Abstract
Endosomes are transportation nodes, mediating selective transport of soluble and transmembrane cargos to and from the Golgi apparatus, plasma membrane and lysosomes. As endosomes mature to become multivesicular bodies (MVBs), ESCRTs (Endosomal Sorting Complexes Required for Transport) selectively incorporate transmembrane cargos into vesicles that bud into the endosome lumen. Luminal vesicles and their cargoes are targeted for destruction when MVBs fuse with lysosomes. Common assays of endosomal luminal targeting, including fluorescence microscopy and monitoring of proteolytic cargo maturation, possess significant limitations. We present a quantitative assay system called LUCID (LUCiferase reporter of Intraluminal Deposition) that monitors exposure of chimeric luciferase-cargo reporters to cytosol. Luciferase-chimera signal increases when sorting to the endosome lumen is disrupted, and silencing of signal from the chimera depends upon luminal delivery of the reporter rather than proteolytic degradation. The system presents several advantages, including rapidity, microscale operation, and a high degree of reproducibility that enables detection of subtle phenotypic differences. Luciferase reporters provide linear signal over an extremely broad dynamic range, allowing analysis of reporter traffic even at anemic levels of expression. Further, LUCID reports transport kinetics when applied to inducible trafficking reporters.
Keywords: endocytosis, endosome, multivesicular body, cargo, vacuole, lysosome, luciferase, vesicle, ESCRT
INTRODUCTION
Endosomes are membrane-bound intracellular compartments that arise from homotypic fusion of endocytic vesicles that originate at the plasma membrane. The process by which endosomes mature and eventually fuse with degradative lysosomal compartments involves several biochemical and morphological changes (1), including an increase in compartment size due to incoming vesicular traffic, acidification of the endosome lumen, exchange of early endosome molecular markers including for late endosome markers (e.g., Rab 5 for Rab7), and the formation of vesicles that bud into the endosome lumen. Endosomes with luminal vesicles are also commonly called multivesicular bodies (MVBs).
Several intracellular vesicular transport pathways intersect at endosomes. While many lipid and transmembrane protein cargos from the plasma membrane or trans-Golgi network are recycled back to their membrane of origin, others are targeted for destruction via selective delivery into luminal vesicles. MVB luminal vesicles and their protein cargos are degraded when endosomes fuse with lysosomes (metazoans) or vacuoles (fungi). Efficient delivery of transmembrane proteins and other substrates to the MVB lumen is an essential process regulating metabolism and cell signaling, such as silencing of activated growth factor receptors (2, 3). Recent studies also highlight the importance of selective transport to the MVB lumen in protein quality control, such as targeting of misfolded transmembrane proteins to the MVB lumen to maintain membrane integrity (4, 5).
The delivery of transmembrane cargoes to endosome luminal vesicles depends on covalent attachment of a ubiquitin sorting signal to the cargo's cytosolic domain and the coordinated action of four protein complexes called ESCRTs (Endosomal Protein Complexes Required for Transport) [reviewed by (6–8)]. ESCRT-0, -I and -II bind ubiquitin tags cargos and cluster these cargos in a membrane domain that will invaginate to form a luminal vesicle, while ESCRT-III assembles in a spiral pattern that helps to sequester cargos and promote membrane curvature toward the lumen. ESCRT-III also provides a scaffold for the recruitment of deubiquitination machinery (Bro1-Doa4) that removes Ub tags from cargos prior to incorporation into vesicles. The ESCRT-III scaffold and associated sorting proteins are recycled to the cytosol by the AAA ATPase Vps4, which is in turn regulated by two protein complexes, Vta1-Vps60 and Did2-Ist1. Disruption of any core component of the ESCRT machinery typically blocks MVB vesicle biogenesis and cargo sorting, leading instead to formation of aberrant, flattened endosomes called 'Class E compartments' (Raymond et al., 1992) that lack luminal membranes and accumulate cargos at the limiting membrane. Disruption of individual Vps4 regulators, such as Vta1 or Did2, results in partial sorting and MVB biogenesis defects.
Enzyme-coupled reporter assays in the yeast system have been especially powerful in elucidating cellular transport pathways, enabling large scale chemical and genetic screens as well as rigorous, quantitative phenotypic comparisons. Notable examples include chimeric reporters of the enzyme sucrase/invertase (9), which enabled early interrogations of cargo transport through the secretory (10) and vacuolar sorting (11) pathways, and chimeras of alkaline phosphatase used for monitoring of both selective and non-selective autophagic transport to the lysosomal vacuole (12, 13). Investigations of MVB luminal targeting have long been handicapped by the absence of enzyme-coupled biochemical assays.
Here, we validate and generalize an enzyme-coupled MVB transport assay called LUCID (LUCiferase reporter of Intraluminal Deposition) that monitors exposure of chimeric luciferase-cargo reporters to cytosol. Luciferase enzymes catalyze the oxidation of their chemical substrates, known generally as luciferins, emitting light as a readily measured byproduct. Luciferases are among the best-characterized and most commonly used reporter proteins owing to their lack of cytotoxicity and rapid, reliable measurement compatible with high throughput screening (reviewed in (14, 15). The LUCID system presents several advantages, including rapidity, microscale operation, and a high degree of reproducibility that enables detection and discrimination of subtle cargo transport defects. When applied to inducible trafficking reporters, LUCID reports transport kinetics. In contrast to alternative assays of MVB transport, silencing of signal from the luciferase chimera depends upon luminal delivery of the reporter rather than proteolytic degradation of the chimera at the lysosome/vacuole. LUCID is quick, quantitative, and offers a unique capacity to monitor cargo delivery to the MVB lumen.
