(See the major article by Montealegre, Singh, and Murray on pages 1914–22.)
Enterococci are commensals of the gastrointestinal tracts (GITs) of animals, from simple invertebrates to humans, and likely evolved early in a primitive GIT-type environment [1–3]. This presumption is supported by their ubiquity and their streamlined genomes (an indicator of host adaptation, which results in a number of nutritional needs that must be fulfilled by the host, the diet of the host, or nutrients produced by other members of the GIT consortium) [1, 3, 4]. As the result of widespread antibiotic use over the past 75 years, Enterococcus faecalis and Enterococcus faecium have spawned distinct sublineages that are now leading causes of multidrug-resistant infection and are endemic to many hospitals [5, 6].
Competition among microbes in the gut is intense [7]. The concept of “fitness cost” arises in discussions of microbes acquiring or evolving antibiotic resistance. Enterococci not only have acquired antibiotic resistance, they also have adapted in other ways to the unique hospital niche. The question, then, is whether an approximately 80-year history of adapting to the unusual opportunities for growth within the ecology of the antibiotic treated patient gut (followed by cycles of shedding into the hospital environment, surviving cleaning and disinfection, surviving desiccation, and recolonizing a new patient) has resulted in multidrug-resistant variants that are now ill tuned to compete effectively in a normal human gut. The answer could explain why vancomycin-resistant Enterococcus (VRE) strains and infections are common in hospitals but infection and carriage rare in the community [1, 8]. The article by Montealegre et al in this issue of The Journal of Infectious Diseases [9] takes an important step in addressing this question.
What traits confer fitness in an environment? The most obvious are those that contribute to the abilities to survive and proliferate. The ability to proliferate depends on many factors, among which are acquiring energy to drive cell division and acquiring or making nutrients necessary to assemble the constituents of a new cell (which, for enterococci, means importing about a dozen essential amino acids and vitamins [10]). In nature, nutrient and energy availability fluctuate, so it is equally important to persist when not proliferating, and then to quickly take advantage of growth opportunities when they arise (ie, before a competing microbe does). This adaptability includes not only surviving troughs in nutrition availability but, for hospital-adapted enterococci, surviving in the harsh environment of the hospital between transmissions.
For commensal enterococci of the healthy human GIT, essential nutrients are provided by the diet and by other microbes within the normal human microbiome [10]. The precise source of energy that drives cell division is unknown, but a recent report [6] shows that operons for the use of different carbohydrates are among the greatest genetic differences between commensal E. faecium and those associated with infection. One carbohydrate pathway characteristic of infection-derived E. faecium was shown to provide a colonization advantage in the antibiotic-treated mouse gut [11]. Possession of operons for use of different carbohydrate use is good evidence that, in the guts of antibiotic-treated patients, hospital strains of E. faecium obtain energy from a source different from that for commensals.
The genomes of E. faecalis and E. faecium have been refined over millions (and potentially hundreds of millions [2]) of years of evolution to maximize efficiency. To protect this finely tuned molecular machine, the fidelity of the genomes in stable ecologies is defended by a variety of mechanisms, including CRISPR systems [6, 12, 13]. Recent widespread human and agricultural use of antibiotics, however, has selected for strains that can rapidly evolve their genomes and adapt [6, 13]. Where antibiotics have eliminated competitors from habitats previously inaccessible to enterococci, rapidly evolving E. faecalis and E. faecium variants able to acquire resistance and adapt seized the opportunity and now proliferate in these new man-made ecologies. This selective pressure has led to the outgrowth of enterococcal strains that almost uniformly lack CRISPR defenses [6, 13, 14]. This occurrence opens the door to entry of many types of mobile elements, and, as a result, these strains are filled with plasmids and insertion elements that confer resistance, virulence, and other phenotypes, as well as phages [6, 15, 16]. For example, >25% of the genome of the prototype E. faecalis VRE strain V583 consists of mobile elements [15].
E. faecalis strain V583, the first VRE isolated in the United States [17], represents an extreme example of the maladaptation that can occur in a hospital environment with little competition. V583 is actually inadvertently killed when exposed to normal commensal strains of E. faecalis [18]. It was found that 2 of the many mobile elements harbored by V583 become incompatible when one was induced by the mere presence of a plasmid-free commensal strain [18]. This observation highlights that hospital and commensal strains of enterococci lead separate lives and underscores that maintaining the competing normal flora of patients plays an important role in limiting the evolution and outgrowth of multidrug-resistant enterococci and other bacteria [18, 19].
