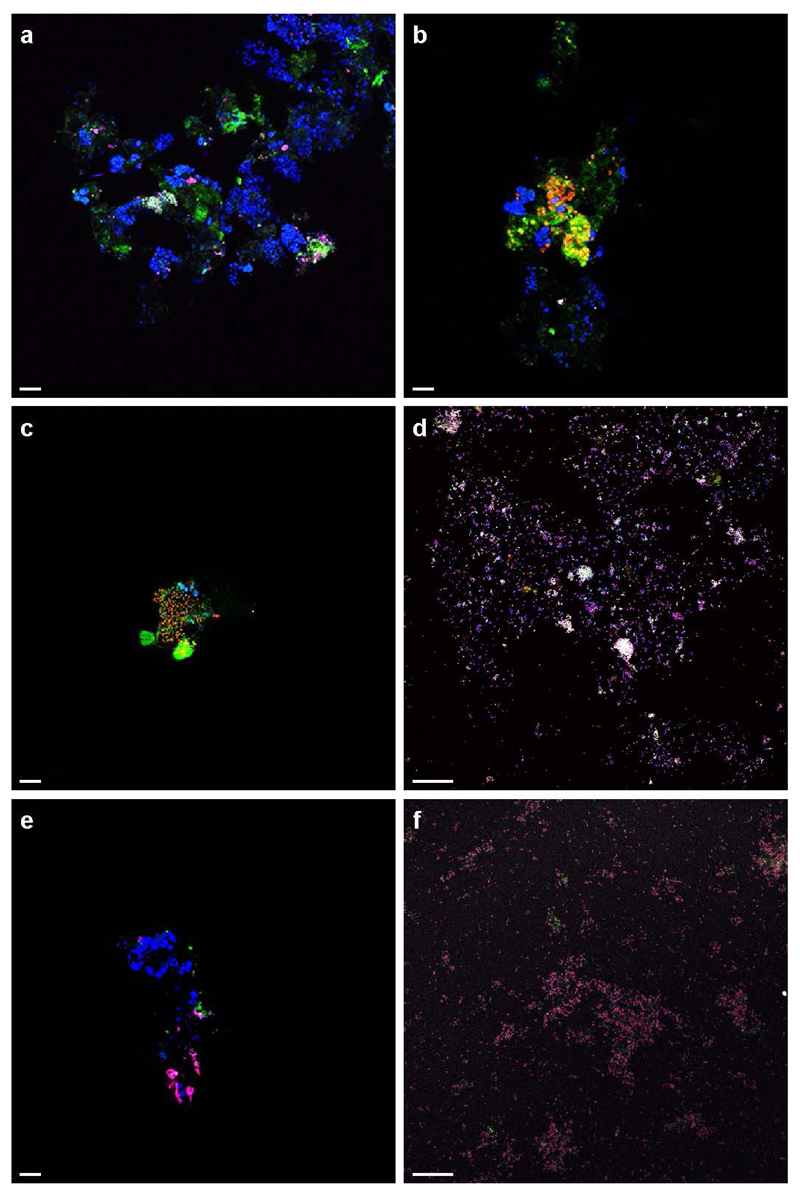
Figure 2. In situ detection of Nitrospira and their ammonia-oxidizing capacity.
a, Co-aggregation of Nitrospira and Brocadia in the enrichment. Cells are stained by FISH with probes for all bacteria (EUB338mix, blue), and specific for Nitrospira (Ntspa712, green, resulting in cyan) and anammox bacteria (Amx820, red, resulting in magenta). b, AMO labelling by FTCP (green). Nitrospira was counterstained by FISH (probes Ntspa662 (blue) and Ntspa476 (red), resulting in white). c, Ammonium-dependent CO2 fixation by Nitrospira shown by FISH-MAR. Silver grain deposition (black) above cell clusters indicates 14CO2 incorporation. Nitrospira was stained by FISH (probes Ntspa476 (red) and Ntspa662 (blue), resulting in magenta). Images in b and c are representative of two individual experiments, with three (b) or two (c) technical replicates each. Scale bars in all panels represent 10 µm.
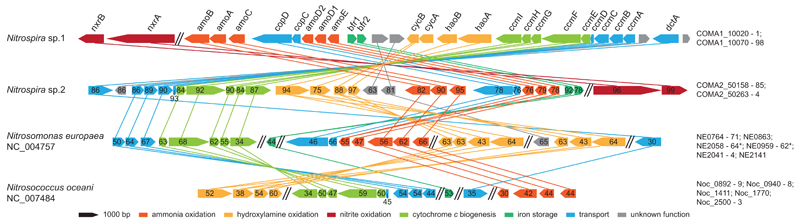
To determine the function of Nitrospira in the community, we extracted and sequenced total DNA from the enrichment culture biomass. In total 4.95 Giga base pairs of trimmed metagenomic sequence were obtained and used for de novo assembly. By differential-coverage and sequence composition-based binning5 it was possible to extract high-quality draft genomes of two Nitrospira species. The two strains had genomic pairwise average nucleotide identities (ANI)6 of 75% and thus clearly represented different species (Nitrospira sp.1 and sp.2, Extended Data Fig. 2 and Extended Data Table 1). Surprisingly, both genomes contained the full set of AMO and hydroxylamine dehydrogenase (HAO) genes for ammonia oxidation, in addition to the nitrite oxidoreductase (NXR) subunits necessary for nitrite oxidation in Nitrospira7. In both species all these genes were localized on a single contiguous genomic fragment, along with general housekeeping genes that allowed reliable phylogenetic assignment. Consequently, these Nitrospira species had the genetic potential for the complete oxidation of ammonia to nitrate. No AMO of canonical ammonia-oxidizing bacteria or archaea could be detected in the trimmed metagenomic reads or by amoA-specific PCR8,9 on DNA extracted from reactor biomass, and no other indications for the presence of ammonia-oxidizing microorganisms were found in the metagenome or by FISH analyses. The AMO structural genes (amoCAB) of both Nitrospira species, along with the putative additional AMO subunits amoEDD210,11, formed one gene cluster with haoAB-cycAB (encoding HAO, the putative membrane anchor protein HaoB, electron transfer protein cytochrome c554 and quinone reducing cytochrome cm552, respectively)12 and showed highest similarities to their counterparts in betaproteobacterial AOB (60% average amino acid identity to the Nitrosomonas europaea genes; Fig. 3 and Supplementary Table 1). The same genomic region also contained genes for copper and heme transport, cytochrome c biosynthesis, and iron storage. These accessory genes were highly conserved in ammonia-oxidizing bacteria but not in other Nitrospira7,13, indicating their involvement in AMO and HAO biosynthesis or activation. Nitrospira sp.1 encoded three discrete amoC genes, one of which was clustered with a second, almost identical copy of amoA (97.7% amino acid identity). Nitrospira sp.2 lacked the second amoA, but contained four additional amoC and a second haoA gene (Supplementary Table 1). Unlike other Nitrospira7,13, both species lacked enzymes for assimilatory nitrite reduction, indicating adaptation to ammonium-containing habitats. For ammonium uptake, they encoded low affinity Rh-type transporters most closely related to Rh50 found in Nitrosomonas europea14, in contrast to most AOB and NOB that have the high affinity AmtB-type proteins. Both species encoded ureases and the corresponding ABC transport systems, indicating that urea could be used as an alternative ammonium source. Interestingly, Ca. N. inopinata, the moderately thermophilic ammonia-oxidizing Nitrospira described by Daims et al.15, encoded a similar set of AMO, HAO and urease proteins, and also lacked genes for assimilatory nitrite reduction. Unlike the two species described here, however, it contained a periplasmic cytochrome c nitrite reductase (NrfA) that could allow it to conserve energy by dissimilatory nitrite reduction to ammonium (DNRA), but might also provide ammonium for assimilation. The evolutionary divergence of these organisms was also reflected in the low ANI values of 70.3 - 71.6% between Ca. N. inopinata and the two species described here. Concerning their genetic repertoire for nitrite oxidation, sp.2 had four almost identical (>99% amino acid identity) NXR alpha and beta (NxrAB) subunits. Sp.1 had two nxrAB copies encoding identical NxrB subunits, but NxrA subunits with amino acid identities of 89.6%, which were separated into distinct clusters in phylogenetic analyses. One homolog branched with sequences from N. moscoviensis, while the other formed a novel sequence cluster together with the sequences from sp.2 (Extended Data Fig. 3).