RESULTS
LUCID reports degrees of phenotypic defect in characterized MVB mutants
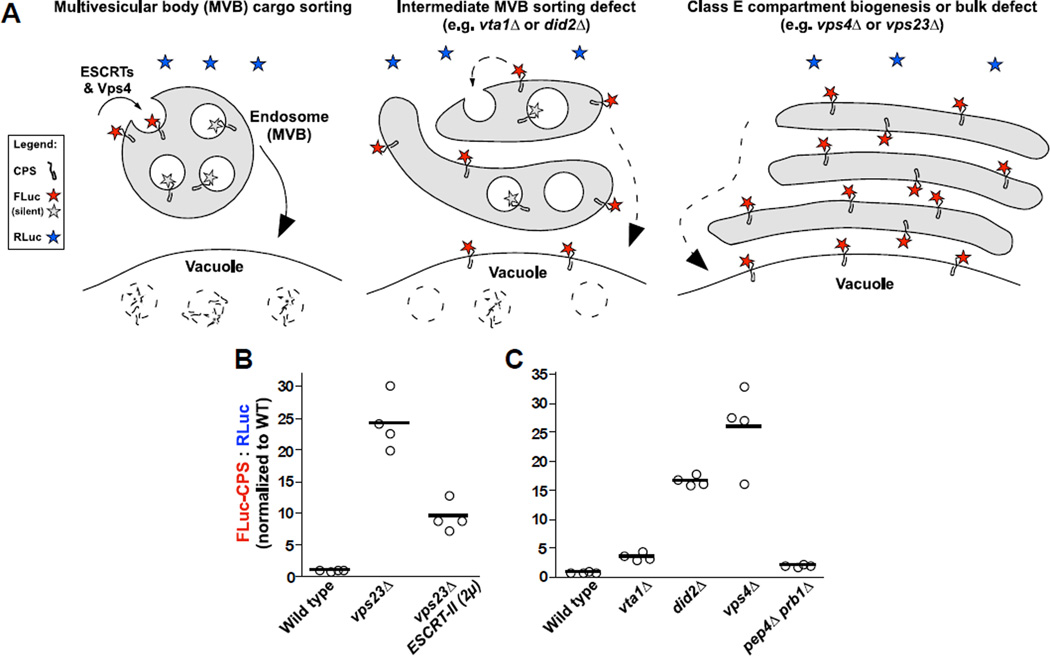
Our initial implementation of LUCID (16) used Sna3, a model cargo for ubiquitin-independent, constitutive transport to endosome luminal vesicles, (17). FLuc was fused to the cytosolic, carboxy-terminal domain of Sna3. This topology exposes the FLuc reporter to the cytoplasm until the ESCRT system sequesters Sna3-FLuc into intraluminal vesicles. The fusion protein is destroyed when MVBs fuse with proteolytic vacuoles (see diagram in Figure 1A). Blocks or kinetic delays in delivering cargo-FLuc to the endosome lumen result in accumulation of active FLuc signal; this signal is ratiometrically normalized to co-expressed soluble RLuc. We now generalize the LUCID technique using another gold-standard MVB cargo, the ubiquitin-dependent vacuole protease carboxypeptidase S (CPS). Using this CPS-FLuc reporter we tested a variety of conditions known to disrupt or rescue MVB cargo sorting. Deletion of the ESCRT-I subunit Vps23 causes formation of aberrant class E compartments and a complete defect in luminal cargo sorting. However, overexpression of the entire ESCRT-II complex can partially suppress the sorting defect in vps23Δ mutant cells (16, 18). We found that FLuc-CPS luminescence was elevated more than 20-fold in vps23Δ cells (Figure 1B). This defect was suppressed by (≥50%) in vps23Δ cells that overproduced ESCRT-II. Several studies have explored incomplete phenotypic defects that arise from misregulation of the Vps4 ATPase and its client complex, ESCRT-III (19–22). Both the Sna3-FLuc (16) and FLuc-CPS (Figure 1C) probes indicate a spectrum of LUCID signal intensity, wild type < vta1Δ < did2Δ < vps4Δ, consistent with known relative intensities of defects in these mutants. Thus, LUCID discriminates among different degrees of missorting defects at MVBs.
Figure 1. LUCID measures degrees of MVB cargo missorting upon disruption and rescue of ESCRT functions.
(A) Model of LUCID assay function in wild type and mutant cells. Firefly luciferase (FLuc) is attached to the cytosolic domain of a transmembrane MVB cargo (carboxypeptidase S, CPS). Luciferase assays detect FLuc signal on pre- or mis-sorted cargo while the tag faces the cytosol (shown in red), but FLuc signal is silenced (shown as gray) when cargo is packaged into luminal vesicles. Vesicles and luciferase-tagged cargos are destroyed when MVBs fuse with the vacuole/lysosome. Inefficient delivery of cargo to the endosome lumen results in increased FLuc signal. Signal from FLuc-cargo is normalized versus soluble Renilla luciferase (RLuc, shown as blue) expressed from the same plasmid. (B and C) LUCID assays of FLuc-CPS sorting in wild type and mutant cells. Bars each represent the mean of 4 biological replicates. (B) Overexpression of ESCRT-II partially rescues cargo missorting defect resulting from disruption of ESCRT-I. (C) Mutants with escalating degrees of disrupted ESCRT-III dynamics and cargo missorting show corresponding escalation in FLuc-CPS signal.
LUCID vector design
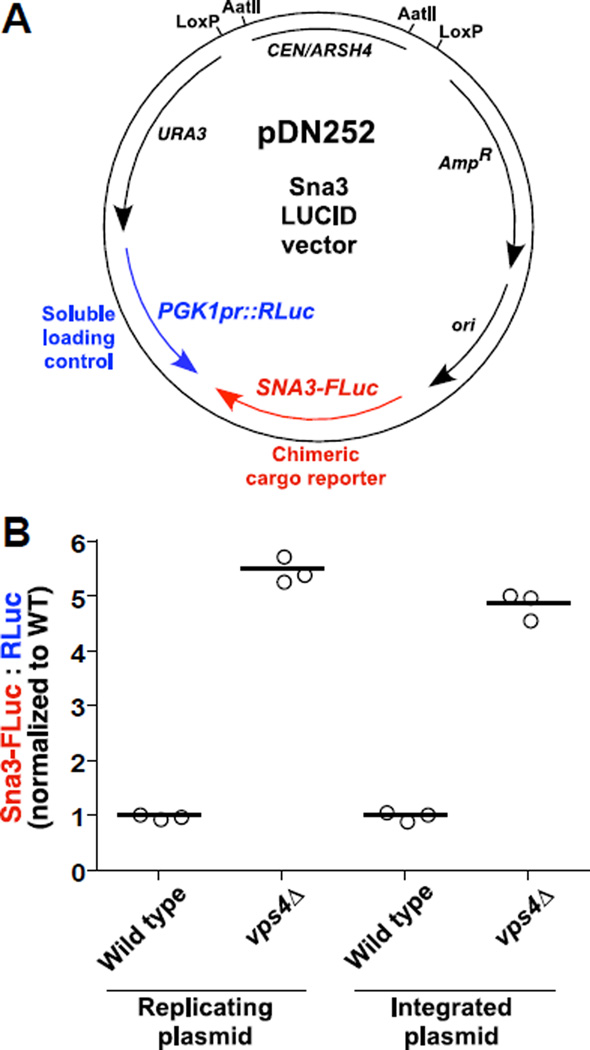
Ratiometric luciferase assay systems generally place the RLuc and FLuc reporters on separate plasmid backbones, and previous work in yeast suggested that chromosomal integration of luciferase reporters improves experimental reproducibility (e.g., (23)). To minimize variance in relative copy numbers of reporter genes, we designed LUCID vectors to encode both reporters on the same low-copy plasmid (Figure 2A). RLuc expression was driven by the constitutive PGK1 promoter since PGK1 transcript levels are stable across a wide range of growth conditions and PGK1 transcripts are commonly used as baseline controls in gene expression studies (24). Anticipating that chromosomal integration of LUCID vectors might reduce experimental variability, we designed LUCID vectors to include parallel LoxP sequences flanking the CEN-ARS replication locus, enabling rapid conversion of the episomal plasmid into an integrating vector (16). Even so, we found that co-expression of RLuc and FLuc-cargo from a replicating plasmid resulted in consistent ratiometric profiles across biological replicates, with reproducibility comparable to that achieved using chromosomal integrants (Figure 2B).
Figure 2. LUCID vector design.
(A) Representative LUCID vector showing low copy plasmid for co-expression of FLuc-tagged cargo under native promoter (SNA3-FLuc) and soluble RLuc driven by the constitutive PGK1 promoter (PGK1pr::RLuc). ori, bacterial origin of replication. URA3, yeast selectable marker. CEN/ARSH4, yeast centrosomal sequence and origin of replication. AmpR, bacterial ampicillin resistance gene. Parallel LoxP sequences or AatII cut sites enable removal of the CEN/ARSH4 locus, converting the plasmid from replicating to integrating. (B) Demonstration of LUCID assay using either replicating or chromosome-integrated plasmids. Bars each represent the mean of 3 biological replicates.