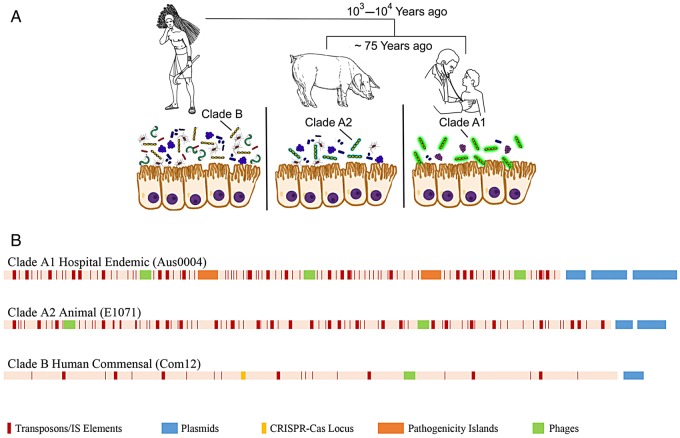
Interestingly, the impact of human intervention on the evolution of enterococci is not limited to the antibiotic era. Close examination of the genomes of E. faecium representing all known clonal lineages found that human commensal strains diverged from animal strains at about the time of the development of agriculture and urbanization—that is, as humans became physically and hygienically insulated from their animals [6]. As for hospital-adapted strains, human commensal E. faecium differ from animal-adapted lineages with respect to the carbohydrate use pathways they possess [6]. In fact, hospital strains (termed clade A1) are very closely related to animal strains, mainly from the guts of agricultural animals (clade A2), and both of these clades differ substantially from human commensal strains (clade B; Figure 1) [6].
Figure 1.
Human impact on the evolution of Enterococcus faecium. A, The species E. faecium split into 2 distinct species in parallel with the urbanization of humans and the spread of animal domestication, and then the animal lineage split again coincident with the application of antibiotics in agriculture and human medicine. Human commensal strains evolved to exist in the complex native consortium of the human gastrointestinal tract. Animal strains largely derive from domesticated animals, to which the majority of antibiotics produced in the United States are applied at low levels for most of the life of the animal, with enterococcal strains adapting to fill opportunities in simplified consortia [20]. Hospital-endemic strains are well adapted to life in the simplified antibiotic-treated patient consortium and to survival outside of the patient in hospitals. B, Linear depiction of representative genomes of E. faecium from clades A1 (Aus004 [16]), A2 (E1071 [6]), and B (Com12 [21]), showing the impact of human intervention on their rapid evolution, which highlights the accretion of mobile genetic elements in clades A1 and A2 that are rare in human commensal strains largely possessing CRISPR/CAS loci.
In a collaboration initiated with the Broad Institute of MIT and Harvard, we have now determined the DNA sequence of >500 enterococcal genomes (available at: https://olive.broadinstitute.org/projects/enterogenome). Several years ago, we [21] and others [22] noted this deep evolutionary divide between commensal and hospital E. faecium in this genome collection. A metric termed “average nucleotide identity” (which compares DNA sequences of genes that 2 strains have in common) was used to quantify the magnitude of this divergence [21]. The evolutionary distance observed hovers around the distance that usually defines separate species [21]; that is, human commensal E. faecium are, or are nearly, a different species from those occurring in the hospital. This observation was further supported and refined in an expanded study that included additional clinical isolates and many animal strains [6].
Montealegre et al [9] now probe the rates at which clade A1 hospital-endemic strains, closely related clade A2 animal strains, and the near species-separated clade B human commensal strains either colonize or are eliminated from the mouse gut as it reestablishes an ecological equilibrium following antibiotic perturbation. The mouse gut differs from the human gut in terms of flora, diet, and specific antimicrobial factors [23] but, nevertheless, has been usefully employed as a tractable model for examining factors that promote colonization by multidrug-resistant enterococci [24, 25]. In the current work, following antibiotic destabilization of the mouse gut, strains of the major E. faecium lineages were introduced and their fates followed. Strains of all E. faecium types were found to be eliminated over the 14-day course of experimentation [9]. No differences among strain types were noted that reached statistical significance. The rate of elimination of enterococci added in this model appeared to follow first-order kinetics, with between 50%–67% of the residual population lost each day over 14 days, irrespective of lineage.
