Abstract
Sézary syndrome is a leukemic and aggressive form of cutaneous T-cell lymphoma (CTCL) resulting from the malignant transformation of skin-homing central memory CD4 positive T cells. Here we performed whole-exome sequencing of tumor-normal sample pairs from 25 Sézary syndrome and 17 other CTCL patients. These analyses revealed a distinctive pattern of somatic copy number alterations in Sézary syndrome including highly prevalent chromosomal deletions involving the TP53, RB1, PTEN, DNMT3A and CDKN1B tumor suppressors. Mutation analysis identified a broad spectrum of somatic mutations in key genes involved in epigenetic regulation (TET2, CREBBP, MLL2, MLL3, BRD9, SMARCA4 and CHD3) and signaling, including MAPK1, BRAF, CARD11 and PRKG1 mutations driving increased MAPK, NFκB and NFAT activity upon T-cell receptor stimulation. Collectively, our findings provide new insights into the genetics of Sézary syndrome and CTCL and support the development of personalized therapies targeting key oncogenically activated signaling pathways for the treatment of these diseases.
Mycosis fungoides and Sézary syndrome are primary cutaneous T-cell malignancies derived from CD4 positive skin-homing T cells1,2. Mycosis fungoides cases with limited skin involvement have a favorable prognosis, however the median survival for cases with cutaneous tumors and generalized erythroderma is of approximately four years and Sézary syndrome patients fare even worse with survival rates of about two years 2,3.
To investigate the genetic mechanisms of aggressive CTCLs we performed whole exome sequencing of 42 CTCL cases, including 25 Sézary syndrome and 8 mycosis fungoides (Supplementary Table 1). For each sample we generated an average of 115 million reads per sample resulting in an average coverage of 99.91% with over 95.3% of targeted regions showing >30x coverage (Supplementary Table 2).
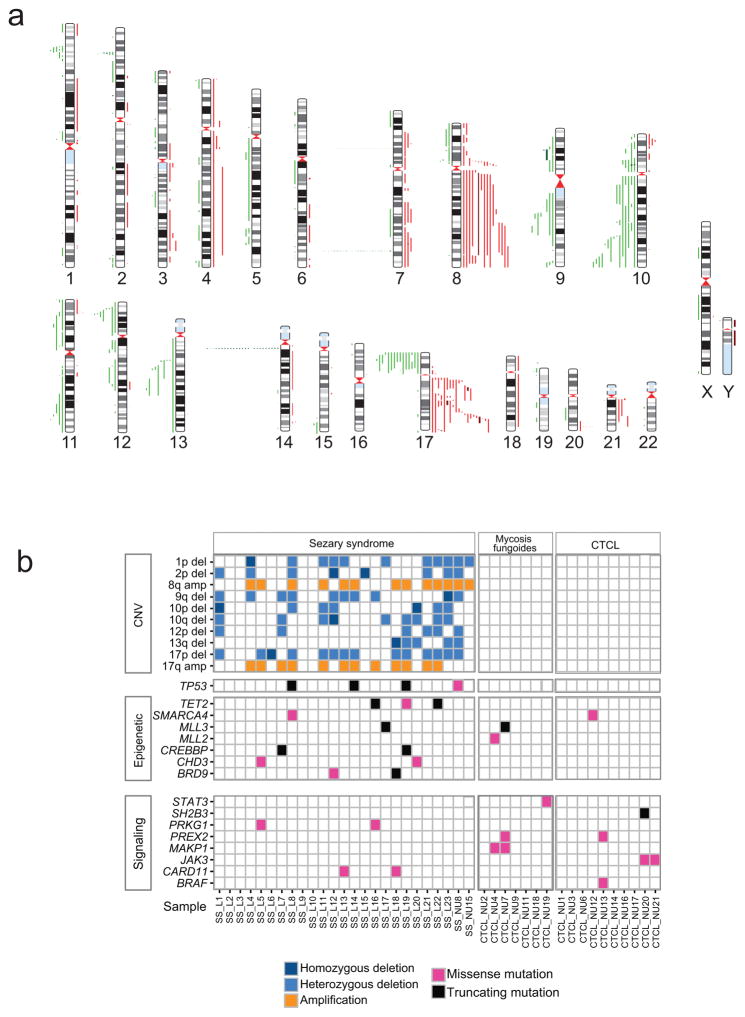
In agreement with previous studies4–8, copy number analysis from exome data identified a median of 21 copy number alterations per sample (range 0–56) in Sézary syndrome with characteristic recurrent gains in chromosome 7 (5/25; 20%), 8q (13/25; 52%) and 17q (2/25; 8%), as well as recurrent deletions involving tumor suppressor genes in 17p13.1 (TP53; 13/25; 52%), 13q14.2 (RB1; 4/25; 16%), 10q23.3 (PTEN; 5/25; 20%) and 12p13.1 (CDKN1B; 5/25; 20%) (Fig. 1 and Supplementary Table 3). Focal chromosome 2p23.3 deletions encompassing the DNMT3A locus were observed in five Sézary patients (5/25; 20%), including two cases with focal homozygous deletion of this epigenetic tumor suppressor gene. Notably, expression analysis of DNMT3A and TP53 showed reduced or complete absence of expression of these tumor suppressors in Sézary samples harboring 2p23.3 and 17p13.1 deletions, respectively (Supplementary Fig. 1). In contrast, non-leukemic mycosis fungoides cases showed lower number of copy number alterations (median 1, range 0–2) (Supplementary Table 4).
Figure 1.
Somatic copy number variants and mutations in Sézary syndrome and CTCL. (a) Human chromosomal ideograms show the areas of genetic gain and loss identified by whole exome sequencing in Sézary syndrome and CTCL samples. Red bars to the right of the chromosomes represent areas of gain with dark red indicating regions of amplification. Green bars to the left of the chromosomes represent areas of heterozygous loss and dark green bars indicate homozygous deletions. (b) Schematic representation of the distribution of recurrent somatic copy number alterations and mutations in epigenetic factors and signaling genes in Sézary syndrome and CTCL.