Optimizations of LUCID reproducibility and dynamic range
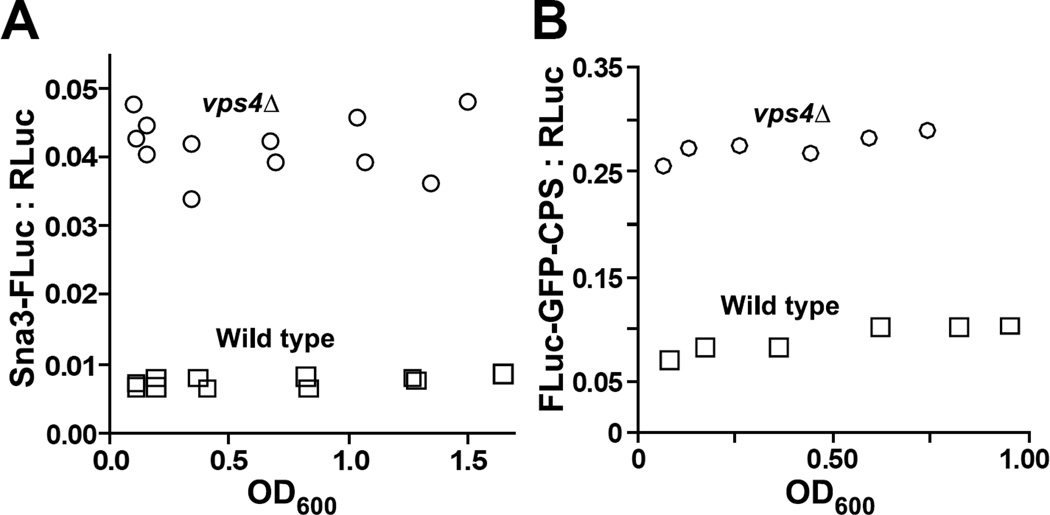
Results from many assays of cellular function vary depending on cellular growth conditions or culture density. Yeast modulate many cellular functions as cultures transition from maximal growth in log phase and to late log phase (OD600 > 1.0), including increased protease expression (25) and autophagy (26). In principle, ratiometric comparison of signals from FLuc-cargo and a co-expressed, soluble RLuc allows immediate correction for variations in 1) plasmid copy number; 2) number or mass of cells analyzed; 3) rates of global protein synthesis and non-specific protein turnover. We tested whether culture density influences LUCID signals from two reporters, Sna3-FLuc (Figure 3A) and FLuc-GFP-CPS (Figure 3B), by growing diluted cultures in parallel to early-, mid-, and late-log phase before collecting the cells for lysis and analysis. In both wild type and MVB-defective cells (vps4Δ), LUCID signals exhibited only minor variation across the range of densities tested. While we would encourage LUCID users to analyze approximately similar masses of cells harvested from cultures grown to similar densities, these data suggest that the assay tolerates differences in these variables, still yielding consistent results. This robustness may make LUCID especially suitable for high-throughput screening.
Figure 3. LUCID operates consistently over a broad range of cell culture densities and cell masses analyzed.
LUCID analysis of cell cultures expressing Sna3-FLuc (A) or FLuc-GFP-CPS (B) diluted to various densities and grown in parallel to indicated culture densities.
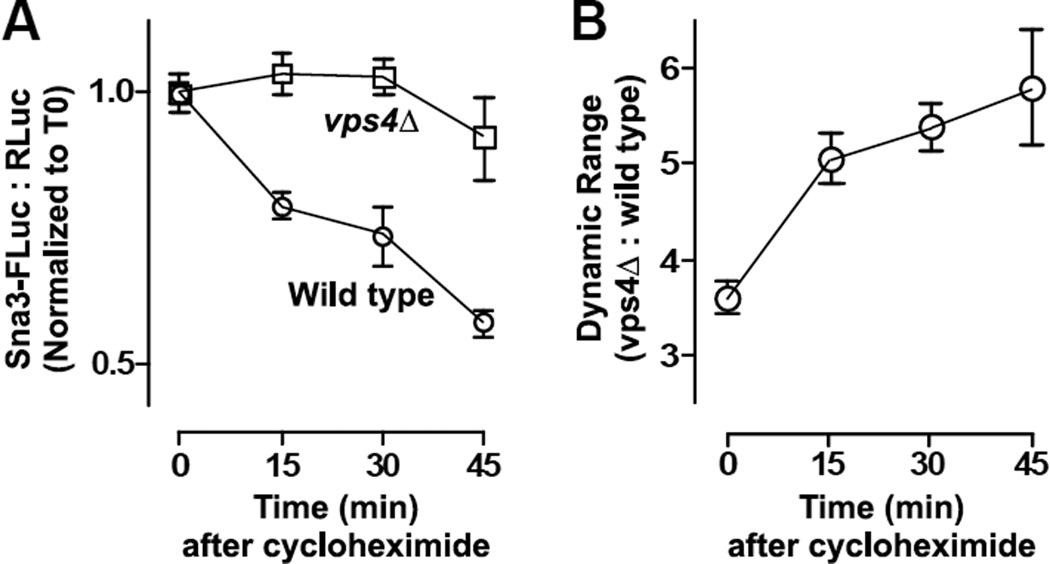
We might expect an intrinsic background from FLuc-cargo traversing the biosynthetic pathway because, unlike fluorescent reporters, FLuc and RLuc enzymes become catalytically active immediately (or nearly so) following synthesis. Thus, we predicted that LUCID would produce a larger dynamic range to discriminate between transport-competent and -defective cells if de novo protein synthesis was halted prior to lysis, as cells with functioning transport would flush recently-synthesized reporter into the MVB. Using the Sna3-FLuc LUCID probe, we subjected cultures of wild type and mutant (vps4Δ) cells to cycloheximide chases of various lengths prior to analysis (Figure 4A). Cycloheximide treatment of wild type cells resulted in the progressive loss of Sna3-FLuc signal relative to RLuc. This specific reduction in wild type LUCID signal over time increases the assay dynamic range when compared to identically treated vps4Δ cells (Figure 4B). We have standardized most LUCID protocols to include a cycloheximide chase of 15 to 20 min prior to collecting cultures for analysis.
Figure 4. Cycloheximide chase increases the dynamic range of the LUCID assay.
(A) Cycloheximide chase reveals loss of LUCID signal in wild type cells but not cells defective in MVB luminal transport (vps4Δ). Points are mean ± S.D. of 3 replicate samples. (B) Dynamic range of vps4Δ samples versus the wild type mean at indicated timepoints. Points represent mean ± S.D. of 3 replicate samples.
The dynamic range between wild type and trafficking-defective mutants for a given LUCID reporter is characteristic of the unique cargo tagged and promoter driving expression of the construct. Of cargos we have examined (including a standard cycloheximide chase), CPS under its native promoter produces the highest typical dynamic range (15- to 25-fold) (Figure 1; (27)), while the pheromone receptor Ste3 has the lowest (about 2-fold) (28).
LUCID reports cargo packaging into the MVB, not cargo destruction at the lysosome
Transmembrane proteins delivered to luminal vesicles are destroyed by proteolytic enzymes when endosomes fuse with lysosomes or vacuoles. When designing the LUCID system, we initially hypothesized that luminscent signal from FLuc-cargo chimeras would be terminated by proteolysis by vacuolar luminal hydrolases. Instead, however, the following experiments argue that luminal packaging of the chimera into the MVB is sufficient to quench FLuc activity under our assay conditions.