Figure 3. Schematic illustration of the AMO genomic region in Nitrospira and selected AOB.
The AMO locus in Nitrospira sp.1 in comparison to sp.2 and the beta- and gammaproteobacterial AOB Nitrosomonas europaea and Nitrosococcus oceani, respectively. The position of NXR on the AMO-containing Nitrospira contigs is also indicated. Homologous genes are connected by lines. Functions of the encoded proteins are represented by colour, the arrow shows direction of transcription. Numbers specify amino acid identities to Nitrospira sp.1. Parallel double lines designate a break in locus organization. Locus tags for each organism are listed on the right. Genes are drawn to scale. amo, ammonia monooxygenase; bfr; bacterioferritin; ccm, cytochrome c biogenesis; cop, copper transport; cyc, cytochrome c; dct, sodium:dicarboxylate symporter; hao, hydroxylamine dehydrogenase; nxr, nitrite oxidoreductase.
To ascertain that ammonia oxidation occurred under hypoxic conditions in the enrichment culture, we supplied the bioreactor with 15N-labelled ammonium. While the anammox bacteria consumed 15NH4+ and converted it into 29N2, a steady increase of 30N2 was also observed (Fig. 1a). This formation of 30N2 could only be explained by the production of 15N-labelled nitrite derived through aerobic ammonium oxidation. As metagenomic analyses confirmed that the Nitrospira species were the only organism in the enrichment harbouring AMO and HAO, this clearly showed that they were able to perform this reaction even at O2 concentrations lower than 3.1 µM. To unambiguously link this activity to Nitrospira, we visualized the AMO protein in situ using batch incubations with reactor biomass and fluorescein thiocarbamoylpropargylamine (FTCP), a fluorescently labelled acetylene analogue that acts as suicide substrate for AMO16 and covalently binds to the enzyme17. When counterstained with Nitrospira-specific FISH probes, including a newly designed probe specifically targeting the 16S rRNA-defined phylogenetic group comprising spp.1 and 2 (Extended Data Table 2 and Extended Data Fig. 4), strong FTCP labelling of Nitrospira cells was observed, providing strong support for the presence of the ammonia-oxidizing enzyme at single-cell level (Fig. 2b and Extended Data Fig. 5).
Batch incubations were performed at ambient oxygen concentrations to determine conversion rates of ammonium and nitrite, the level of inhibition by allylthiourea (ATU; a potent inhibitor of bacterial ammonia oxidation18,19), and the use of urea as ammonium source for nitrification. Flocs were mechanically disrupted to ensure complete exposure of the biomass to oxygen, which inhibits the anammox and denitrification processes20,21. This inhibition was confirmed by the lack of labelled N2 formation in incubations with 15NH4+. In these incubations (Fig. 1 and Extended Data Fig. 6), the culture oxidized ammonium (6.0 ± 1.0 µM NH4+/h) and nitrite (23 ± 4.7 µM NO2-/h) to nitrate. ATU selectively inhibited ammonia oxidation, but did not affect nitrite oxidation rates. Urea was converted to ammonium, which was subsequently oxidized to nitrate (7.8 ± 1.1 µM nitrate/h) suggesting that these Nitrospira species were capable of using urea as source of ammonia to drive nitrification, as was also reported for some AOA22 and AOB23. This trait could enable them to thrive in environments like fertilized soils, wastewater treatment plants (wwtps), and many aquatic systems where urea is often present at micromolar levels24. However, it should be noted that the two Nitrospira spp. were not the only organisms in the enrichment culture that encoded ureases.
To investigate substrate-dependent inorganic carbon fixation as a proxy for energy conservation from ammonia and nitrite oxidation, we used FISH in combination with microautoradiography (FISH-MAR)25. Aerobic incubations with mechanically disrupted flocs were performed in the presence of 500 µM ammonium, 500 µM ammonium with 100 µM ATU, or 500 µM nitrite. Nitrospira incorporated carbon from 14C-labelled bicarbonate in the presence of either ammonium or nitrite, and ammonia-dependent carbon fixation was strongly inhibited by the addition of ATU (Fig. 2c and Extended Data Fig. 7). Only flocs containing Nitrospira were labelled during all incubations, suggesting that these were the only chemolithoautotrophic nitrifying organisms present in the culture and indeed could conserve energy from the oxidation of ammonia and nitrite.
In 16S rRNA-based phylogenetic analyses, the two ammonia-oxidizing Nitrospira species from our enrichment culture formed two separate lineages within one strongly supported sequence cluster affiliated with Nitrospira sublineage II26 (Extended Data Fig. 4). They both grouped with highly similar sequences (>99% nucleotide identity) from a diverse range of habitats, including soil, groundwater, RAS, wastewater treatment plants (wwtps) and drinking water distribution systems. The formation of distinct clusters containing sp.1 and sp.2 indicated that the last common ancestor encoded genes for complete nitrification and that this lifestyle might be conserved in most organisms affiliated with this sequence group.
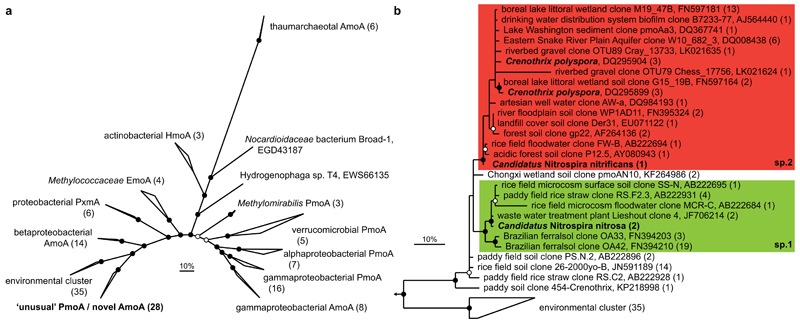
To explore the environmental relevance of these Nitrospira, we searched the NCBI nr database27 for closely related amoA genes. Surprisingly, we found the AmoA proteins of the two Nitrospira species to be phylogenetically divergent from the described bacterial AmoA sequences. Nitrospira sp.2 AmoA was 97-98% identical to the so-called “unusual” methane monooxygenase (PMO) proteins of Crenothrix polyspora28. The two AmoA copies from Nitrospira sp.1 had lower similarities to Crenothrix PmoA (90-91% identity), but also affiliated with this group (Fig. 4). Sequences within this group cannot be amplified by standard amoA primers, but only by pmoA primers when used at reduced stringency29. Therefore the public databases only contain few closely related sequences, which mainly were derived from habitats studied for their bacterial methane-oxidizing (MOB) communities. Highly similar sequences derived from wwtps and drinking water systems, however, indicated occurrence of ammonia-oxidizing Nitrospira in a range of engineered and natural environments. We furthermore screened all publicly available shotgun datasets on MG-RAST30. Indeed, 168 metagenomes (out of 6255) and 28 metatranscriptomes (out of 1051) contained at least two reads affiliated with this amoA group, yielding a total of 3727 reads that were obtained mainly from soil, sediments and wwtps (Extended Data Table 3). Thus, our results showed that the Crenothrix sequence group consists of so far unrecognized AMO sequences overlooked in nitrification studies based on amoA gene detection. Based on these findings, it is highly likely that the PCR-based determination of the Crenothrix pmoA gene from an environmental sample28 was erroneous, and this cluster only contains genes encoding AMOs. Nevertheless, with the currently available information it cannot be excluded that certain Crenothrix species attained an amoA gene through lateral gene transfer and use the encoded protein as a surrogate PMO.
Figure 4. Phylogenetic analysis of the AmoA/PmoA sequence family.
Bayesian interference tree (s.d.=0.01) showing the affiliation of the Nitrospira AmoA. Posterior probabilities ≥70% and ≥90% are indicated by open and filled circles, respectively. Scale bars indicate 10% sequence divergence. a, Radial tree indicating the localisation of the novel AmoA/’unusual’ PmoA sequence group in relation to the main functional groups within the sequence family. Numbers in brackets indicate sequences per group (137 sequences in total). Amo, ammonia monooxygenase; Emo, ethane monooxygenase; Hmo, hydrocarbon/butane monooxygenase; Pmo/Pxm, particulate methane monooxygenase. b, Cladogram detailing the affiliation of the Nitrospira sp.1 (green box) and sp.2 (red box) AmoA sequences within this sequence group. Nitrospira and Crenothrix sequences are depicted in bold. One representative sequence per study is shown for highly similar sequences; numbers in brackets indicate the number of sequences represented.