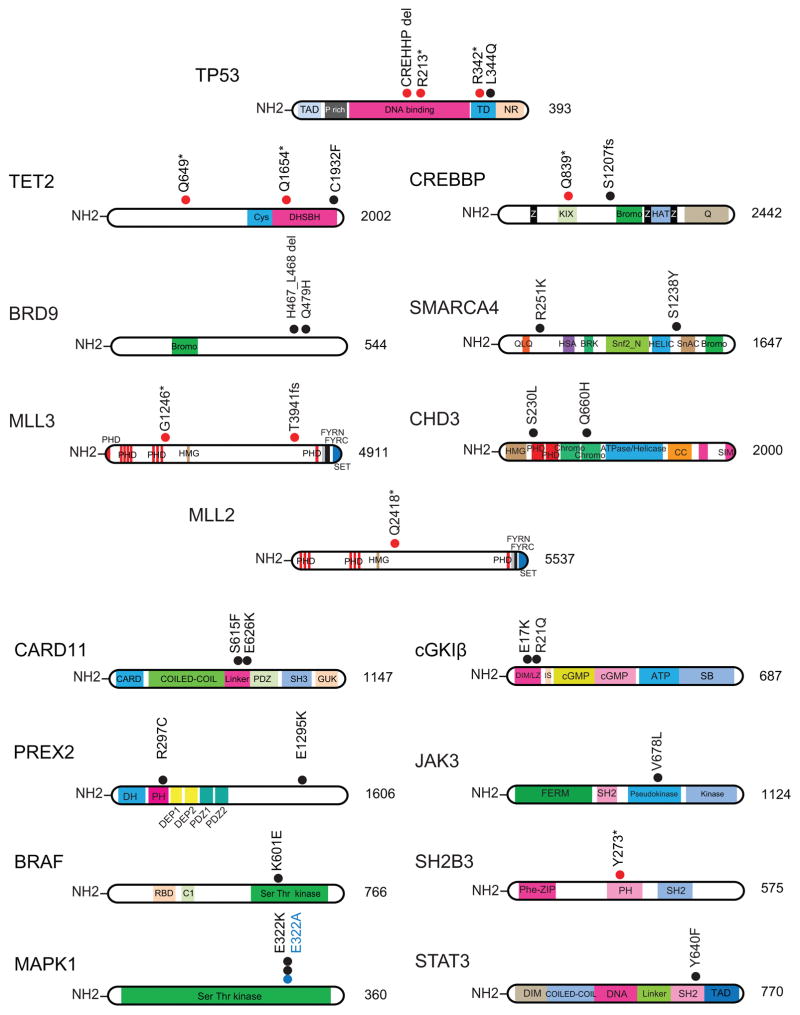
Mutation analysis showed a median of 39 non synonymous somatic mutations per sample (range of 1–182) in Sézary syndrome cases and 62 (range of 2–419) in mycosis fungoides (Supplementary Table 5). Overall we identified 1,261 candidate high confidence somatic mutations in Sézary syndrome affecting 1,123 different genes. Analysis of mutational processes 9 revealed the presence of a mutational signature characterized by C>T substitutions at NpCpG trinucleotides, as well as a high frequency of C>A substitutions at CpCpN trinucleotides and C>T substitutions at CpCpN and TpCpN trinucleotides (Supplementary Fig. 2). Mutations in Sézary syndrome included loss of function lesions in TP53 (p.Arg213*; p.Arg342*; p.Pro177_Cys182del and p.Leu344Gln) and three mutations in TET2 (p.Gln1654*, p.Cys1932Phe and p.Gln649*), an epigenetic tumor suppressor gene frequently mutated in myeloid malignancies and angioimmunoblastic T-cell lymphoma10. Additional epigenetic mutations included loss of function mutations in the CREBBP histone acetyl transferase (p.Gln839* and p.Ser1207fs); the MLL3 histone H3K4 methyl transferase (p.Thr3941fs) and mutations in components of the SWI/SNF (BRD9 p.Gln479His and pHis467_Leu468del; SMARCA4 p.Ser1238Tyr) and NuRD (CHD3 p.Gln660His and p.Ser230Leu) chromatin remodeling complexes (Supplementary Table 6) (Fig. 2). Analysis of mycosis fungoides revealed 958 somatic mutations in 866 genes. These included a mutation in SMARCA4 (p.Arg251Lys) and two truncating mutations in the MLL2 and MLL3 histone H3K4 methyl transferase genes (MLL2 p.Gln2418* and MLL3 p.Gly1246*) (Fig. 2 and Supplementary Table 7). The functional significance of epigenetic mutations in Sézary syndrome and CTCL is evidenced by the strongly deleterious alleles resulting in protein truncations identified in TET2 (p.Arg213*; p.Arg342*); CREBBP (p.Gln839* and p.Ser1207fs) and MLL3 (p.Thr3941fs and p.Gly1246*). In addition the TET2 p.Cys1932Phe mutation locates in the critical C terminal region of the DHSBH domain, which is recurrently disrupted by missense mutations in myeloid tumors and peripheral T-cell lymphoma11,12. The CHD3 p.Gln660His mutation is located in the second chromodomain of this protein and has a strongly damaging prediction score (PolyPhen 2 score = 0.99), while the p.Ser230Leu mutation is located within the first PHD domain, which is implicated in the reading of repressive histone marks13. Similarly, both the SMARCA4 p.Ser1238Tyr and the BRD9 p.Gln479His mutation have a very strong PolyPhen 2 damaging score of 0.99.
Figure 2.