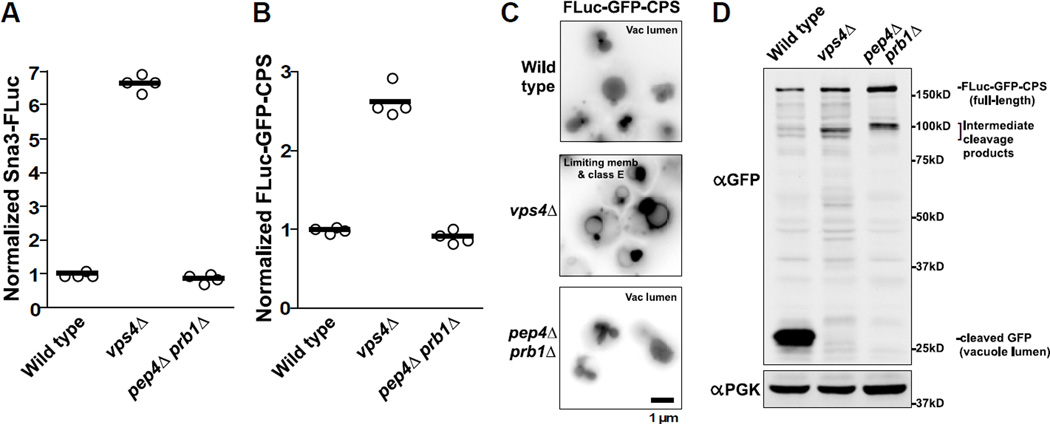
Proteases and other hydrolases are delivered to the vacuole lumen as inactive precursors that are activated upon arrival by Pep4/protease A (25) or Prb1/protease B (29). Double pep4ΔS prb1ΔS mutant cells have severely compromised vacuolar hydrolase activity and degrade luminal contents far more slowly than wild type (30), but have little or no defect in MVB cargo delivery. If luminal hydrolysis were required for termination of the LUCID signal, pep4Δ prb1Δ double mutants would be predicted to have elevated LUCID signals similar to those seen with ESCRT mutants. However, loss of vacuole proteases produced no obvious increase in LUCID signal for FLuc-CPS (Figure 1D), Sna3-FLuc (Figure 5A), nor a CPS construct that included a cytosolic GFP epitope to permit microscopic examination of reporter localization (Figure 5 B). Note that unlike FLuc-CPS, which is expressed under the weak, native CPS1 promoter, the FLuc-GFP-CPS reporter is expressed under a strong, non-native PGK1 promoter to aid visualization. As expected, FLuc-GFP-CPS localized to the vacuole lumen in both wild type and pep4Δ prb1Δ mutant cells (Figure 5 C), while vps4Δ cells mistargeted FLuc-GFP-CPS to the vacuole limiting membrane and peri-vacuolar Class E compartments that accumulate MVB cargo (30, 31). In western blot analyses (Figure 5 D), FLuc-GFP-CPS was efficiently degraded in wild type cells, leaving behind a characteristic ~25 kDa cleavage product that corresponds to soluble GFP. In contrast, vps4Δ and pep4Δ prb1Δ mutants accumulated full-length FLuc-GFP-CPS but not the ~25 kDa soluble GFP band, indicating a failure to proteolytically process the cargo.
Figure 5. Silencing of LUCID reporter correlates with delivery to luminal vesicles, but not with vacuolar proteolysis.
(A–B) LUCID assay of Sna3-FLuc (A) or FLuc-GFP-CPS (B) in cells with no luminal cargo sorting (vps4Δ) or with luminal cargo sorting but with reduced vacuole protease activity (pep4Δ prb1Δ). Bars each represent the mean of 4 biological replicates. (C) Fluorescence microscopy of GFP signal from FLuc-GFP-CPS. Correctly sorted MVB cargo is indicated by GFP signal from the vacuole lumen, whereas missorted cargo instead appears at the vacuole limiting membrane and adjacent puncta called class E compartments. (D) Western blot analysis of yeast cell lysates. Delivery of FLuc-GFP-CPS to the MVB/vacuole lumen results in proteolytic processing in wild type cells, indicated by soluble GFP that resists degradation in the vacuole. Mutants show no soluble GFP, but instead accumulate full-length and high MW partial degradation products.
As silencing of the luciferase signal from the FLuc-GFP-CPS reporter corresponds with its trafficking itinerary into the MVB lumen but not its proteolytic destruction in the vacuole, our data indicate that LUCID reports the ESCRT-mediated sequestration of FLuc-cargo reporters into the MVB, rather than the subsequent fusion of the MVB with the vacuole or subsequent proteolysis of the LUCID reporter within the vacuole lumen. This conclusion is also consistent with our previous examinations of yeast lacking Rab7/Ypt7, the Rab GTPase that regulates endosome/MVB fusion with the vacuole/lysosome: LUCID indicated yeast lacking Ypt7 accumulated no more Sna3-FLuc signal than wild type cells (16), despite the fact that these cells hyperaccumulate morphologically correct MVBs due to delayed fusion kinetics with the vacuole (32).
Kinetics of MVB targeting
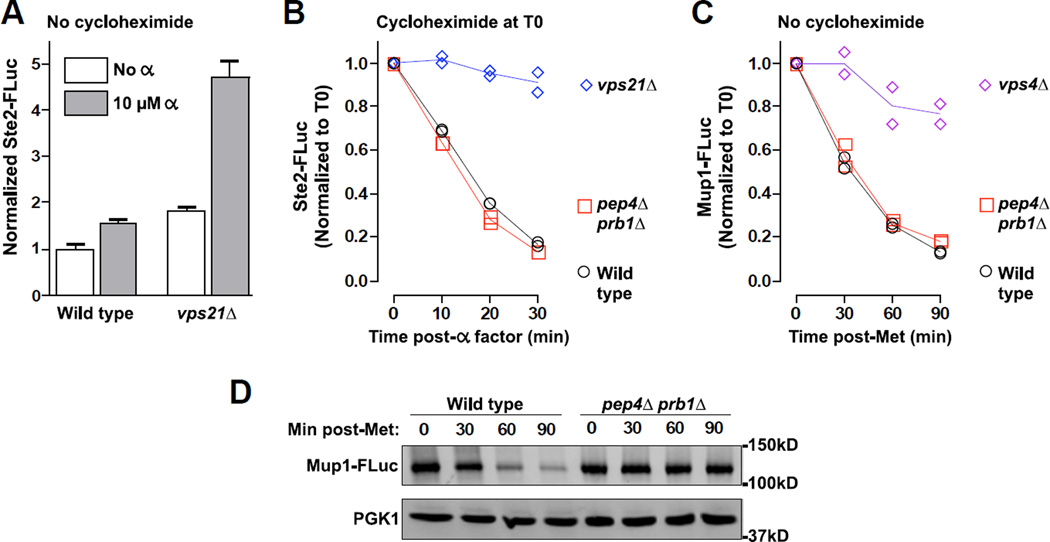
The G-protein-coupled pheromone receptor Ste2 is a standard, classic cargo used to follow signal-mediated endocytosis and MVB targeting in yeast. When Ste2 at the cell surface binds its pheromone ligand, alpha-factor, Ste2 is rapidly ubiquitinated, endocytosed, and shuttled to the vacuole lumen via the MVB (33). We constructed a Ste2-FLuc LUCID reporter and monitored the kinetics of its endocytic downregulation by performing a cycloheximide chase and applying alpha factor (Figure 6 A). While cells impaired in early endocytosis (vps21Δ) showed little reduction in Ste2-FLuc LUCID signal, wild type cells down-regulated Ste2-FLuc with rapid kinetics that closely approximated kinetics reported using alternative assays (33). We underscore the importance of arresting translation with cycloheximide when monitoring down-regulation of Ste2 after alpha-factor stimulation. Because the Ste2-FLuc reporter is expressed under control of the native STE2 promoter, it is upregulated upon activation of the mating signaling pathway. Despite rapid downregulation of Ste2-FLuc in transport-competent cells, alpha-factor-stimulated synthesis of new Ste2-FLuc produced a net increase in reporter signal when no cycloheximide was added (Figure 6 B). As expected, cells defective in endocytic silencing of Ste2-FLuc (vps21Δ) displayed a considerably larger increase in reporter levels 1 h after alpha-factor addition.