In conclusion, here we demonstrated the existence of complete nitrification in a single organism (comammox) and identified two Nitrospira species capable of catalysing this process (equation (3)). In 16S rRNA or amoA/pmoA-based studies these organisms would have been classified as NOB or MOB, respectively. Hence, our results show that a whole group of ammonia-oxidizing organisms was previously overlooked. Our findings furthermore disprove the long-held assumption that nitrification (ammonia oxidation via nitrite to nitrate) is catalysed by two distinct functional groups, thus redefining a key process of the biogeochemical nitrogen cycle.
Based on their physiology, differences in genome content, and separation in different phylogenetic groups in 16S rRNA-based analyses, we propose tentative names for both Nitrospira species present in our enrichment: “Candidatus Nitrospira nitrosa” (Etymology: L. fem. adj. nitrosa, full of natron; the nitrite and nitrate forming Nitrospira) for sp.1 and “Candidatus Nitrospira nitrificans” (N.L. part. adj. nitrificans, nitrifying; the nitrifying Nitrospira) for sp.2. Both species are chemolithoautotrophic and fully oxidize ammonia via nitrite to nitrate.
Methods
Enrichment and cultivation
A bioreactor was inoculated with biomass from a RAS biofilter (3.5 l, obtained from the anoxic part of the trickling filter compartment) connected to an aquaculture system. The system accommodated common carp (Cyprinus carpio, approximately 3.5 kg total weight) and had a total volume of 900 l. The bioreactor (Applikon Biotechnology BV, Schiedam, The Netherlands) consisted of stainless steel and glass, had a 7 l working volume, was equipped with pH and dissolved oxygen sensors (Applikon Dependable Instruments BV Applisens, Schiedam, The Netherlands) and connected to an ADI1030 biocontroller (Applikon Biotechnology BV, Schiedam, The Netherlands). It was operated as a sequencing batch reactor (SBR) with 12 h or 24 h cycles. In the first 5 months, the reactor was operated with a 24 h cycle that consisted of 23 h 15 min filling, 15 min settling (no stirring) and 30 min removal of the supernatant. Afterwards, in 12 h cycles, each filling cycle consisted of 11 h 15 min, followed by 15 min settling and 30 min removal of the supernatant. During every filling period, the reactor was supplied with 600 ml of medium (0.83 ml/min). The reactor and the medium were flushed constantly with Ar/CO2 (95%/5% v/v, 10 ml min-1). The temperature was kept at 23 ± 1°C with a heating blanket and pH was maintained at 6.99 ± 0.1 using a 1 M KHCO3 solution. The reactor was stirred at 200 rpm. Medium was prepared using aquaculture water taken from the RAS biofilter. This water contained 300 - 1,848 µM NO3-, 0 - 29 µM NO2- and 0 - 75 µM NH4+. The water was filter-sterilized (polysulfone filter HF80S, Fresenius Medical Care, Bad Homburg, Germany) and supplemented with 100 - 500 µM NH4+, 100 - 450 µM NO2- and 500 µM NO3-.
DNA extraction and genome sequencing
DNA was extracted using the PowerSoil DNA isolation kit (MoBio, Carlsbad, CA) or a CTAB-based extraction method31. 1 µg of DNA was used to prepare paired-end sequencing libraries using the TruSeq PCR-free kits (Illumina, San Diego, CA, USA) following the manufactures recommendation except that the 550 bp protocol was used with 1 µg of input DNA. Mate-pair libraries were prepared using the Nextera Mate-pair kit (Illumina) using the gel-free approach. The prepared libraries were sequenced using an Illumina MiSeq with MiSeq Reagent Kit v3 (2x301 bp; Illumina).
Bioinformatics
Data generation and binning of metagenome scaffolds to individual genome bins was conducted as described in the mmgenome workflow32 which builds on the multi-metagenome principles5. Paired-end Illumina reads in FASTQ format were imported to CLC Genomics Workbench v. 8.0 (CLCBio, Aarhus, Denmark) and trimmed using a minimum phred score of 20, a minimum length of 50 bp, allowing no ambiguous nucleotides and trimming off Illumina sequencing adaptors. Mate-pair reads in FASTQ format were trimmed using NextClip33 and only reads in class A were used for assembly. Passing reads were co-assembled using CLCs de novo assembly algorithm, using a kmer of 63 and a minimum scaffold length of 1 kbp. The trimmed metagenome reads were afterwards independently mapped to the assembled scaffolds using CLCs “map reads to reference” algorithm, with a minimum similarity of 95% over 80% of the read length.
Open reading frames were predicted in the assembled scaffolds using the metagenome version of Prodigal34. A set of 107 HMMs of essential single-copy genes35 were searched against the predicted open reading frames using HMMER336 with default settings, except for the use of the trusted cutoff (-cut_tc). Identified proteins were taxonomically classified using BLASTP against the RefSeq (version 52) protein database with a maximum e-value cutoff of 1e-5. MEGAN37 was used to extract class level taxonomic assignments from the BLAST .xml output file. The script network.pl was used to extract paired-end read connections between scaffolds using a SAM file of the read mappings to the metagenome.
Individual genome bins were extracted using the multi-metagenome principles5 and refined using tetranucleotide frequencies, as implemented in the mmgenome R package32. The script extract.fastq.reassembly.pl was used to extract paired-end reads from the binned scaffolds, which were used for re-assembly using SPAdes 3.5.038. Paired-end and mate-pair connections were used to manually refine the extracted genome bins. For all genomes quality was assessed using coverage plots through the mmgenome R package and by the use of QUAST39 and CheckM40 (see Supplementary Table 2 for CheckM counts of single-copy genes). Manual inspection of potential misassemblies was done using Circos41 as described32. In addition, key regions were manually inspected in CLC Genomics Workbench.
The Nitrospira draft genomes were integrated into the MicroScope annotation platform42. The automatic annotation of genes in key metabolic pathways was manually refined using the respective tools in MaGe43 as described previously7. Genomic pairwise average nucleotide identity values were calculated using BLAST (ANIb) in JSpecies6.
Absence of canonical bacterial or archaeal amoA sequences in the metagenome data was confirmed by searching a set of reference sequences against a BLAST database containing all trimmed metagenome reads.
Code availability
The Rmarkdown files used for extracting the genome bins are available for download32.
Activity assays
For activity assays, the reactor was supplied with medium containing labelled ammonium (15NH4+). The medium flow was kept at normal operating rate (0.83 ml/min) and the biomass was stirred continuously. Isotopic composition of the nitrogen gas produced was analysed using gas chromatography (Agilent 6890 equipped with a Porapak Q column at 80°C and a TCD detector at 300°C; Agilent Technologies, Santa Clara, CA, USA) combined with mass spectrometry (Agilent 5975c, quadruple inert MS).
For batch assays, 150 ml biomass was taken from the reactor and harvested by centrifugation (300 x g, 10 min). Flocs were disrupted by resuspending the biomass in 1.5 ml mineral medium44, followed by rigorous horizontal shaking in the presence of a ¾″ glass sphere for 10 minutes. Subsequently, biomass was washed twice in mineral medium and resuspended in 150 ml mineral medium containing no N-source. 12 ml biomass per incubation was transferred to 30 ml serum bottles and ammonium, nitrite or urea was added (200 µM final concentration). To test for anammox activity and denitrification 15NH4+ was used and the headspace analysed for labelled dinitrogen gas production as described above. For inhibition experiments ATU was added to a final concentration of 100 µM and biomass was preincubated for 10 min before substrate addition. Bottles were sealed with rubber stoppers and 10 ml air was added to the headspace to ensure slight overpressure. Incubations were performed at room temperature in the dark with mild agitation (50 rpm). At each time point, 0.5 ml sample was taken and stored at -20°C for further analysis.
Analytical methods
Ammonium was determined colorimetrically using a modified orthophatal-dialdehyde assay45 (detection limit 10 µM) and nitrite (≥5 µM) by the sulfanilamide reaction46. Nitrate (≥1 µM) was measured by converting it into nitric oxide at 95°C using a saturated solution of VCl3 in HCl. Nitric oxide was than measured using a Nitric Oxide Analyser (NOA280i, GE Analytical Instruments, Manchester, UK). To determine the total organic carbon (TOC) concentration of the medium, medium was first acidified to remove inorganic carbon. After 6.5x dilution with ultrapure water, samples were measured using a TOC-L CPH/CPN analyser (Shimadzu, Duisburg, Germany).