Somatic mutations in Sézary syndrome and CTCL. Schematics of TP53, epigenetic factors TET2, BRD9, MLL3, MLL2, CREBBP, SMARCA4 and CHD3 and the CARD11, BRAF, MAPK1, PREX2, cGK1β, JAK3, SH2B3 and STAT3 signaling proteins showing mutations identified in this study. Black and blue circles indicate amino acid substitutions; red circles truncating mutations. Cys, cysteine rich domain; DHSBH, double-stranded beta helix fold domain; Bromo, bromo domain; PHD, Plant-homeodomain; HMG, High mobility group domain; FYRN “FY-rich” domain N-terminal; FYRC “FY-rich” domain C-terminal; SET, Su(var), Enhancer of zest, and Trithorax; Z, zinc finger domain; KIX, binding site of CREB; HAT, histone acetyl transferase domain; Q, glutamine-rich domain; QLQ, Gln, Leu, Gln motif; HAS, helicase/SANT-associated domain; BRK, Brahma and Kismet domain, SNF2_N, SF2 family N-terminal domain; Hel_C, Helicase superfamily C-terminal domain; SnAC, Snf2-ATP coupling, chromatin- remodeling complex; CC-coiled-coil domain; SIM, SUMO interacting domain; CARD, caspase recruitment domain; PDZ, PSD-95 (95 kDa protein involved in signaling in the post-synaptic density), Drosophila disc large tumor suppressor (Dlg1), Zonula occludens-1 protein (zo-1); SH3, Src homology 3 domain; GUK, guanylate kinase domain; TAD, transactivation domain; P rich, proline rich domain; TD, tetradimerization domain; NR, negative regulation domain; RBD, Ras binding domain; C1; protein kinase C-conserved region 1 domain; DH, DBL homology domain; PH, plekstrin homology domain; DEP domain, Dishevelled, Egl-10 and Pleckstrin domain; DIM/LZ, dimerization and leucine zipper domain; IS, auto-inhibitory domain; cGMP, cGMP binding domain; ATP, ATP binding domain; SB, substrate binding domain; FERM, 4.1protein/Ezrin/Radixin/Moesin domain; SH2, Src homology 2 domain; Phe-ZIP, phenylalanine zipper.
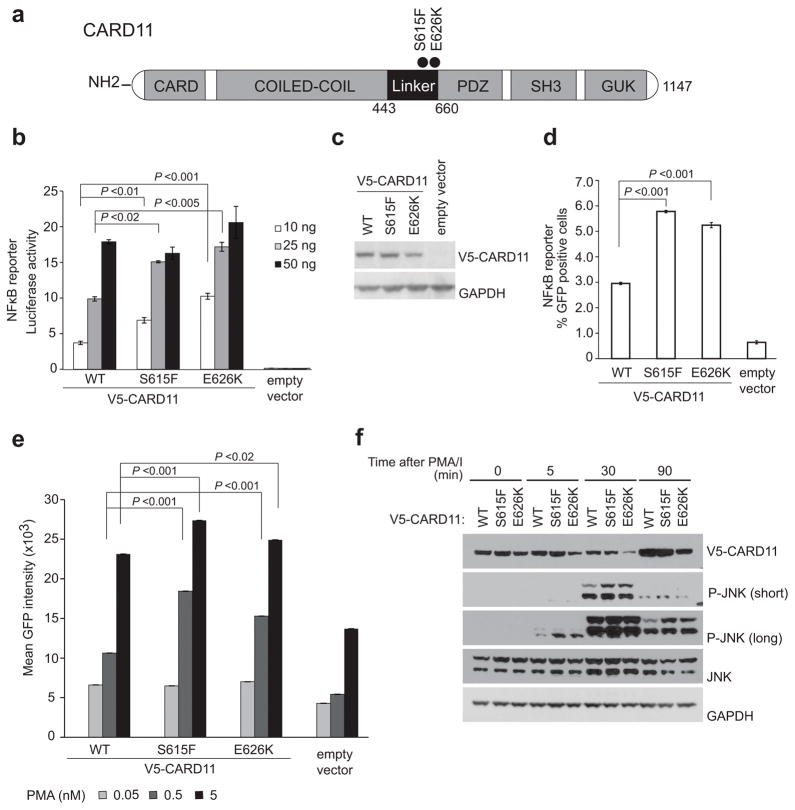
Signaling factor mutations included an activating mutation in the BRAF gene (p.Lys601Glu)14 in a CD30- CTCL sample and three mutations involving position E322 in MAPK1 (p.Glu322Ala and p.Glu322Lys) in two mycosis fungoides patients. Notably, the MAPK1 p.Glu322Lys mutation has been reported as an activating allele 15 and expression of both MAPK1 E322K and E322A resulted in increased ERK1/2 activation (Supplementary Fig. 3). We also found a JAK3 mutation previously reported in prolymphocytic leukemia (p.Val678Leu)16 co-occurring with a loss of function mutation in SH2B3 (p.Tyr273*), a negative regulator of JAK signaling; and an activating mutation in STAT3 (p.Tyr640Phe)17 in two mycosis fungoides cases. In addition we identified two mutations in PREX2, which encodes a phosphatidylinositol 3,4,5-trisphosphate RAC exchange factor involved in the regulation of PTEN tumour suppressor18 (Fig. 2). Notably, the PREX2 p.Arg297Cys mutation present in a mycosis fungoides sample is recurrently found in melanoma 19 and the PREX2 p.Glu1295Lys allele found in a CD30- CTCL has been reported in melanomas, cutaneous squamous cell carcinoma and head and neck cancer19. Finally, we identified three point mutations in Sézary syndrome in PLCG1, an oncogene described as carrying gain-of function mutations in CTCL8,20. One of the alleles is a novel variant p.Gly722Val, while the other two, p.Arg48W and p.Glu1163Lys have been previously reported8. The NFkB pathway is known to be recurrently activated in CTCL and Sézary syndrome21. Alterations in TNFR2 (tumor necrosis factor receptor 2) leading to NFkB activation have been recently identified in mycosis fungoides and Sézary syndrome 7. Consistently, we identified two mutations in TNFR2 in our Sézary cohort, p.Gly256Cys and p.Thr377Ile7. However, these analyses also revealed the presence of two closely located mutations in the CARD11 gene (p.Ser615Phe and p.Glu626Lys), another important mediator of NFκB activation, in two Sézary syndrome cases. Both CARD11 mutations identified in our analysis are located in the so called linker domain region of the protein. This is in contrast with diffuse large B cell lymphoma where CARD11 mutations are typically located in the coiled-coil domain 22. In basal conditions CARD11 adopts an inactive closed configuration, which upon