Figure 6. LUCID reports kinetics of transport to the endosome lumen.
(A–B) Effect of alpha-factor stimulation on Ste2-FLuc levels with and without simultaneous cycloheximide treatment. (A) Without cycloheximide treatment, pheromone treatment stimulates increased synthesis of Ste2-FLuc that is counterbalanced by endocytic downregulation of Ste2-FLuc in wild type cells but not vps21Δ mutants. Bars represent mean ± S.D. of 4 replicate samples each. (B) Cycloheximide treatment allows monitoring of pheromone-stimulated endocytic transport to the MVB lumen. Each of 2 replicate samples is plotted with lines connecting the mean value at each timepoint. (C) Timecourse of endocytic downregulation of the Mup1 methionine permease upon addition of 1 mM methionine to culture media. Each of 2 replicate samples is plotted with lines connecting the mean value at each timepoint. (D) Western blot analysis of whole cell lysates from indicated samples and timepoints in panel C. Note that silencing of signal from Ste2-FLuc (B) or Mup1-FLuc (C) upon induced transport to the endosome lumen persists without efficient vacuolar proteolysis (pep4Δ prb1Δ).
We further examined LUCID performance by monitoring the kinetics of endocytic transport to the MVB using a LUCID reporter for the high-affinity methionine (Met) permease Mup1 (28). Mup1 accumulates at the plasma membrane as cells are starved of Met. In response to a pulse of exogenous Met, Mup1 is ubiquitinated, endocytosed, and sorted into the MVB en route to the vacuole (34, 35). Met-triggered Mup1 transport to the vacuole was previously studied using fluorescent-tagged Mup1 reporters (28, 35–37). Addition of 1 mM Met resulted in loss of Mup1-FLuc signal in wild type cells (Figure 6 C) (28), in contrast to a mutant that cannot sequester Mup1-FLuc into the MVB lumen, such as vps4Δ (Figure 6 C), or mutants with impaired early endocytic transport, such as cells lacking Vps21/Rab5 or its primary Rab5 GEF, Vps9 (28).
Importantly, for induced transport of both Ste2-FLuc (Figure 6 A) and Mup1-FLuc (Figure 6 C) we observed no obvious delay in the kinetics of Mup1-FLuc signal loss when comparing pep4Δ prb1Δ vacuolar protease-deficient cells and protease-replete wild type cells, again demonstrating that LUCID reports luminal targeting rather than reporter destruction in the vacuole lumen. Indeed, Western blot analysis of whole cell lysates derived from cells that had been subjected to a 1 mM Met pulse confirmed that while wild type cells and pep4Δ prb1Δ mutants have indistinguishable kinetics of Mup1-FLuc signal reduction in LUCID experiments (Figure 6 C), pep4Δ prb1Δ mutants fail to proteolytically degrade Mup1-FLuc (Figure 6 D).
Considerations on the native firefly luciferase peroxisomal import signal
Native Firefly luciferase contains a canonical carboxyterminal SKL (Ser-Lys-Leu) peroxisomal targeting signal (PTS) that directs soluble FLuc into the peroxisome lumen (38, 39). In order to discern whether the FLuc PTS could interfere with trafficking kinetics of transmembrane proteins on an endolysosomal itinerary, we compared the performance of LUCID probes either with ('SKL') or without ('ΔKL') the FLuc carboxyterminal PTS (Figure S1). Monitoring the constitutively trafficked cargo Sna3-FLuc, we observed no difference in steady state levels of SKL and ΔKL cargos in wild type cells nor vps4Δ mutants lacking MVB transport (Figure S1 A). Similarly, pheromone-induced transport kinetics of Ste2-FLuc reporters with or without the PTS were essentially indistinguishable (Figure S1 B). The observed lack of effect of the FLuc PTS in the LUCID assay is hardly surprising given that cargo reporter constructs are not soluble, but are all transmembrane proteins with no peroxisomal transport itineraries. Nonetheless we recommend that operators of the LUCID system check for the presence or absence of the FLuc PTS in their cloning templates and take steps to omit it if necessary.
DISCUSSION
Two previous studies have used the Sna3-FLuc LUCID probe to discriminate intermediate degrees of MVB cargo missorting, one exploring functional significance of Vps4 interactions with each ESCRT-III subunit (22), the other identifying Rab GTPase signaling requirements for MVB maturation and luminal transport (16). Both studies correlated cargo sorting defects measured via LUCID with defects in cargo sorting and endosome morphology as observed by fluorescence microscopy or electron microscopy. Here, we show that LUCID is robust across a range of conditions, can be generalized to numerous substrates, and specifically reports cargo sequestration into the MVB, rather than proteolytic processing or destruction of cargo at the vacuole. We conclude that LUCID is a fast, sensitive and reproducible assay to gauge the fidelity of endosome maturation and cargo transport, and that the assay is particularly useful in analyses of intermediate or subtle defects.
Why might FLuc signal go silent upon luminal delivery? It has long been observed that FLuc does not tolerate luminal targeting in the secretory pathway (14, 15). Also, luminal vesicles at endosomes might sequester FLuc away from its luciferin substrate. Luminal vesicles are thought to arise from detergent-resistent membrane microdomains (40). While the composition of the commercially-supplied passive lysis buffer (PLB) used in the LUCID assay is proprietary, its bulk detergent component is ≤ 0.2% BigCHAP (N,N-Bis(3-D-gluconamido-propyl)cholamide, a non-ionic detergent. All attempts so far to expose or liberate sequestered or inactivated FLuc signal from pep4Δ prb1Δ luminal vesicles, including freeze-thaw cycles and addition of ionic- and non-ionic detergents to lysis buffer, have yielded either no improvement or substantial decreases in FLuc signal (data not shown). Hence, we conclude that under the standard assay conditions presented here, signal from FLuc-cargo reporters is effectively quenched upon its delivery into luminal vesicles.
Luciferase enzymes rank among the most popular reporters for cellular analysis (15), and several features the LUCID assay (summarized in Table II) persuade us that luciferase reporters are a valuable addition to the membrane trafficking toolkit. LUCID is rapid and reproducible when performed at microscale. For constitutively expressed/trafficked cargos (e.g. Sna3 or CPS), plasmid transformation and culturing of yeast cells represent the major investments of time; collecting cells, lysis and luminometry can typically be performed in 15 min to 1h, depending on the number of samples. Quantitative results from numerous replicate samples are returned quickly without the many extra steps of sample processing and analysis that introduce error in common alternatives like Western blotting and radiolabeling. As LUCID monitors delivery of luciferase-tagged cargo to luminal vesicles, it offers a versatile complement to alternative assays that monitor MVB luminal transport indirectly as their reporters lose or change signal upon arrival at the acidic, proteolytic vacuolar lysosome.