Fluorescence in situ hybridization (FISH)
For FISH analysis, samples from the reactor were fixed with 4% (v/v) paraformaldehyde (PFA), followed by hybridization with fluorescently labelled oligonucleotides as described elsewhere47. FISH probes used in this study (Extended Data Table 2) were 5’ labelled with the dyes FLUOS (5(6)-carboxyfluorescein-N-hydroxysuccinimide ester), Cy3 or Cy5 (Thermo Electron Corporation, Ulm, Germany). After hybridization, slides were air-dried and embedded in Vectashield (Vector Laboratories Inc., Burlingame, CA). Probe-conferred fluorescence was recorded on an Zeiss Axioplan 2 (Carl Zeiss AG, Oberkochen, Germany) equipped with a HBO 100 light source and specific filter sets for the detection of FLUOS, Cy3 and Cy5, a Leica TCS SP2 AOBS (Leica Microsystems, Wetzlar, Germany) or a Zeiss LSM510 META (Carl Zeiss AG) confocal laser scanning microscope (CLSM), both equipped with one argon ion (450–514nm) and two helium neon lasers (543 and 633 nm). Images were recorded with 63x glycerol or oil immersion objectives at a resolution of 1024 x 1024 pixels and 8 bit depth.
For quantifying relative biovolume fractions, PFA-fixed reactor biomass was hybridized to probes Ntspa662, Amx820 and EUB338mix (Extended Data Table 2) as described above. Subsequently, 45 image pairs were recorded at random fields of view using the Leica TCS SP2 AOBS CLSM. The images were imported into the image analysis software daime48 and evaluated as described elsewhere49.
AMO-labelling
Washed and disrupted (see above) biomass was incubated for 30 minutes at room temperature with freshly prepared fluorescein thiocarbamoylpropargylamine (FTCP, synthesized as described elsewhere16). After incubation, cells were harvested, washed, PFA-fixed and hybridized to specific FISH probes as described above.
FISH combined with microautoradiography (FISH-MAR)
FISH-MAR experiments were performed as described before50. 150 ml biomass was taken from the reactor and flocs were disrupted as described above. After harvesting and washing, the biomass was resuspended in mineral medium44 and transferred to serum bottles. Ammonium or nitrite was added to a final concentration of 500 µM. As controls, incubations with ammonium and ATU (100 µM), without nitrogen source and a dead control (PFA-fixed biomass) were performed. 10 µCi [14C]-labelled bicarbonate were added to all samples, bottles were sealed with rubber stoppers and incubated at room temperature in the dark for 18 h. After incubation, the biomass was harvested by centrifugation (20,000 x g, 10 min), PFA-fixed and FISH was performed on coverslips as described above. Hybridized samples were dipped in preheated (48°C) and diluted (1:1 with deionised water) film emulsion (Ilford Nuclear Emulsion K5, Harman Technology, UK). After overnight drying at room temperature, samples were exposed for 6 days at 4°C and developed in Kodak D19 developer as described before50. Images were recorded on a Zeiss LSM510 META CLSM as detailed above. To correct for the different levels of unspecific silver grain deposition in the incubations, the degree of silver grain formation in areas without biomass was compared to the amount of silver grains above biomass flocs. Only cell clusters which showed grain deposition clearly above background level were considered positive.
Phylogenetic analyses
16S rRNA sequences with nucleotide identities ≥98% and amoA sequences with identities ≥70%, to the respective sequences of Nitrospira sp.1 or sp.2 were identified in the NCBI nr database by BLAST27. 16S rRNA sequences were imported into the SILVA51 small subunit ribosomal RNA database release 119, amoA sequences in a custom-made database containing a reference set of amoA and pmoA sequences. nxrA sequences were imported in a custom-made database containing all published sequences from Nitrospira, Nitrospina and anammox organisms. Sequence alignments for all datasets where generated and manually refined using ARB 5.552. Bayesian interference trees were calculated using MrBayes 3.2.353 until a standard deviation <0.01 was reached. For 16S rRNA analyses the GTR substitution model and a 50% conservation filter resulting in 1463 valid alignment positions were used. amoA genes were translated into their amino acid sequence and a 10% conservation filter resulting in 264 alignment positions in combination with the WAG substitution model were used for tree calculation. nxrA trees were calculated from nucleic acid sequences with the GTR substitution model and without conservation filter, resulting in 2660 distinct alignment patterns. For all trees 50% majority rule consensus trees are shown.
Database mining
All 7306 public shotgun metagenomes and metatranscriptomes available in MG-RAST54 were searched for the presence of the diagnostic amoA gene. Datasets were downloaded and searched against a small set of characteristic amoA sequences using DIAMOND55 with the default settings. The resulting 44993 hits were filtered using a BLAST score ratio56 of the initial alignment score versus the alignment score against the NCBI nr.
Extended Data
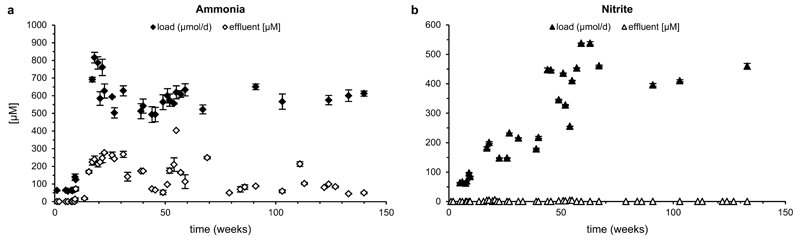
Extended Data Figure 1. Ammonium and nitrite conversion by the enrichment culture.
Inorganic nitrogen load of the enrichment culture per 24 h cycle (filled symbols) and effluent concentrations (open symbols) for (a) ammonium (diamonds) and (b) nitrite (triangles). Effluent nitrite concentrations were below the detection limit (<5 µM) at all time points. Data points represent the mean of three technical replicates, error bars the standard deviations of these triplicates. Nitrate concentration in the medium varied between 0.5 and 2.0 mM and total organic carbon (TOC) content between 1.30 and 1.44 ppm, which was due to medium preparation with water obtained directly from the RAS.
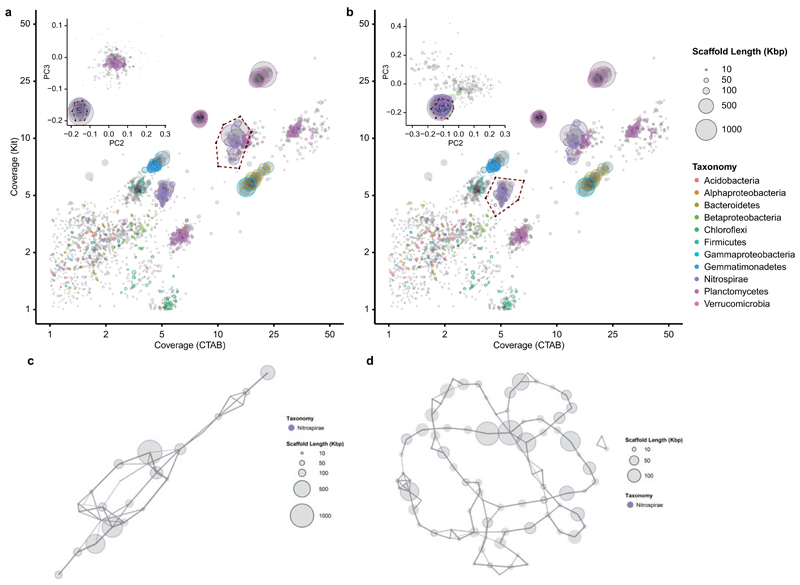
Extended Data Figure 2. Metagenome binning.
Extraction of the Nitrospira sp.1 (a) and sp.2 (b) genome sequences from the metagenome using differential coverage binning. Each circle represents a metagenomic scaffold, with size proportional to scaffold length; the plots contain a total of 47584 scaffolds. The inlay of each figure shows the secondary binning based on tetranucleotide frequencies, with a total of 331 (a) and 281 (b) scaffolds included. Taxonomic classification is indicated by colour; a total of 3158 essential marker genes were detected. The extracted bins are enclosed by a dashed line. Genome contaminations were excluded by generating linkage maps of the final bins of sp.1 (c, 25 scaffolds) and sp.2 (d, 86 scaffolds) using mate-pair sequencing data.