T-cell receptor (TCR) activation and consequent PKCθ kinase phosphorylation of the linker domain, turns into an open active scaffold structure that recruits factors such as BCL10, MALT1, TRAF6, TAK1, CASP8 and IKKγ in a complex that activates downstream NFκB signaling23. To test their functional significance we first analyzed the effects of CARD11 wild type, CARD11 S615F and CARD11 E626K expression in NFκB luciferase reporter assays in transfected HEK293-T cells (Fig. 3b). Expression of both CARD11 S615F and CARD11 E626K resulted in increased NFκB luciferase reporter activity compared with wild type CARD11 transfected cells. Moreover, lentiviral transduction of CARD11 S615F and CARD11 E626K in JURKAT T-cells expressing GFP under the control of an NFκB promoter resulted in increased NFκB-driven GFP levels compared with wild-type CARD11 transduced controls (Fig. 3c). Treatment of CARD11 S615F- and CARD11 E626K-expressing JURKAT cells with phorbol 12-myristate 13-acetate (PMA) plus ionomycin to mimic TCR activation, resulted in further increased NFκB-driven GFP expression (Fig. 3e). Finally, CARD11 S615F- and CARD11 E626K-expressing JURKAT cells showed earlier, increased and more sustained JNK phosphorylation after PMA plus ionomycin treatment in comparison with wild-type CARD11 expressing controls (Fig. 3f), a result consistent with increased CARD11 activation downstream of TCR signaling. Next we compared the activity of CARD11 linker domain mutations with that of well characterized diffuse large B cell lymphoma CARD11 coiled-coil mutations with weak (M183L), strong (D230N) and very strong (L251P) NFκB activating effects 22. In the absence of Ionomycin+PMA stimulation CARD11 linker domain mutations showed moderate NFκB luciferase reporter activity intermediate between that of the CARD11 M183L and the D230N coiled-coil mutations (Supplementary Fig. 4). However, in cells treated with PMA plus ionomycin, CARD11 linker mutations showed stronger NFκB activation, similar to that observed in cells expressing the strong D230N and L251P CARD11 coiled-coil mutants (Fig. 3e and Supplementary Fig. 4).
Figure 3.
Functional characterization of Sézary syndrome CARD11 linker domain mutations. (a) Schematic representation of the structure of the CARD11 protein. CARD11 mutations identified are indicated with circles. (b) NFκB luciferase reporter activity in HEK293-T cells transfected with increasing amounts of V5-CARD11 wild-type, S615F and E626K or empty vector. Data is representative of 3 independent experiments. (c) Analysis of the levels of V5-CARD11 protein in JURKAT NFκB-GFP reporter cell line infected with lentiviruses driving the expression of CARD11 wild type, CARD11 mutants S615F and E626K or empty vector. (d) NFκB-dependent GFP reporter activity in non-stimulated JURKAT cells expressing CARD11 wild type, CARD11 mutants S615F and E626K or empty vector. Bar graphs indicate the percentage of GFP positive cells analyzed by flow cytometry. Data is representative of 3 independent experiments. (e) NF-κB-dependent GFP reporter activity in JURKAT cells, after stimulation for 6h with 1 μg/ml ionomycin and increasing concentration of PMA (0.05, 0.5 and 5 nM), in JURKAT cells expressing CARD11 wild type, CARD11 mutants S615F and E626K or empty vector. Bar graphs indicate mean GFP intensity measured by flow cytometry across 3 biological replicates. (f) Western blot analysis of JNK phosphorylation in JURKAT cells infected with lentivirus driving the expression of V5-CARD11 wild type, V5-CARD11 mutants S615F and E626K or empty vector, after stimulation with 1 μg/ml Ionomycin and 25 nM PMA. The bar graphs in b, d and e show the mean values and error bars represent the s.d. P values were calculated using Student’s t test. WT, wild type
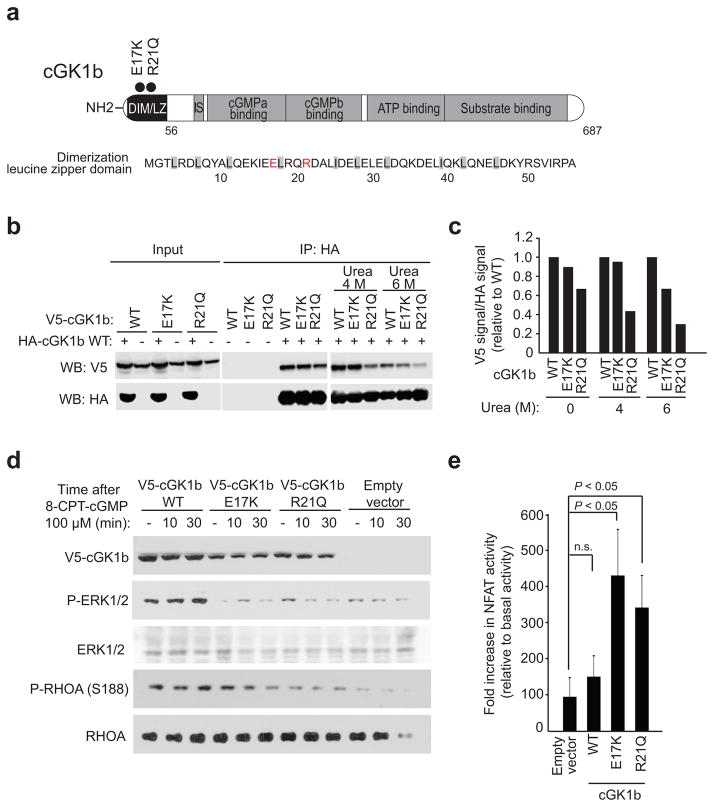