When strains were added in pairs, which allowed the numbers of one type to be normalized against the number of the other type, subtler differences that achieved statistical significance could be observed. In 3 of 4 experiments in which human commensal clade B strains were mixed with hospital clade A1 isolates, by day 14 the clade B strain was more abundant (by 3–4-fold in 2 cases and approximately 100-fold in the third case). In the case in which the greatest difference was observed, the ratios of the 2 strains fluctuated widely, inverting during the course of the experiment. In the fourth case, the hospital isolate was more abundant at each time point measured. This pattern shows that human commensals and infection-derived isolates of E. faecium were eliminated from the ecologically recovering mouse gut at broadly similar rates; the commensals won in 75% of instances when tested in pairs, with differences generally within an order of magnitude of each other. Over two 14-day experiments, and one 7-day experiment, rates of elimination of animal versus human commensal strains were compared [9]. In each case, the human commensal strain was more slowly eliminated, resulting generally in a 10-fold difference after 14 days. When rates of elimination of animal versus hospital isolates were compared, in 2 of 3 cases the hospital strain was eliminated more slowly, and in the third case the animal strain was eliminated more slowly [9]. In aggregate, these findings indicate that, during elimination in this model, there are ecological dynamics occurring of an unknown type and that the rates, although broadly similar, show strain-dependent differences for reasons that will be interesting to determine.
Commensal strains have evolved over eons to compete with other flora in the native GIT ecology, and the mobile elements that confer antibiotic resistance and other adaptations important in the hospital (or agricultural) environment are superfluous in the native gut and, presumably, also in the model murine gut as tested. In nature, competition between enterococci involves many factors that are challenging to simulate in models. For example, human commensal enterococci likely exhibit some degree of host adaptation that includes optimization for (1) coexistence with other human flora and cross-feeding relationships with them; (2) adaptation to the pressures of human variants of innate defenses (eg, timing and magnitude of secretion of human bile conjugates around meals, human-specific antimicrobial factors secreted into the mucosa, and human alleles of iron sequestration functions); (3) composition of carbon sources, including those contained within human mucins and those abundant in the modern human diet; and (4) the continual challenge posed by an influx of new enterococcal strains and phages constantly entering the open digestive system. The effect of subtle selective pressures may not be evident in the span of days or even weeks.
The current article incorporates many ecological complexities by assessing fitness in the mammalian murine system. Why were significant differences between strains only measurable when strains were paired? There are 2 possible explanations: (1) the added strain competes directly with the test strain, depriving it of nutrients or other factors needed for survival and replication (or, as for V583 in the presence of commensals, being killed by it [18]); or (2) the added strain serves as an internal control, which normalizes for animal-to-animal variation, allowing subtler differences to be detected. As there was no deviation from first-order elimination, such as a finding that might suggest growth or establishment during the course of experimentation, the latter seems more likely.
The results generated in this substantial undertaking provide additional support for the collective body of work that indicates that enterococci that are adapted to survival and proliferation in hospital (and agricultural) settings are less well adapted for life outside of those environments. Precisely defining the ecological space that multidrug-resistant hospital-endemic strains of enterococci occupy and how this space differs from that of commensal strains is important for understanding how to manage patients in a way that limits VRE colonization and the subsequent overgrowth that leads to infection [1, 4, 8, 11, 3]. Detecting subtle but potentially important differences may require the incorporation of human flora, a human diet, studies in animals anatomically more similar to humans, and/or studies over longer times to allow for establishment of the added flora. In some humans, the presence of microbes identified as Barnesiella species aids in resistance to VRE colonization [21]. The extent to which there are homologs for such specific antagonisms in the murine model is unclear. Nevertheless, important steps were taken by Montealegre et al in their use of a tractable experimental model [9], which provides insights into how we may impede the spread and proliferation of hospital-endemic multidrug-resistant enterococci, a leading concern in an era when antibiotic resistance is outstripping the introduction of new drugs.
Notes
Acknowledgments. We thank François Lebreton, Daria Van Tyne, Anthony Gaca, Elizabeth Selleck Fiore, Julia Schwartzman, Eric Pamer, Willem van Schaik, and Lou Rice for editorial assistance and advice in preparation of the manuscript.