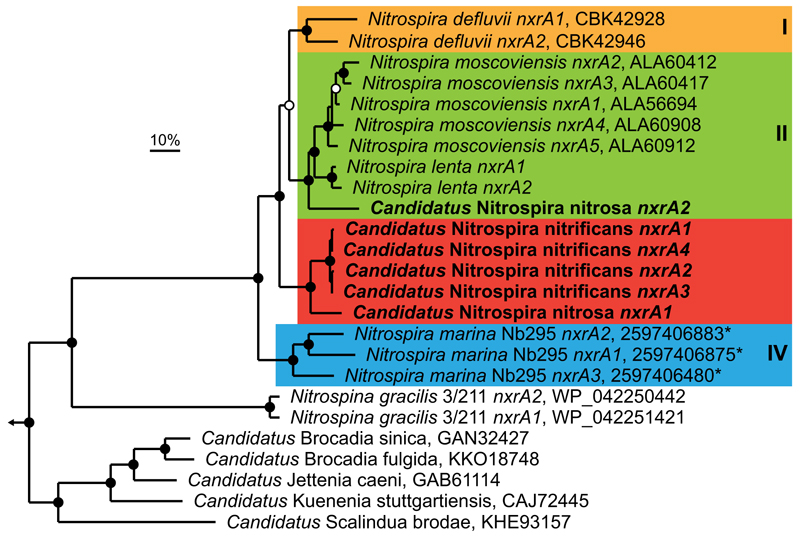
Extended Data Figure 3. Phylogenetic analysis of NXR.
Bayesian interference tree (s.d.=0.0099) showing the affiliation of the Nitrospira sp.1 and sp.2 nxrA sequences in comparison to other genome-sequenced Nitrospira, Nitrospina and anammox bacteria. Posterior probabilities ≥70% and ≥90% are indicated by open and filled circles, respectively. NCBI protein accession numbers for all publicly available sequences are indicated, numbers with an asterisk are IMG gene IDs. The described Nitrospira sublineages are indicated by coloured boxes and roman numbers. The scale bar represents 10% sequence divergence. Note the different affiliation of the “Ca. N. nitrosa” (sp.1) nxrA sequences. The tree contains 25 sequences from 12 species, belonging to 3 different phyla. Sequences from closely related bacterial putative nitrate reductases were used as outgroup (n=4); the outgroup position is indicated by the arrow.
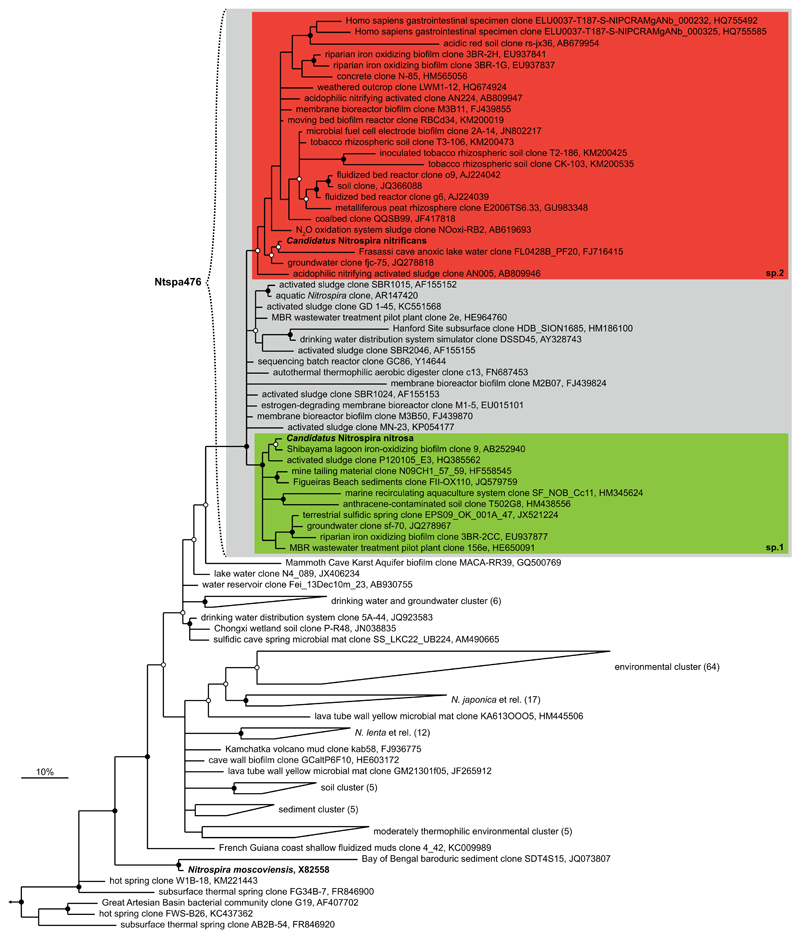
Extended Data Figure 4. 16S rRNA-based phylogenetic analysis.
Bayesian interference tree (s.d.=0.0098) showing the affiliation of the Nitrospira sp.1 and sp.2 16S rRNA sequences within Nitrospira sublineage II. Posterior probabilities ≥70% and ≥90% are indicated by open and filled circles, respectively. The strongly supported sequence group containing the novel Nitrospira spp. catalysing complete nitrification is shaded in grey, the two subgroups containing Nitrospira sp.1 and sp.2 (in bold) are highlighted by green and red boxes, respectively. N. moscoviensis is depicted in bold for comparison. The curly bracket indicates the target group of the newly designed FISH probe Ntspa476 (see Extended Data Table 2). Scale bar indicates 10% sequence divergence. The tree contains a total of 181 sequences; the size of sequence groups is indicated in brackets. Sequences from members of Nitrospira sublineages I and IV were used as outgroup (n=24); the outgroup position is indicated by the arrow.
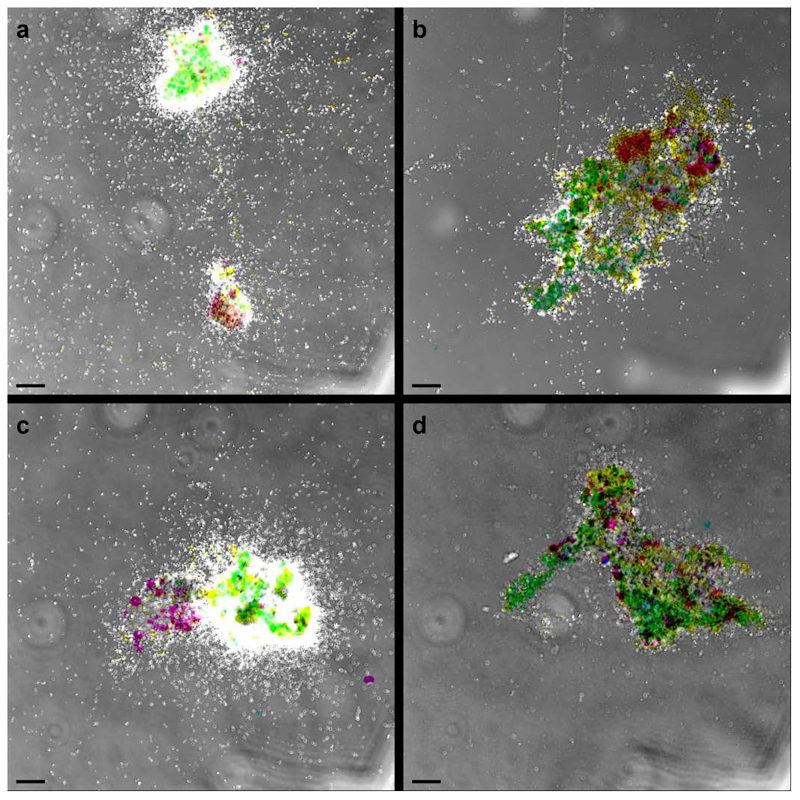
Extended Data Figure 5. Control experiments of AMO-labelling.
a, Cells incubated with the fluorescent dye FTCP (green) were stained by FISH using probes specific for Nitrospira (Ntspa662, red) and all bacteria (EUB338mix, blue). A small cell cluster was stained by FTCP and targeted by both probes (resulting in a white overlay signal), while all other bacteria (in blue) were not or only slightly stained by FTCP. The green signal is due to autofluorescence and unspecific FTCP binding to the floc matrix. b, Anammox cells (Amx820, blue) showed minor staining by FTCP (green), but to a much lesser degree than Nitrospira (Ntspa662, red; yellow overlay). c and d, Positive controls: (c) ammonium oxidizing bacteria (Nso1225 and Nso190, red) in an aerobic enrichment culture and (d) a Nitrosomonas europaea pure culture (NEU, red, and EUB338mix, blue) were stained by FTCP (resulting in yellow and white overlays, respectively). e and f, Negative controls: (e) canonical Nitrospira in an aerobic enrichment culture (Ntspa662, blue) and (f) a Nitrospira moscoviensis pure culture (Ntspa662, red, and EUB338mix, blue; magenta overlay) did not show any labelling with FTCP (green). The two bright green structures in (c) and the bright pink signal in (e) are due to autofluorescence. Images are representative of two (a and b) or one (c to f) individual experiments, with three technical replicates each. Scale bars in all panels represent 10 μm.