Finally, we also identified two mutations in the cGKIβ protein (p.Glu17Lys and p.Arg21Gln) one of the two cyclic GMP-dependent protein kinases (cGKIα and cGKIβ) generated by alternative splicing of the PRKG1 gene24, in two Sézary syndrome samples (Fig. 4a). In T-cells, cGKIβ antagonizes TCR activation-induced interleukin 2 release and proliferation25,26. Structurally, the two PRKG1 mutations in our series are located in the N-terminal leucine/isoleucine zipper dimerization domain of cGKIβ27 (Fig. 4a), which mediates the formation of homodimers implicated in cGK activation25,28. Urea dissociation experiments of immunoprecipitated cGKIβ complexes revealed reduced stability of wild-type cGKIβ-cGKIβ E17K and wild-type cGKIβ-cGKIβ R21Q dimers compared with wild-type cGKIβ homodimer controls (Fig. 4b,c). Moreover, JURKAT cells expressing cGKIβ E17K and cGKIβ R21Q showed decreased cGKIβ-mediated ERK1/2 activation29 after stimulation with the cGMP analog 8-CPT-cGMP, in comparison with wild-type cGKIβ expressing controls (Fig. 4d). Additionally, JURKAT cGKIβ E17K- and cGKIβ R21Q-expressing cells treated with 8-CPT-cGMP also failed to phosphorylate RHOA at position S188 30 (Fig. 4d). These results support that N-terminal cGKIβ mutations identified in Sézary samples are loss of function alleles with reduced signal transduction ability. Consistently, JURKAT cells expressing cGKIβ E17K and cGKIβ R21Q showed increased NFAT activity after PMA plus ionomycin stimulation compared with wild type controls, supporting a positive role for cGKIβ mutations in enhancing the TCR signaling response (Fig. 4e). These results are in agreement with data describing mutations in genes activating NFκB and NFAT signaling in CTCL20.
Figure 4.
Functional characterization of Sezary syndrome cGKIβ mutations. (a) Schematic representation of the structure of the cGKIβ protein and sequence of the leucine zipper domain. Mutations are indicated with circles and mutated residues highlighted in red. (b,c) Western blot analysis (b) and quantification (c) of cGKIβ complex stability (urea dissociation) for wild type cGKIβ-HA/cGKIβ-V5, wild type cGKIβ-HA/cGKIβ-V5 E17K and wild type cGKIβ-HA/cGKIβ-V5 R21Q immunoprecipitates. (d) Western blot analysis of cGKIβ signaling (ERK activation and RhoA S188 phosphorylation) after 8-CPT-cGMP stimulation in JURKAT cells infected with virus driving the expression of wild type cGKIβ-V5, cGKIβ-V5 E17Q and cGKIβ-V5 R21Q mutants. (e) NFAT luciferase reporter assays in JURKAT cells expressing wild-type V5-cGKIβ, V5-cGKIβ E17K and V5-cGKIβ R21Q mutants 6 hours post stimulation with PMA (1μM) plus ionomycin (1 μg/ml). The bar graphs in (e) show the mean values and error bars represent the s.d. Data is representative of triplicate samples from two independent experiments. P values were calculated using two-tailed Student’s t test. WT, wild type
Western blot analysis of signaling pathways revealed high levels of P-STAT3, P-ERK, P-JNK and nuclear p50 NFKB1 in most CTCL lines (Supplementary Fig. 5). Therapeutically, inhibition of JAK signaling with tofacitinib and ruxolitinib was active HUT78 cells harboring a JAK3-p.Ala573Val mutation. Inhibition of NFκB signaling with Mi-2 and bortezomib was broadly and highly active across all 4 cell lines (HH, HUT78, HUT102 and SeAX). In contrast MEK1/2 inhibition with U0126 and inhibition of NFAT with FK506 showed only modest antitumor effects (Supplementary Fig. 6).
Our results identify new genetic drivers in the pathogenesis of Sézary syndrome and point to TCR-controlled signaling pathways as potential therapeutic targets for the treatment of this disease. Extended analysis of clinically annotated series and further genetic characterization including whole genome sequencing and transcriptomic analysis will be instrumental to elucidate the complete repertoire of genetic driver alterations in Sézary syndrome and CTCL.
Online Methods
Patient samples
Tumor and matched normal samples were provided by the departments of Dermatology at Northwestern University (Chicago, USA) and the Leiden University Medical Center (Leiden, The Netherlands). Samples were obtained with informed consent and analysis was conducted under the supervision of the Columbia University Medical Center Institutional Review Board. We selected samples for sequencing based on the availability of sufficient matched tumor and normal DNA. Tumor samples were CTCL skin biopsies or CD4+ T-cells cells from peripheral blood for Sézary syndrome cases. Normal DNA samples were obtained from buccal swabs or granulocytes in the case of Sézary syndrome patients.