Table II.
Summary of LUCID features
|
In comparison to GFP- and pHluorin-based assays of cargo transport, two features of LUCID warrant particular mention. Unlike luciferases, GFP/pHluorin reporters require post-translational maturation (tens of minutes) to emit signal. As a consequence, freshly synthesized GFP-tagged cargo evades optical detection, obscuring some kinetic features of trafficking. For example, yeast lacking Vps21/Rab5 showed no obvious mislocalization of Sna3 tagged with conventional GFP (16, 32), but LUCID analysis using Sna3-Fluc revealed cargo accumulation in these cells (16, 28). Fluorescence imaging and quantification of GFP/pHluorin signals is often complicated, especially in yeast, by cellular autofluorescent background, and pHluorin quenching depends on the presence of a trasmembrane ΔpH gradient. Consequently, pHluorin reporters of MVB packaging have the potential to be biased by genetic lesions, phamacological treatments, or physiological conditions (e.g., replicative aging; Henderson and Gottschling, 2014) that alter endolysosoml pH. LUCID, like most luciferase-based assays, has negligible background and is highly sensitive, obviating a common need to overexpress cargo reporters for analysis—even when expression under a native promoter is anemic. A relative advantage of GFP/pHluorin assays is their ability to perform in vivo and single cell analyses including, in the case of pHluorin-based sensors, flow cytometry. In contrast, luciferase assays in yeast require cell permeabilization or lysis due to impermeability of yeast cells to luciferase substrates (41).
We anticipate several future improvements and elaborations to the LUCID method. While LUCID performs reliably using proprietary commercial reagents, we aspire to optimize the system using entirely open-source, non-proprietary reagents. We note that non-proprietary buffer systems for sequential analysis of FLuc and RLuc reporters have been reported (42, 43), but these systems still employ a proprietary passive lysis buffer for cell extracts (42) or describe lysis buffer deficient in reducing luciferase-independent autoluminescence (43). We envision several advantages to combining LUCID probes with pHluorin reporters for endocytic traffic (36), which might enable resolution of luminal cargo delivery at MVBs and the subsequent fusion of MVBs with the acidic vacuole/lysosome. We also anticipate testing the performance of LUCID probes in nematodes and mammalian cells. LUCID assays in mammalian cell culture should be especially appealing from both a chemico-genomic screening standpoint as well as for the likely compatibility of LUCID with compartment-specific bioluminescence resonance energy transfer-acceptors (44). Such reporters could in principle monitor passage of luciferase-tagged cargos through specific subcellular locales as they progress through the endolysosomal system.
MATERIALS & METHODS
Strain and plasmid construction
Yeast strains and plasmids used in this study are summarized in Table I. Strains DNY527 and DNY528 were constructed by cutting within the URA3 locus of non-replicating LUCID vector pDN249 to integrate at ura3-2 chromosomal loci of SEY6210 and MBY3, respectively. Knockout of VPS21 in SEY6211 and subsequent removal of the URA3 auxotrophic marker via Cre-recombinase to create DNY439 was performed as described (45).
Table I.
Plasmids and strains used in this study
| Plasmid/strain | Genotype/description | Source |
|---|---|---|
| Plasmids | ||
| pDN616 | URA3 LoxP::CEN/ARSH4::LoxP AmpR | (16) |
| pDN606 | URA3 LoxP AmpR | (16) |
| pDN252 | PGK1pr::RLuc SNA3pr::SNA3-FLuc (pDN616) | (16) |
| pDN263 | PGK1pr::RLuc SNA3pr::sna3Y109A-FLuc (pDN616) | (16) |
| pDN249 | PGK1pr::RLuc SNA3pr::SNA3-FLuc (pDN606) | This study |
| pDN360 | PGK1pr::RLuc SNA3pr::SNA3-FLucΔKL (pDN616) | This study |
| pDN268 | PGK1pr::RLuc MUP1pr::MUP1-FLuc (pDN616) | (28) |
| pDN278 | PGK1pr::RLuc CPS1pr::FLuc-CPS1 (pDN616) | (27) |
| pDN312 | PGK1pr::RLuc STE3pr::STE3-FLuc (pDN616) | (28) |
| pDN320 | CDC28pr::RLuc PGK1pr::FLuc-GFP-CPS1 (pDN616) | This study |
| pDN326 | PGK1pr::RLuc STE2pr::STE2-FLuc (pDN616) | This study |
| pDN371 | PGK1pr::RLuc STE2pr::STE2-FLucΔKL (pDN616) | This study |
| pDN614 | TRP1 LoxP::CEN/ARSH4::LoxP AmpR | (27) |
| pDN336 | PGK1pr::RLuc CPS1pr::FLuc-CPS1 (pDN614) | (27) |
| pMB175 | LEU2 AmpR 2µVPS22 VPS25 VPS36 (pRS425) | (18) |
| S. cerevisiae | ||
| SEY6210 | MATα leu2-3,112 ura3-52 his3-200 trp1-901 lys2-801 suc2-9 | (11) |
| SEY6210a | MATa leu2-3,112 ura3-52 his3-200 trp1-901 lys2-801 suc2-9 | (11) |
| DNY439 | SEY6211 vps21Δ0 | This study |
| BHY10 | SEY6210 CPY-Invertase::LEU2 (pBHY11) | (48) |
| MBY3 | SEY6210 vps4Δ::TRP1 | (49) |
| GOY23 | SEY6210 prb1Δ::LEU2 pep4Δ::LEU2 | (50) |
| GOY24 | SEY6211 prb1Δ::LEU2 pep4Δ::LEU2 | Gift of G. Odorizzi |
| DNY38 | BHY10 did2Δ::KAN | (51) |
| DNY40 | BHY10 vta1Δ::KAN | (20) |
| DNY527 | SEY6210 RLuc SNA3-FLuc::URA3 (pDN249) | This study |
| DNY528 | MBY3 RLuc SNA3-FLuc::URA3 (pDN249) | This study |
| EEY6-2 | SEY6210 vps23Δ::HIS3 | (52) |
| GOY223 | BHY10 vps9Δ::HIS3 | (16) |
| DNY516 | BHY10 muk1Δ::KAN | (28) |
| DNY206 | BHY10 vps21Δ::KAN | (16) |
Plasmid pDN326 was constructed by gap repair cloning of a PCR product containing the STE2 coding sequence plus 500 bp of upstream promoter into SacII-digested LUCID vector pDN251 (16). pDN320 was constructed via 3 cloning steps: first, sequence overlap extension (SOE) PCR generated 840 bp of PGK1 promoter driving expression of FLuc with an engineered SacII-site preceding the stop codon, which was cloned via gap repair into SmaI/SacII-digested low copy vector pDN616; the GFP-CPS1 gene cassette was PCR amplified from pGO45 (30) and inserted in-frame via gap repair into SacII-digested PGK1pr::FLuc vector; finally, SOE PCR generated 980 bp of CDC28 promoter driving expression of RLuc with a GCY1 terminator, which was cloned into KpnI/HindIII-digested PGK1pr::FLuc-GFP-CPS1 vector via gap repair. CDC28 promoter was selected to drive expression of the RLuc loading control due to the stable expression profile of CDC28 mRNA (46). Mutation of the native peroxisome targeting (SKL) sequence in FLuc was performed by mutating terminal codons encoding Lys and Leu to a pair of Stop codons (ΔKL) via SOE PCR.