Extended Data Figure 6. Batch incubations with nitrite, urea and without substrate.
a and b, Nitrite (triangles) oxidation by the enrichment culture to nitrate (squares) (a) in the absence and (b) in the presence of ATU. The ammonia (diamonds) in (b) presumably stems from biomass decay and is not oxidized due to ATU inhibition. c, Urea conversion to ammonium (diamonds) and subsequent oxidation to nitrate (squares). d, No-substrate control; minor amounts of ammonium (diamonds) presumably stem from mineralisation of degrading biomass, leading subsequently to nitrate (squares) formation. Symbols in all plots represent averages of three independent incubations; ammonium was determined in single measurements, nitrite and nitrate in duplicate (a and b) or triplicate (c and d). Error bars represent standard deviations of three biological replicates.
Extended Data Figure 7. FISH-MAR.
FISH with probes for all bacteria (EUB338mix, blue), and probes specific for Nitrospira (Ntspa662, red; resulting in magenta) and anammox bacteria (Amx820, green; resulting in cyan). a, Ammonia-dependent carbon fixation. Only Nitrospira cells were active, as indicated by silver grain deposition. Note the inactive anammox cells on the left side of the smaller floc, co-localizing with highly active Nitrospira cells on the right side of the same floc. b, Inhibition of ammonia-dependent carbon fixation by ATU. c, Nitrite-dependent carbon fixation. Only Nitrospira cells incorporated 14CO2. d, No-substrate control. Images are representative of two individual experiments, with two technical replicates each. Scale bars in all panels represent 10 μm.
Extended Data Table 1.
General genomic characteristics of Nitrospira sp.1 and sp.2.
| Ca. N. nitrosa (sp.1) | Ca. N. nitrificans (sp.2) | |||
|---|---|---|---|---|
| Bin | initial | final | initial | final |
| Genome size (bp) | 4413075 | 4422398 | 4088547 | 4117083 |
| Contigs | 25 | 15 | 86 | 36 |
| Largest contig (bp) | 1073143 | 1804237 | 335390 | 475968 |
| N50 | 659693 | 727365 | 103850 | 174194 |
| # Ns per 100 Kbp | 355 | 0 | 420 | 0 |
| Completeness* | 99% (97%) | >99% (97%) | >95% (97%) | >95% (97%) |
| Contamination* | 0% (2.3%) | 0% (2.3%) | <1% (2.8%) | <1% (2.7%) |
| Coverage (CTAB)† | -‡ | 13.0 | -‡ | 4.9 |
| Coverage (Kit)† | -‡ | 10.0 | -‡ | 5.0 |
| Average G+C content | -‡ | 54.8 | -‡ | 56.6 |
| Number of coding sequences (CDS) | -‡ | 4309 | -‡ | 4502 |
| rRNA operons | -‡ | 1 | -‡ | 1 |
| tRNAs | -‡ | 46 | -‡ | 43 |
Extended Data Table 2.
FISH probe specifications.
| Probe name | Probe full name* | Sequence (5'-3') | Binding position† | FA%‡ | Specificity | Ref. |
|---|---|---|---|---|---|---|
| Amx820 | S-*-Amx-0820-a-A-22 | AAA ACC CCT CTA CTT AGT GCC C | 820 - 841 | 40 | Genera Brocadia, Kuenenia | 57 |
| Arch915 | S-D-Arch-0915-a-A-20 | GTG CTC CCC CGC CAA TTC CT | 915 - 934 | nd§ | Domain Archaea | 58 |
| Eub338║ | S-D-Bact-0338-a-A-18 | GCT GCC TCC CGT AGG AGT | 338 - 355 | 0 - 50 | Domain Bacteria | 59 |
| Eub338II║ | S-*-Bact-0338-b-A-18 | GCA GCC ACC CGT AGG TGT | 338 - 355 | 0 - 50 | Order Planctomycetales | 60 |
| Eub338lll║ | S-*-Bact-0338-c-A-18 | GCT GCC ACC CGT AGG TGT | 338 - 355 | 0 - 50 | Order Verrucomicrobiales | 60 |
| NEU | S-*-Nsm-0651-a-A-18 | CCC CTC TGC TGC ACT CTA | 653 - 670 | 40 | Nitrosomonas spp. | 61 |
| cNEU | - | TTC CAT CCC CCT CTG CCG | 659 - 676 | - | Competitor to NEU | 61 |
| NmV | S-S-Nmob-0174-a-A-18 | TCC TCA GAG ACT ACG CGG | 174 - 191 | 35 | Nitrosococcus mobilis lineage¶ | 62 |
| Nso190 | S-F-bAOB-0189-a-A-19 | CGA TCC CCT GCT TTT CTC C | 189 - 207 | 55 | Betaproteobacterial AOB | 63 |
| Nso1225 | S-F-bAOB-1224-a-A-20 | CGC CAT TGT ATT ACG TGT GA | 1224 - 1243 | 35 | Betaproteobacterial AOB | 63 |
| Ntspa662 | S-G-Ntspa-662-a-A-18 | GGA ATT CCG CGC TCC TCT | 662 - 679 | 35 | Genus Nitrospirae | 26 |
| cNtspa662 | - | GGA ATT CCG CTC TCC TCT | 662 - 679 | - | Competitor to Ntspa662 | 26 |
| Ntspa712 | S-*-Ntspa-712-a-A-21 | CGC CTT CGC CAC CGG CCT TCC | 712 - 732 | 35 | Phylum Nitrospirae | 26 |
| cNtspa712 | - | CGC CTT CGC CAC CGG TGT TCC | 712 - 732 | - | Competitor to Ntspa712 | 26 |
| Ntspa476 | S-*-Ntspa-0476-a-A-22 | CTG CAG GTA CCG TCC GAA | 476 - 494 | 20 | Ca. N. nitrosa, Ca. N. nitrificans | This study |
| cNtspa476 | - | CTG GAG GTA CCG TCC GAA | 476 - 494 | - | Competitor to Ntspa476 | This study |
Probe nomenclature according to Alm et al.64
probe binding position according to Escherichia coli 16S rRNA gene numbering.
Percent formamide (v/v) added to the hybridization buffer for optimal hybridization stringency.
Not determined.
Probes where used in a equimolar mixture (EUB338mix) to detect all Bacteria.
Probe targets N. mobilis, which is affiliated with the betaproteobacterial Nitrosomonas lineage and not the gammaproteobacterial genus Nitrosococcus.
Extended Data Table 3.