Whole exome capture and nextgen sequence analysis
We used matched tumor and normal DNA samples (Supplementary Table 1) for exome capture with the SureSelect 50 Mb All Exon kit (Agilent Technologies) following standard protocols. We performed paired-end sequencing (2 × 100 bp) using HiSeq2000 sequencing instruments at Centrillion Biosciences (Palo Alto, CA). Illumina HiSeq analysis produced between 84.5 and 137.94 million paired-end reads per sample (Supplementary Table 2). We mapped reads to the reference genome hg19 using the Burrows-Wheeler Aligner (BWA) alignment tool version 0.5.9. Mean depth (defined as mean number of reads covering the captured coding sequence of a haploid reference) was 143.51x with 95.3% of the genome covered more than 30x. We identified sites that differ from reference (called here variants) in each sample independently. The average number of germline SNPs were 19,631 comparable with previous reports 12. Most of the candidate germline SNPs (~90% of germline variants) were reported in the dbSNP database. We used the SAVI algorithm (Statistical Algorithm for Variant Identification) 31 developed at Columbia University, and constructed empirical priors for the distribution of variant frequencies in each sample. Following SAVI’s Bayesian framework, we obtained a corresponding high-credibility interval for the frequency of each variant (posterior probability ≥1–10−5) and tested the null hypothesis that variants were due to sequencing errors. We found that for those variants observed with frequency of 0 to 2%, it was not possible to reject the null hypothesis and distinguish true mutations from sequencing errors. Consequently, we considered variants observed with a frequency of 0 to 2% as absent. Also using the SAVI algorithm, we obtained high-credibility intervals for the corresponding change in frequency between tumor and normal samples (posterior probability ≥1–10−5). Consistent with Sanger sequencing sensitivity and based on our previously published specificity assessment (false positive rate of 2.9% for variants called in >20% of the reads and 18.3% for variants called in 3–20% of the reads) 32, we retained candidate somatic variants in CTCL samples as those absent in normal and present in tumor in ≥15% of the reads with at least 1% change in frequency from the normal with high posterior probability (≥1–10−5). We excluded variants in regions with low coverage depth (<10x) or in regions with extremely high coverage depth (>300x), associated with genomic repetitive elements. We also removed all variants that were found present in any of this study’s normal cases, or in any one of 220 exomes from unaffected individuals that were analyzed at our institution. Additionally, for Sézary samples in which control DNA was obtained from blood after CD4 depletion (SS_L1–23) we retained as candidate somatic variants those present in control DNA at frequencies <20% if their variant allele frequency increased >2 fold in tumor DNA to account for contaminating tumor lymphocytes in the control preparation.
For mutation process analysis we used the Wellcome Trust Sanger Institute’s framework9 implemented in MATLAB scripts. To identify mutational signatures in Sézary syndrome, we used all synonymous and non-synonymous variants. We assessed signature stability and computed average Frobenius reconstruction error for k=1 to k=10 number of signatures. Significant reduction in reconstruction error until k=3, as well as stable reproducibility between k=1 and k=3, indicated three recognized signatures by the algorithm.
Identification of recurrent and focal copy number variants
Somatic copy number variants (CNV) were identified from the depth of coverage using EXCAVATOR33 with default parameters. Samples for which the matching normal was collected from buccal swabs were analyzed with EXCAVATOR pooled mode, which compares tumor depth of coverage with a compendium of all normal samples. Normal compendium for chromosome X and Y was computed according to the patient gender to avoid diploid biases. Samples for which the matching normal was collected from granulocytes were analyzed with EXCAVATOR somatic mode, which performs a pairwise tumor-normal comparison for each case. For these cases, we obtained additional statistical significance for these CNV with the following approach: for all sequenced exons, we computed the ratio of the depth of coverage in tumor to the total depth of coverage in both tumor and normal. These ratios, by definition, have values between 0 and 1, which we assumed follow a beta distribution. Following the Bayesian workflow described in the SAVI algorithm, we constructed an empirical prior for the ratios taken from all exons, and calculated a posterior distribution for the value of each ratio. The p-values for the null hypothesis that an exon has no copy number alterations were therefore computed. Finally, we obtained the statistical significance for each CNV by taking the geometric mean of the p-values of the exons within the altered region. We included the CNVs with confidence of at least 95% in the final list.
Quantitative real-time PCR (qPCR)
We performed reverse transcription reactions with the ThermoScript RT-PCR system (Invitrogen) and analyzed the resulting cDNA products by quantitative real-time PCR (FastStart Universal SYBR Green Master Mix, Roche) using a 7300 Real-Time PCR system (Applied Biosystems). geNorm pairwise variation (V) analysis was performed to determine optimal number of reference genes for data normalization. Primer sequences can be found in Supplementary Table 8.
Plasmid and vectors
We obtained the pLX304-blasV5 MAPK1, CARD11 and PKG1/cGKIβ constructs from the Broad Lentiviral Expression Library. We introduced specific variants by site-directed mutagenesis using the QuikChange II XL Site-Directed Mutagenesis kit (Stratagene) according to the manufacturer’s instructions. pGL3-NFAT luciferase (Plasmid #17,870) and PKG1-HA pHACE (Plasmid #16,394) were obtained from Addgene.
Cell lines
We cultured HEK293T (Thermo Scientific) in DMEM supplemented with 10% FBS, 100 U/ml penicillin G and 100 μg/ml streptomycin at 37 °C in a humidified atmosphere under 5% CO2. We grew Jurkat (ATCC) and the NF-κB/Jurkat/GFP Transcriptional Reporter cells (System Biosciences) and the CTCL cell lines HH, HUT78, HUT102, SeAX (ATCC under similar conditions in RPMI 1640 supplemented with 10% FBS and 100 U/ml penicillin G and 100 μg/ml streptomycin. Cell lines were regularly tested for mycoplasma contamination.
Lentivirus production and infection
We transfected the lentiviral constructs pLX304-blasV5 CARD11 wild type, CARD11 S615F, E626K, D230N, L215P, M183L and the empty vector with pCMV ΔR8.91 and pMD.G VSVG in HEK293T cells using Fugene 6 (Promega), according the manufacturers protocol. We collected viral supernatants after 48 h and used them for infection of JURKAT cells and the JURKAT-NF-κB-GFP cells by spinoculation. After infection, we selected cells for 2 weeks in medium containing 1 μg/ml Blasticidine.
Western Blot analysis
In different experiments we stimulated JURKAT cells with Ionomycin (1 μg/ml, EMD Millipore) and PMA (25 nM, Sigma Aldrich) or with 8-CPT-cGMP (100 μM, Sigma C240). For immunoblotting we washed cells in cold PBS 1X supplemented with phosphatase inhibitors, lysed them for 30 min in RIPA buffer (20 mM Tris pH7.5. 150 mM NaCl, 1% NP40, 0.1 % Sodium deoxycholate, supplemented with protease inhibitors and phosphatase inhibitors) and clear the lysates by 10 min centrifugation at 15,000 g at 4°C before SDS-PAGE. For subcellular fractionation, we washed CTCL cells with PBS and resuspended them in buffer 1 (25 mM Hepes, pH 7.9, 5 mM KCl, 0.5 mM MgCl2, supplemented with protease and phosphatase inhibitors). We lysed cells by adding equal volume of buffer 2 (buffer 1 supplemented with 1% Nonidet 40) and incubating for 15 min end-over-end at 4ºC. Nuclei were pelleted at 500 g for 5 min and washed once with buffer 3 (1:1 mixture of buffer 1 and 2). We collected the cytoplasmic fraction and lysed the nuclei with nuclear extraction buffer (10 mM Hepes pH 7.9, 400 mM NaCl, 1.5 mM MgCl2 and 1% Triton X-100 supplemented with protease and phosphatase inhibitors). Cytosolic and nuclear fractions were subjected to SDS-PAGE.