Financial support. This work was supported by the Public Health Service (grants AI072360 and AI108710) and the Harvard-wide Program on Antibiotic Resistance (grant AI083214).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Lebreton F, Willems RJL, Gilmore MS. Enterococcus Diversity, Origins in Nature, and Gut Colonization. In: Gilmore MS, Clewell DB, Ike Y et al., eds. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet]. Boston: Massachusetts Eye and ear Infirmary, 2014. [PubMed] [Google Scholar]
- 2.Van Tyne D, Gilmore MS. Friend Turned Foe: Evolution of Enterococcal Virulence and Antibiotic Resistance. Annu Rev Microbiol 2014; 68:337–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilmore MS, Lebreton F, van Schaik W. Genomic transition of enterococci from gut commensals to leading cause of multidrug-resistant hospital infection in the antibiotic era. Curr Opin Microbiol 2013; 16:10–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mundy LM, Sahm DF, Gilmore M. Relationships between Enterococcal Virulence and Antimicrobial Resistance. Clin Microbiol Rev 2000; 13:513–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McBride SM, Fischetti VA, LeBlanc DJ, Moellering RC, Gilmore MS. Genetic Diversity among Enterococcus faecalis (Diversity of E. faecalis). PLoS One 2007; 2:p.e582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lebreton F, van Schaik W, McGuire AM et al. Emergence of Epidemic Multidrug-Resistant Enterococcus faecium from Animal and Commensal Strains. MBio 2013; 4:e00534–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sassone-Corsi M, Raffatellu M. No vacancy: how beneficial microbes cooperate with immunity to provide colonization resistance to pathogens. J Immunol 2015; 194:4081–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arias CA, Murray BE. The rise of Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 2012; 10:266–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montealegre MC, Singh KV, Murray BE. Gastrointestinal tract colonization dynamics by different Enterococcus faecium lineages. J Infect Dis 2016; 213:1914–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramsey M, Hartke A, Huycke M. The physiology and metabolism of enterococci. In: Gilmore MS, Clewell DB, Ike Y et al., eds. Enterococci: from commensals to leading causes of drug resistant infection. Boston: Massachusetts Eye and Ear Infirmary, 2014. [Google Scholar]
- 11.Zhang X, Top J, de Been M et al. Identification of a genetic determinant in clinical Enterococcus faecium strains that contributes to intestinal colonization during antibiotic treatment. J Infect Dis 2013; 207:1780–6. [DOI] [PubMed] [Google Scholar]
- 12.Murray BE, Singh KV, Ross RP, Heath JD, Dunny GM, Meinstock GM. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J Bacteriol 1993; 175:5216–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmer KL, Gilmore MS. Multidrug-resistant enterococci lack CRISPR-cas. MBio 2010; 1:e00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hullahalli K, Rodrigues M, Schmidt BD, Li X, Bhardwaj P, Palmer KL. Comparative Analysis of the Orphan CRISPR2 Locus in 242 Enterococcus faecalis Strains. PLoS One 2015; 10:e0138890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulsen IT, Banerjei L, Myers GS et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 2003; 299:2071–4. [DOI] [PubMed] [Google Scholar]
- 16.Lam MM, Seemann T, Bulach DM et al. Comparative analysis of the first complete Enterococcus faecium genome. J Bacteriol 2012; 194:2334–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahm DF, Kissinger J, Gilmore MS et al. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother 1989; 33:1588–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilmore MS, Rauch M, Ramsey M et al. Pheromone killing of multidrug-resistant strains of Enterococcus faecalis V583 by commensal strains. Proc Natl Acad Sci U S A 2015; 112:7273–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ubeda C, Bucci V, Caballero S et al. Intestinal Microbiota Containing Barnesiella Species Cures Vancomycin-Resistant Enterococcus faecium Colonization. Infect Immun 2013; 81:965–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Summary Report On Antimicrobials Sold or Distributed for Use in Food-Producing Animals. Rockville, MD: Center for Veterinary Medicine, Food and Drug Administration, Department of Health and Human Services, 2014. http://www.fda.gov/downloads/ForIndustry/UserFees/AnimalDrugUserFeeActADUFA/UCM338170.pdf. [Google Scholar]
- 21.Palmer KL, Godfrey P, Griss A et al. Comparative genomics of enterococci: variation in Enterococcus faecalis, lineage structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. mBio 2012; 3:30031811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galloway-Peña J, Roh JH, Latorre M, Qin X, Murray BE. Genommic and SNP analyses demonstrate a distant separation of the hospital and community-associated clade of Enterococcus faecium. PLoS One 2012; 7:e30187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen TL, Vieira-Silva S, Liston A, Raes J. How informative is the mouse for human gut microbiota research? Dis Model Mech 2015; 8:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donskey CJ, Chowdhry TK, Hecker MT et al. Effect of Antibiotic Therapy on the Density of Vancomycin-Resistant Enterococci in the Stool of Colonized Patients. N Engl J Med 2000; 343:1925–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stiefel U, Pultz NJ, Helfand MS, Donskey CJ. Increased susceptibility to vancomycin-resistant Enterococcus intestinal colonization persists after completion of anti-anaerobic antibiotic treatment in mice. Infect Control Hosp Epidemiol 2004; 25:373–9. [DOI] [PubMed] [Google Scholar]