Metagenome screening for Nitrospira-like amoA sequences.
| Source | Geographical location | Number of hits* | Total reads† | Project name | Dataset ID‡ |
|---|---|---|---|---|---|
| Metagenome projects | |||||
| River sediment | Tongue river, Montana, USA | 1327 | 556,961,375 | Tongue_all_2011 | 4481956-57; 63-72; 74-86 |
| Soil | Houston, Texas, USA | 367 | 321,988,632 | Metagenomic investigation for a ethanol-blended fuel spill | 4519753-58; 60-64, 67-76 |
| Prairie soil | Auburn, Ilinois, USA | 119 | 1,075,325,181 | ISA-SMC-2011 | 4502539-2541; 2543; 2923-2924; 2926; 2928; 2930; 2932-33; 2935 |
| Soil | Ha Noi, Vietnam | 94 | 246,030,284 | Rice field | 4626743-47; 53-54 |
| Garden soil | Xiamen, Fujian, China | 80 | 46,831,964 | 13C labeling Soil Metagenome | 4635904-5 |
| Air | Beijing, China | 68 | 978,592,643 | Bejijing PM2.5 and MP10 Pollutants | 4516402-6403; 6455; 6459; 6637; 6651; 6802-6803; 6910-6911; 6952; 7064 |
| Agricultural soil | Amazonia, Brazil | 63 | 254,067,071 | Amazon Soil metagenome 2_mendes | 4497370-371; 376; 391-393; 395-396; 407-409; 411-412 |
| Marine sediment | Gulf of Mexico, USA | 45 | 2,425,926,864 | BP_Sediments | 4510162-66; 68-69; 71; 73-74 |
| Marine sediment | Plum Island, Massachusetts, USA | 33 | 38,370,475 | IGERT Reverse Ecology 2011-2013 | 4519628; 19632; 19636; 20031 |
| Activated sludge | Stanley wwtp, Hong Kong | 26 | 16,663,946 | Stanley wwtp activated sludge sample | 4467420 |
| Soil | Danum, Malaysia | 24 | 43,344,688 | Effect of logging on soil microbial community in tropics | 4582264-267; 270; 798; 802-803; 805 |
| Agricultural soil | Richmond, Indiana, USA | 23 | 70,731,826 | EarlhamMetagenomes2012 | 4508937-38; 40 |
| wwtp sludge | Malaysia | 23 | 40,000,000 | UTM waste water treatment plant project A | 4544292-4293; 4301; 4307;5190; 6367-6368; 6370; 6373; 6375 |
| Activated sludge | Switzerland | 20 | 9,455,087 | Swiss wwtp metatranscriptomics | 4491800 |
| Soil | Cologne, Germany | 20 | 46,128,675 | Barley | 4529836; 30504 |
| Alkaline travertine water | Voltri Massif, Liguria, Italy | 19 | 42,594,481 | Microbial Biogeography of Serpentinites | 4537864-69 |
| Soil | Iowa, USA | 16 | 790,560,095 | GP corn unassembled | 4539519; 21; 23; 28 |
| Sports facility soil | Norman, Oklahoma, USA | 15 | 10,247,092 | Natural products | 4573678; 83 |
| River water | Minnesota, USA | 14 | 60,806,478 | M3P 2012 | 4534334-35; 45-47 |
| Ochard soil | Haifa, Israël | 13 | 27,265,311 | Revital_aft_qc | 4631721; 24 |
| Freshwater sediment | Rifle, Colorado, USA | 8 | 236,916,472 | Subsurface Rifle | 4465820;4465822 |
| Rizosphere | Golm, Germany | 8 | 32,897,323 | Barley_Rhizomicrobiomics_test_ B_PE | 4524591; 96 |
| Coral reef | Xisha island, China | 8 | 125,160,089 | S_TS_MG | 4580696-698; 702 |
| Soil | Basque Country, Spain | 6 | 3,293,845 | Metal_soil | 4510865 |
| Mine soil | Coto Txomin, Spain | 6 | 196,440 | Pb-Zn-Mine | 4580863; 73 |
| River biofilm | West Virginia, USA | 6 | 3,487,276 | MTR_GeMS_DNA | 4589540-1 |
| Marine sediment | Santa Barbara, California, USA | 5 | 96,123,985 | Scott_Nitro | 4537093 |
| Cave microbial mat | Weebubbie cave, Eucla, Australia | 4 | 475,608 | Weebubbie Cave Slime Curtain Metagenome | 4448052 |
| Groundwater | Tulum, Quintana Roo, Mexico | 4 | 59,482,508 | Yucatan Groundwater | 4536382-3 |
| Grassland soil | Bethel, Minnesota, USA | 4 | 71,162,444 | CedarCreek_minsoil_june2013 | 4541645 |
| Soil | Amazonia, Brazil | 3 | 23,648,292 | Amazon Soil metagenome 1 | 4493652 |
| Freshwater microbial mat | Hot creek, Colorado, USA | 3 | 6,877,377 | International geobiology course 02014 PreTrip | 4549766 |
| River sediment | Athabasca, Alberta, Canada | 2 | 2,524,335 | Athabasca-biofilms | 4482887 |
| Metatranscriptome projects | |||||
| River microbial mat | West Virginia, USA | 523 | 174,983,655 | MTR_GeMS_RNA | 4597881-86 |
| Oil contaminated soil | Varennes, Quebec, Canada | 164 | 234,156,703 | GenoRem_GH_MT | 4512573; 576-580; 586; 590; 592; 608 |
| Soil | Kalamazoo, Michigan, USA | 28 | 205,252,966 | Miscanthus Metatranscriptome | 4554103 |
| Marine sediment | Gulf of Mexico, USA | 9 | 152,742,090 | MG-Core_Metat_Merged | 4508038; 41; 53 |
| Paddy soil | Jiangdu, China | 6 | 52,988,024 | paddy soil | 4553284-5 |
Number of sequences affiliated with the novel AmoA/unusual PmoA sequence group.
Total number of metagenomic reads in the respective MG-RAST project.
For retrieving these datasets from MG-RAST '.3' must be added to the respective dataset ID.
Supplementary Material
Supplementary Information is available in the online version of this paper.
Acknowledgements
We would like to thank Karin Stultiens, Theo van Alen, Jeroen Frank, Peter Klaren, Liesbeth Pierson and Lieke Claessens-Joosten for technical assistance, Tom Spanings for biofilter maintenance and Craig Herbold for the ANI analysis. We are grateful for the use of the confocal microscope from the Microscopic Imaging Centre (MIC, Radboud UMC, Nijmegen) and would like to thank Huib Croes and Marieke Willemse for technical assistance. The LABGeM team and the National Infrastructure “France Genomique” are acknowledged for support within the MicroScope annotation platform. We are thankful to Chris Dupont, Alyson Santoro and Mak Saito for consenting to our use of the Nitrospira marina nxrA sequences, which were produced by the US Department of Energy Joint Genome Institute.
M.A.H.J.v.K was supported by the Technology Foundation STW (grant 13146), D.R.S by the BE-Basic Foundation (grant fs7-002), M.A. and P.H.N. by the Danish Council for Independent Research (DFF 4005-00369), M.S.M.J. by the European Research Council (ERC Advanced Grant projects anammox 232937 and Eco_MoM 339880) and the Dutch Ministry of Education, Culture and Science (Gravitation grant SIAM 024002002), B.K. and S.L. by the Netherlands Organization for Scientific Research (NWO VENI grants 863.11.003 and 863.14.019, respectively). The Radboud Excellence Initiative is acknowledged for support to S.L.
Footnotes
Author Contributions
M.A.H.J.v.K and S.L. executed experiments and analysed data. D.R.S. and M.A. contributed to metagenomic data analyses. M.A. and P.H.N. performed sequencing, assembly and binning. M.A.H.J.v.K., H.J.M.O.d.C., B.K., M.S.M.J. and S.L. planned research. M.A.H.J.v.K., B.K. and S.L. wrote the paper. All authors discussed results and commented on the manuscript.
Author information
Metagenomic data is available in the European Nucleotide Archive (ENA) under accession numbers CZQA01000001-CZQA01000015 and CZPZ01000001-CZPZ01000036. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper. Correspondence and requests for materials should be addressed to S.L. (s.luecker@science.ru.nl).