Yeast media and growth
All yeast analyzed in luciferase/LUCID experiments were shaken overnight at 30°C in minimal synthetic media with 2% (w/v) glucose, supplemented with all amino acids except those whose absence ensured plasmid selection. Except for experiments in Figures 1 B, 6 C, and 6 D, selective media was further supplemented with 0.15% (w/v) casamino acids. On the day of analysis, cells were diluted in identical media and shaken at 30°C until entering mid- or late- log phase, which typically represented two doublings of the diluted culture density. All LUCID plasmids presented here use either URA3 or TRP1 as their yeast auxotrophic markers. While we have successfully used other markers (e.g. LEU2, (47)), we prefer Trp- and Ura-selections because these are compatible with media supplementation with casamino acids, the addition of which reduces growth rate disparities between wild type cells and many endolysosomal trafficking mutants.
LUCID: lysis, sample analysis & statistics
Log phase cells (OD600 = 0.4 to 0.8) were treated with cycloheximide (50 µg/mL culture) 20 minutes prior to collection of 0.5 to 1.4 mL cell culture by centrifugation at 1000×g. To aid pelleting of yeast from minimal media, 50 µL of YPD (yeast peptone dextrose) media was added to each sample. Cell pellets were resuspended in 500 µL 1X passive lysis buffer (PLB) from a dual luciferase assay system (cat. no. E1910, Promega), except for cells expressing FLuc-CPS driven from the native CPS1 promoter, which were lysed in 200 µL PLB. Glass beads were added to a bed volume of ~100 µL and samples were vortexed at RT 5 to 10 minutes. Five µL aliquots of lysate were loaded into round-bottomed wells of an opaque 96-well plate. Samples were spaced with an empty well between each in order to eliminate contaminating signal from neighboring samples (maximum of 24 samples analyzed per plate). Analysis was performed using a PerkinElmer Victor Light Model 1420 luminometer equipped with automated pump-injectors for delivery of dual luciferase assay reagents. FLuc and RLuc signals were analyzed sequentially for each individual sample with a standard protocol: FLuc analysis entailed injection of 25 µL Luciferase Assay Reagent (Promega), 2 sec of plate shaking, 2 sec delay, then 10 seconds of luminometric monitoring to determine counts per second; quenching of FLuc signal and simultaneous RLuc analysis entailed injection of 25 µL Stop & Glow® reagent (Promega), 2 sec of plate shaking, 2 sec delay, then 10 seconds of luminometric monitoring. The normalized cargo-FLuc signal (Fnorm) in a sample is a ratiometric expression of FLuc signal (F0) versus soluble RLuc signal (R0) after first subtracting the luminometer background signal (B) from each:
Western blotting and microscopy
Protein extracts of yeast cells expressing FLuc-GFP-CPS were made from cells grown to log phase in selective, synthetic media. Cells were pelleted and chilled on ice before resuspension (40 µL per OD600 × mL equivalent) in ice cold Laemmli sample buffer supplemented with 5 mM EDTA, 1 mM PMSF and 1X protease inhibitor cocktail (EDTA-free; Roche). Cell suspensions were boiled, chilled on ice, and vortexed with beads before pelleting insoluble debris via centrifugation. Protein extracts from cells expressing Mup1-FLuc were generated by the same procedure, omitting the boiling step to prevent aggregation of Mup1. 0.2 (FLuc-GFP-CPS) or 0.05 (Mup1-FLuc) OD600 × mL equivalent of cell suspension were resolved by SDS-PAGE and transferred to nitrocellulose. Monoclonal anti-GFP and -PGK1 antibodies were obtained from Roche and Invitrogen, respectively. Polyclonal anti-FLuc was obtained from Sigma-Aldrich. Western blots were imaged using an Odyssey fluorescence scanner (LI-COR Biosciences). Fluorescence microscopy of log phase cells expressing FLuc-GFP-CPS was performed as described (16).
Supplementary Material
Synopsis.
Selective targeting of transmembrane cargo proteins to luminal vesicles at endosomes is a process essential for correct regulation of cell signaling pathways and maintenance of cellular health. We present a convenient, microscale, quantitative assay system that monitors exposure of chimeric luciferase-transmembrane cargoes to cytosol. Luciferase-chimera signal increases when sorting to the endosome lumen is disrupted, and silencing of signal depends upon luminal delivery of the reporter rather than proteolytic degradation.
ACKNOWLEDGMENTS
We thank Vivian Mackay (UW) and members of the Merz Lab for helpful discussions. DPN was supported by an American Cancer Society (ACS) postdoctoral fellowship (PF-10-024-01-CSM). The Merz group's work on endolysosomal traffic is supported by by NIH GM077349.
Abbreviations
- MVB
multivesicular body
- ILV
intraluminal vesicle
- FLuc
firefly luciferase
- RLuc
Renilla luciferase
- LUCID
luciferase reporter of intraluminal deposition
- CPS
carboxypeptidase S
- ESCRT
endosomal sorting complex required for transport
Footnotes
REFERENCES
- 1.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30(17):3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wegner CS, Rodahl LM, Stenmark H. ESCRT proteins and cell signalling. Traffic. 2011;12(10):1291–1297. doi: 10.1111/j.1600-0854.2011.01210.x. [DOI] [PubMed] [Google Scholar]
- 3.Babst M, Odorizzi G. The balance of protein expression and degradation: an ESCRTs point of view. Curr Opin Cell Biol. 2013;25(4):489–494. doi: 10.1016/j.ceb.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Y, Macgurn JA, Liu M, Emr S. The ART-Rsp5 ubiquitin ligase network comprises a plasma membrane quality control system that protects yeast cells from proteotoxic stress. eLife. 2013;2:e00459. doi: 10.7554/eLife.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacGurn JA. Garbage on, garbage off: new insights into plasma membrane protein quality control. Curr Opin Cell Biol. 2014;29:92–98. doi: 10.1016/j.ceb.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henne WM, Stenmark H, Emr SD. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harbor perspectives in biology. 2013;5(9) doi: 10.1101/cshperspect.a016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuh AL, Audhya A. The ESCRT machinery: from the plasma membrane to endosomes and back again. Critical reviews in biochemistry and molecular biology. 2014;49(3):242–261. doi: 10.3109/10409238.2014.881777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat Rev Mol Cell Biol. 2010;11(8):556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darsow T, Odorizzi G, Emr SD. Invertase fusion proteins for analysis of protein trafficking in yeast. Methods Enzymol. 2000;327:95–106. doi: 10.1016/s0076-6879(00)27270-4. [DOI] [PubMed] [Google Scholar]
- 10.Pelham HR, Hardwick KG, Lewis MJ. Sorting of soluble ER proteins in yeast. EMBO J. 1988;7(6):1757–1762. doi: 10.1002/j.1460-2075.1988.tb03005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Molecular and cellular biology. 1988;8(11):4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;210(1):126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- 13.Noda T, Klionsky DJ. The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol. 2008;451:33–42. doi: 10.1016/S0076-6879(08)03203-5. [DOI] [PubMed] [Google Scholar]
- 14.Inouye S. Fusions to imidazopyrazinone-type luciferases and aequorin as reporters. Methods Enzymol. 2000;326:165–174. doi: 10.1016/s0076-6879(00)26054-0. [DOI] [PubMed] [Google Scholar]