We performed Western blot detection using standard procedures with the following antibodies: V5 (Cell Signaling Technology, 13202s, 1:2,000), HA (Roche, 11867423001, 1:500), GAPDH (Cell Signaling Technology, 5174, 1:5,000), SAPK/JNK (Cell Signaling Technology, 9252, 1:1,000), Phospho-SAPK/JNK (Thr183/Tyr185) (Cell Signaling Technology, 4668, 1:1,000), ERK total (Santa Cruz, SC-271270, 1:250), Phospho-p44/p42 (Cell signaling technologies, 4377, 1:1,000), RhoA total (67B9) (Cell Signaling Technology, 2117, 1:1,000) and phospho-RhoA (S188) (Abcam, ab41435, 1:1,000), Phospho-STAT3 (Y705) (Cell Signaling Technology, 9145, 1:1,000), STAT3 total (Cell Signaling Technology, 9132, 1:1,000), NFKB1 (p50/p105) (Cell Signaling Technology, 3035, 1:1,000), HSP90 (H-114, Santa Cruz Biotechnology, sc-7947, 1:250), Lamin B (M-20, Santa Cruz Biotechnology, sc-6217, 1:250).
cGKIβ complex dissociation assays
We transfected HEK293-T cells were with pHACE cGKIβ-HA wild type together with pLX304-blas-V5 cGKIβ wild-type, pLX304-blas-V5 cGKIβ E17K or pLX304-blas-V5 cGKIβ R21Q mutants. After 24 hours we lysed the cells in co-immunoprecipitation buffer (50 mM Tris pH7.5, 150 mM NaCl, 0.5% NP40) and immunoprecipitated with EZview Red Anti-HA Affinity Gel (Sigma, E6779). After washing the beads three times with co-immunoprecipitation buffer, we performed a 1 h incubation at room temperature with co-immunoprecipitation buffer containing urea (4M and 6M), followed by two additional washes with co-immunoprecipitation buffer. We then eluted the immunoprecipitated cGKIβ dimers from the beads with Laemmli sample buffer and analyzed them by SDS-PAGE and Western blot.
Luciferase and GFP reporter assays
We transfected HEK293-T cells with pLX304-blasV5 CARD11 wild-type, or S615F, E626K, D230N, L215P, M183L mutant constructs together with 200ng of a mix (40:1) of the inducible NFκB Luciferase reporter and constitutively expressing Renilla luciferase construct (Cignal reporter assay, Qiagen). After 24 hours we analyzed luciferase activity using the Dual-Luciferase Reporter assay kit (Promega).
We analyzed the percentage of GFP positive cells and the mean of GFP intensity of Jurkat-NF-κB-GFP reporter cells stably expressing CARD11 wild-type and mutants by flow cytometry in basal conditions or 6 h after stimulation with ionomycin 1μg/ml and different amounts of PMA (0.05, 0.2, 0.5 and 5 nM).
We electroporated 5 million JURKAT cells with 5 μg of cGKIβ wild type, cGKIβ E17K and cGKIβ R21Q pLX304 constructs, together with 5 μg of pGL3-NFAT luciferase reporter and 1 μg of Renilla luciferase expressing vectors. After 24 h we serum-starved the cells for 24 h, stimulated them for 6 h with PMA (1μM) plus ionomycin (1 μg/ml) and analyzed the NFAT driven luciferase activity using the Dual-Luciferase Reporter assay kit.
Drug treatments and cell viability
We analyzed cell viability and proliferation with the Cell Proliferation Kit I (Roche) after treating HH, HUT78, HUT102 and SeAX cell lines for 72 hours with a range of concentration of specific compounds. Ruxotilinib was kindly provided by Dr. Ross Levine. Tofacitinib, Mi-2 and Bortezomib were obtained from Shelleck Chemicals. U0126 was purchased from Sigma-Aldrich and FK506 from Invivogen.
Statistical analyses
Analyses of significance were performed using Student’s t test assuming equal variance. Continuous biological variables were assumed to follow a normal distribution. A P value of <0.05 was considered to indicate statistical significance.
Supplementary Material
Supplementary Table 1
Suplementary Table 2. Exome Sequencing parameters
Supplementary Table 3: CNV in SS
Supplementary Table 4: CNV in CTCL samples
Supplementary Table 5: Number of SNV per sample
Supplementary Table 6: Somatic SNVs in Sezary Syndrome samples
Supplementary Table 7: Somatic SNVs in CTCL samples
Supplementary Table 8: Sequence of primers used in the study
Acknowledgments
This work was supported by a Leukemia & Lymphoma Society Translational Research Grant (A.F.), a Herbert Irving Comprehensive Cancer Center inter-programmatic pilot project grant (A.F. & R.R.) and a Dutch Cancer Society UL2013-6104 grant (C.P.T and M.H.V). ACdSA is supported by a Lady Tata Memorial Trust fellowship.
Footnotes
Accession codes
Data has been deposited in dbGaP under accession number phs000994.v1.p1.