References
- 1.Winogradsky S. Recherches sur les organismes de la nitrification. Ann Inst Pasteur. 1890;4:213–231. [Google Scholar]
- 2.Vlaeminck SE, Hay AG, Maignien L, Verstraete W. In quest of the nitrogen oxidizing prokaryotes of the early Earth. Environmental Microbiology. 2011;13:283–295. doi: 10.1111/j.1462-2920.2010.02345.x. [DOI] [PubMed] [Google Scholar]
- 3.Costa E, Pérez J, Kreft JU. Why is metabolic labour divided in nitrification? Trends Microbiol. 2006;14:213–219. doi: 10.1016/j.tim.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Crab R, Avnimelech Y, Defoirdt T, Bossier P, Verstraete W. Nitrogen removal techniques in aquaculture for a sustainable production. Aquaculture. 2007;270:1–14. [Google Scholar]
- 5.Albertsen M, et al. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 2013;31:533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- 6.Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lücker S, et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. PNAS. 2010;107:13479–13484. doi: 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rotthauwe JH, Witzel KP, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Könneke M, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 10.El Sheikh AF, Poret-Peterson AT, Klotz MG. Characterization of Two New Genes, amoR and amoD, in the amo Operon of the Marine Ammonia Oxidizer Nitrosococcus oceani ATCC 19707. Appl Environ Microbiol. 2008;74:312–318. doi: 10.1128/AEM.01654-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berube PM, Stahl DA. The Divergent AmoC3 Subunit of Ammonia Monooxygenase Functions as Part of a Stress Response System in Nitrosomonas europaea . J Bacteriol. 2012;194:3448–3456. doi: 10.1128/JB.00133-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klotz MG, Stein LY. Nitrifier genomics and evolution of the nitrogen cycle. FEMS Microbiol Lett. 2008;278:146–156. doi: 10.1111/j.1574-6968.2007.00970.x. [DOI] [PubMed] [Google Scholar]
- 13.Koch H, et al. Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proceedings of the National Academy of Sciences. 2015;112:11371–11376. doi: 10.1073/pnas.1506533112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lupo D, et al. The 1.3-A resolution structure of Nitrosomonas europaea Rh50 and mechanistic implications for NH3 transport by Rhesus family proteins. PNAS. 2007;104:19303–19308. doi: 10.1073/pnas.0706563104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daims H, et al. Complete nitrification by Nitrospira bacteria. Nature. 2015 doi: 10.1038/nature16461. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McTavish H, Fuchs JA, Hooper AB. Sequence of the gene coding for ammonia monooxygenase in Nitrosomonas europaea. J Bacteriol. 1993;175:2436–2444. doi: 10.1128/jb.175.8.2436-2444.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyman MR, Arp DJ. 14C2H2- and 14CO2-labeling studies of the de novo synthesis of polypeptides by Nitrosomonas europaea during recovery from acetylene and light inactivation of ammonia monooxygenase. J Biol Chem. 1992;267:1534–1545. [PubMed] [Google Scholar]
- 18.Taylor AE, et al. Use of aliphatic n-alkynes to discriminate soil nitrification activities of ammonia-oxidizing thaumarchaea and bacteria. Appl Environ Microbiol. 2013;79:6544–6551. doi: 10.1128/AEM.01928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ginestet P, Audic J-M, Urbain V, Block J-C. Estimation of Nitrifying Bacterial Activities by Measuring Oxygen Uptake in the Presence of the Metabolic Inhibitors Allylthiourea and Azide. Appl Environ Microbiol. 1998;64:2266–2268. doi: 10.1128/aem.64.6.2266-2268.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zumft WG. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strous M, Kuenen JG, Jetten MS. Key physiology of anaerobic ammonium oxidation. Appl Environ Microbiol. 1999;65:3248–3250. doi: 10.1128/aem.65.7.3248-3250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alonso-Saez L, et al. Role for urea in nitrification by polar marine Archaea. Proc Natl Acad Sci U S A. 2012;109:17989–17994. doi: 10.1073/pnas.1201914109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burton SAQ, Prosser JI. Autotrophic Ammonia Oxidation at Low pH through Urea Hydrolysis. Appl Environ Microbiol. 2001;67:2952–2957. doi: 10.1128/AEM.67.7.2952-2957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Solomon C, Collier J, Berg G, Glibert P. Role of urea in microbial metabolism in aquatic systems: a biochemical and molecular review. Aquat Microb Ecol. 2010;59:67–88. [Google Scholar]
- 25.Wagner M, Nielsen PH, Loy A, Nielsen JL, Daims H. Linking microbial community structure with function: fluorescence in situ hybridization-microautoradiography and isotope arrays. Curr Opin Biotechnol. 2006;17:83–91. doi: 10.1016/j.copbio.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 26.Daims H, Nielsen JL, Nielsen PH, Schleifer KH, Wagner M. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl Environ Microbiol. 2001;67:5273–5284. doi: 10.1128/AEM.67.11.5273-5284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson M, et al. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoecker K, et al. Cohn's Crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase. PNAS. 2006;103:2363–2367. doi: 10.1073/pnas.0506361103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luesken FA, et al. Diversity and enrichment of nitrite-dependent anaerobic methane oxidizing bacteria from wastewater sludge. Appl Microbiol Biotechnol. 2011;92:845–854. doi: 10.1007/s00253-011-3361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glass EM, Wilkening J, Wilke A, Antonopoulos D, Meyer F. Using the metagenomics RAST server (MG-RAST) for analyzing shotgun metagenomes. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5368. pdb prot5368. [DOI] [PubMed] [Google Scholar]
- 31.Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Albertsen M. mmgenome: Tools for extracting individual genomes from metagenomes. 2015 http://madsalbertsen.github.io/mmgenome/
- 33.Leggett RM, Clavijo BJ, Clissold L, Clark MD, Caccamo M. NextClip: an analysis and read preparation tool for Nextera Long Mate Pair libraries. Bioinformatics. 2014;30:566–568. doi: 10.1093/bioinformatics/btt702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hyatt D, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dupont CL, et al. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. 2012;6:1186–1199. doi: 10.1038/ismej.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.HMMER: biosequence analysis using profile hidden Markov models. 2015 http://hmmer.janelia.org/
- 37.Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prjibelski AD, et al. ExSPAnder: a universal repeat resolver for DNA fragment assembly. Bioinformatics. 2014;30:i293–301. doi: 10.1093/bioinformatics/btu266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vallenet D, et al. MicroScope: an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res. 2013;41:D636–D647. doi: 10.1093/nar/gks1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vallenet D, et al. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 2006;34:53–65. doi: 10.1093/nar/gkj406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spieck E, Lipski A. In: Methods Enzymol. Klotz Martin G, editor. Vol. 486. Academic Press; 2011. pp. 109–130. [DOI] [PubMed] [Google Scholar]
- 45.Taylor S, Ninjoor V, Dowd DM, Tappel AL. Cathepsin B2 measurement by sensitive fluorometric ammonia analysis. Anal Biochem. 1974;60:153–162. doi: 10.1016/0003-2697(74)90140-7. [DOI] [PubMed] [Google Scholar]
- 46.Griess P. Bemerkungen zu der Abhandlung der HH. Weselsky und Benedikt „Ueber einige Azoverbindungen”. Berichte der deutschen chemischen Gesellschaft. 1879;12:426–428. [Google Scholar]
- 47.Daims H, Stoecker K, Wagner M. In: Molecular Microbial Ecology. Osborn AM, Smith CJ, editors. Ch. 9. Taylor & Francis Group; 2005. pp. 213–239. [Google Scholar]
- 48.Daims H, Lücker S, Wagner M. daime, a novel image analysis program for microbial ecology and biofilm research. Environ Microbiol. 2006;8:200–213. doi: 10.1111/j.1462-2920.2005.00880.x. [DOI] [PubMed] [Google Scholar]
- 49.Daims H, Wagner M. Quantification of uncultured microorganisms by fluorescence microscopy and digital image analysis. Appl Microbiol Biotechnol. 2007;75:237–248. doi: 10.1007/s00253-007-0886-z. [DOI] [PubMed] [Google Scholar]
- 50.Lee N, et al. Combination of fluorescent in situ hybridization and microautoradiography - a new tool for structure-function analyses in microbial ecology. Appl Environ Microbiol. 1999;65:1289–1297. doi: 10.1128/aem.65.3.1289-1297.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ludwig W, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 54.Meyer F, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 56.Rasko DA, Myers GS, Ravel J. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics. 2005;6:2. doi: 10.1186/1471-2105-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmid M, et al. Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst Appl Microbiol. 2000;23:93–106. doi: 10.1016/S0723-2020(00)80050-8. [DOI] [PubMed] [Google Scholar]
- 58.Stahl DA, Amann R. In: Nucleic Acid Techniques in Bacterial Systematics. Stackebrandt E, Goodfellow M, editors. John Wiley and Sons Ltd; 1991. [Google Scholar]
- 59.Amann RI, et al. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: Development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- 61.Wagner M, Rath G, Amann R, Koops H-P, Schleifer K-H. In situ identification of ammonia-oxidizing bacteria. Syst Appl Microbiol. 1995;18:251–264. [Google Scholar]
- 62.Juretschko S, et al. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol. 1998;64:3042–3051. doi: 10.1128/aem.64.8.3042-3051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mobarry BK, Wagner M, Urbain V, Rittmann BE, Stahl DA. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl Environ Microbiol. 1996;62:2156–2162. doi: 10.1128/aem.62.6.2156-2162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alm EW, Oerther DB, Larsen N, Stahl DA, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.