- 15.Thorne N, Inglese J, Auld DS. Illuminating insights into firefly luciferase and other bioluminescent reporters used in chemical biology. Chemistry & biology. 2010;17(6):646–657. doi: 10.1016/j.chembiol.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nickerson DP, Russell MR, Lo SY, Chapin HC, Milnes JM, Merz AJ. Termination of isoform-selective Vps21/Rab5 signaling at endolysosomal organelles by Msb3/Gyp3. Traffic. 2012;13(10):1411–1428. doi: 10.1111/j.1600-0854.2012.01390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reggiori F, Pelham HR. Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 2001;20(18):5176–5186. doi: 10.1093/emboj/20.18.5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell. 2002;3(2):283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 19.Azmi IF, Davies BA, Xiao J, Babst M, Xu Z, Katzmann DJ. ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev Cell. 2008;14(1):50–61. doi: 10.1016/j.devcel.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 20.Nickerson DP, West M, Henry R, Odorizzi G. Regulators of Vps4 ATPase activity at endosomes differentially influence the size and rate of formation of intralumenal vesicles. Mol Biol Cell. 2010;21(6):1023–1032. doi: 10.1091/mbc.E09-09-0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rue SM, Mattei S, Saksena S, Emr SD. Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol Biol Cell. 2008;19(2):475–484. doi: 10.1091/mbc.E07-07-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adell MA, Vogel GF, Pakdel M, Muller M, Lindner H, Hess MW, Teis D. Coordinated binding of Vps4 to ESCRT-III drives membrane neck constriction during MVB vesicle formation. J Cell Biol. 2014;205(1):33–49. doi: 10.1083/jcb.201310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riezman H, Munn A, Geli MI, Hicke L. Actin-, myosin- and ubiquitin-dependent endocytosis. Experientia. 1996;52(12):1033–1041. doi: 10.1007/BF01952099. [DOI] [PubMed] [Google Scholar]
- 24.Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, Pak DN, Fields S, Kennedy BK, Kaeberlein M. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133(2):292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones EW. Vacuolar proteases and proteolytic artifacts in Saccharomyces cerevisiae. Methods Enzymol. 2002;351:127–150. doi: 10.1016/s0076-6879(02)51844-9. [DOI] [PubMed] [Google Scholar]
- 26.Cheong H, Klionsky DJ. Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 2008;451:1–26. doi: 10.1016/S0076-6879(08)03201-1. [DOI] [PubMed] [Google Scholar]
- 27.Shideler T, Nickerson DP, Merz AJ, Odorizzi G. Ubiquitin binding by the CUE domain promotes endosomal localization of the Rab5 GEF Vps9. Mol Biol Cell. 2015;26(7):1345–1356. doi: 10.1091/mbc.E14-06-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulsel AL, Merz AJ, Nickerson DP. Vps9 family protein Muk1 is the second Rab5 guanosine nucleotide exchange factor in budding yeast. J Biol Chem. 2013;288(25):18162–18171. doi: 10.1074/jbc.M113.457069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merz AJ, Wickner WT. Resolution of organelle docking and fusion kinetics in a cell-free assay. Proc Natl Acad Sci U S A. 2004;101(32):11548–11553. doi: 10.1073/pnas.0404583101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95(6):847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- 31.Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3(12):1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russell MR, Shideler T, Nickerson DP, West M, Odorizzi G. Class E compartments form in response to ESCRT dysfunction in yeast due to hyperactivity of the Vps21 Rab GTPase. J Cell Sci. 2012;125(Pt 21):5208–5220. doi: 10.1242/jcs.111310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84(2):277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 34.Menant A, Barbey R, Thomas D. Substrate-mediated remodeling of methionine transport by multiple ubiquitin-dependent mechanisms in yeast cells. EMBO J. 2006;25(19):4436–4447. doi: 10.1038/sj.emboj.7601330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell. 2008;15(4):578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 36.Prosser DC, Whitworth K, Wendland B. Quantitative analysis of endocytosis with cytoplasmic pHluorin chimeras. Traffic. 2010;11(9):1141–1150. doi: 10.1111/j.1600-0854.2010.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henne WM, Buchkovich NJ, Zhao Y, Emr SD. The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell. 2012;151(2):356–371. doi: 10.1016/j.cell.2012.08.039. [DOI] [PubMed] [Google Scholar]
- 38.Gould SG, Keller GA, Subramani S. Identification of a peroxisomal targeting signal at the carboxy terminus of firefly luciferase. J Cell Biol. 1987;105(6 Pt 2):2923–2931. doi: 10.1083/jcb.105.6.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keller GA, Gould S, Deluca M, Subramani S. Firefly luciferase is targeted to peroxisomes in mammalian cells. Proc Natl Acad Sci U S A. 1987;84(10):3264–3268. doi: 10.1073/pnas.84.10.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wubbolts R, Leckie RS, Veenhuizen PT, Schwarzmann G, Mobius W, Hoernschemeyer J, Slot JW, Geuze HJ, Stoorvogel W. Proteomic biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J Biol Chem. 2003;278(13):10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- 41.Gehret AU, Bajaj A, Naider F, Dumont ME. Oligomerization of the yeast alpha-factor receptor: implications for dominant negative effects of mutant receptors. J Biol Chem. 2006;281(30):20698–20714. doi: 10.1074/jbc.M513642200. [DOI] [PubMed] [Google Scholar]
- 42.Dyer BW, Ferrer FA, Klinedinst DK, Rodriguez R. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Analytical biochemistry. 2000;282(1):158–161. doi: 10.1006/abio.2000.4605. [DOI] [PubMed] [Google Scholar]
- 43.Hampf M, Gossen M. A protocol for combined Photinus and Renilla luciferase quantification compatible with protein assays. Analytical biochemistry. 2006;356(1):94–99. doi: 10.1016/j.ab.2006.04.046. [DOI] [PubMed] [Google Scholar]
- 44.Lan TH, Liu Q, Li C, Wu G, Lambert NA. Sensitive and high resolution localization and tracking of membrane proteins in live cells with BRET. Traffic. 2012;13(11):1450–1456. doi: 10.1111/j.1600-0854.2012.01401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic acids research. 2002;30(6):e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MacKay VL, Li X, Flory MR, Turcott E, Law GL, Serikawa KA, Xu XL, Lee H, Goodlett DR, Aebersold R, Zhao LP, Morris DR. Gene expression analyzed by high-resolution state array analysis and quantitative proteomics: response of yeast to mating pheromone. Mol Cell Proteomics. 2004;3(5):478–489. doi: 10.1074/mcp.M300129-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Lobingier BT, Nickerson DP, Lo SY, Merz AJ. SM proteins Sly1 and Vps33 co-assemble with Sec17 and SNARE complexes to oppose SNARE disassembly by Sec18. eLife. 2014;3:e02272. doi: 10.7554/eLife.02272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horazdovsky BF, Busch GR, Emr SD. VPS21 encodes a rab5-like GTP binding protein that is required for the sorting of yeast vacuolar proteins. EMBO J. 1994;13(6):1297–1309. doi: 10.1002/j.1460-2075.1994.tb06382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16(8):1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luhtala N, Odorizzi G. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J Cell Biol. 2004;166(5):717–729. doi: 10.1083/jcb.200403139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nickerson DP, West M, Odorizzi G. Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J Cell Biol. 2006;175(5):715–720. doi: 10.1083/jcb.200606113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Babst M, Odorizzi G, Estepa EJ, Emr SD. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic. 2000;1(3):248–258. doi: 10.1034/j.1600-0854.2000.010307.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.