Author contributions
ACdSA performed functional assays; FA and HK performed exome and copy number analysis; EM-E, JG, CPT and MHV contributed clinical samples; RR directed sequencing analyses; AF and TP designed the study, directed and supervised research and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests
References
- 1.Querfeld C, Rosen ST, Guitart J, Kuzel TM. The spectrum of cutaneous T-cell lymphomas: new insights into biology and therapy. Curr Opin Hematol. 2005;12:273–8. doi: 10.1097/01.moh.0000166498.64515.03. [DOI] [PubMed] [Google Scholar]
- 2.Wilcox RA. Cutaneous T-cell lymphoma: 2014 update on diagnosis, risk-stratification, and management. Am J Hematol. 2014;89:837–51. doi: 10.1002/ajh.23756. [DOI] [PubMed] [Google Scholar]
- 3.Agar NS, et al. Survival outcomes and prognostic factors in mycosis fungoides/Sezary syndrome: validation of the revised International Society for Cutaneous Lymphomas/European Organisation for Research and Treatment of Cancer staging proposal. J Clin Oncol. 2010;28:4730–9. doi: 10.1200/JCO.2009.27.7665. [DOI] [PubMed] [Google Scholar]
- 4.Cristofoletti C, et al. Comprehensive analysis of PTEN status in Sezary syndrome. Blood. 2013;122:3511–20. doi: 10.1182/blood-2013-06-510578. [DOI] [PubMed] [Google Scholar]
- 5.Mao X, et al. Heterogeneous abnormalities of CCND1 and RB1 in primary cutaneous T-Cell lymphomas suggesting impaired cell cycle control in disease pathogenesis. J Invest Dermatol. 2006;126:1388–95. doi: 10.1038/sj.jid.5700224. [DOI] [PubMed] [Google Scholar]
- 6.Brito-Babapulle V, et al. p53 allele deletion and protein accumulation occurs in the absence of p53 gene mutation in T-prolymphocytic leukaemia and Sezary syndrome. Br J Haematol. 2000;110:180–7. doi: 10.1046/j.1365-2141.2000.02174.x. [DOI] [PubMed] [Google Scholar]
- 7.Ungewickell A, et al. Genomic analysis of mycosis fungoides and Sezary syndrome identifies recurrent alterations in TNFR2. Nat Genet. 2015;47:1056–60. doi: 10.1038/ng.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi J, et al. Genomic landscape of cutaneous T cell lymphoma. Nat Genet. 2015;47:1011–9. doi: 10.1038/ng.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexandrov T, Becker M, Guntinas-Lichius O, Ernst G, von Eggeling F. MALDI-imaging segmentation is a powerful tool for spatial functional proteomic analysis of human larynx carcinoma. J Cancer Res Clin Oncol. 2013;139:85–95. doi: 10.1007/s00432-012-1303-2. [DOI] [PubMed] [Google Scholar]
- 10.Solary E, Bernard OA, Tefferi A, Fuks F, Vainchenker W. The Ten-Eleven Translocation-2 (TET2) gene in hematopoiesis and hematopoietic diseases. Leukemia. 2014;28:485–96. doi: 10.1038/leu.2013.337. [DOI] [PubMed] [Google Scholar]
- 11.Smith AE, et al. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood. 2010;116:3923–32. doi: 10.1182/blood-2010-03-274704. [DOI] [PubMed] [Google Scholar]
- 12.Palomero T, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014;46:166–70. doi: 10.1038/ng.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, et al. CHD3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome. Proc Natl Acad Sci U S A. 2012;109:5773–8. doi: 10.1073/pnas.1203148109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barollo S, et al. Prevalence, tumorigenic role, and biochemical implications of rare BRAF alterations. Thyroid. 2014;24:809–19. doi: 10.1089/thy.2013.0403. [DOI] [PubMed] [Google Scholar]
- 15.Van Allen EM, et al. Genomic Correlate of Exceptional Erlotinib Response in Head and Neck Squamous Cell Carcinoma. JAMA Oncology. 2015 doi: 10.1001/jamaoncol.2015.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergmann AK, et al. Recurrent mutation of JAK3 in T-cell prolymphocytic leukemia. Genes Chromosomes Cancer. 2014;53:309–16. doi: 10.1002/gcc.22141. [DOI] [PubMed] [Google Scholar]
- 17.Koskela HL, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366:1905–13. doi: 10.1056/NEJMoa1114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fine B, et al. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science. 2009;325:1261–5. doi: 10.1126/science.1173569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forbes SA, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–11. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaque JP, et al. PLCG1 mutations in cutaneous T-cell lymphomas. Blood. 2014;123:2034–43. doi: 10.1182/blood-2013-05-504308. [DOI] [PubMed] [Google Scholar]
- 21.Sors A, et al. Down-regulating constitutive activation of the NF-kappaB canonical pathway overcomes the resistance of cutaneous T-cell lymphoma to apoptosis. Blood. 2006;107:2354–63. doi: 10.1182/blood-2005-06-2536. [DOI] [PubMed] [Google Scholar]
- 22.Lenz G, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–9. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 23.Sommer K, et al. Phosphorylation of the CARMA1 linker controls NF-kappaB activation. Immunity. 2005;23:561–74. doi: 10.1016/j.immuni.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann F, Wegener JW. cGMP-dependent protein kinases (cGK) Methods Mol Biol. 2013;1020:17–50. doi: 10.1007/978-1-62703-459-3_2. [DOI] [PubMed] [Google Scholar]
- 25.Francis SH, Busch JL, Corbin JD, Sibley D. cGMP-dependent protein kinases and cGMP phosphodiesterases in nitric oxide and cGMP action. Pharmacol Rev. 2010;62:525–63. doi: 10.1124/pr.110.002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischer TA, et al. Activation of cGMP-dependent protein kinase Ibeta inhibits interleukin 2 release and proliferation of T cell receptor-stimulated human peripheral T cells. J Biol Chem. 2001;276:5967–74. doi: 10.1074/jbc.M009781200. [DOI] [PubMed] [Google Scholar]
- 27.Casteel DE, et al. A crystal structure of the cyclic GMP-dependent protein kinase I{beta} dimerization/docking domain reveals molecular details of isoform-specific anchoring. J Biol Chem. 2010;285:32684–8. doi: 10.1074/jbc.C110.161430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richie-Jannetta R, Francis SH, Corbin JD. Dimerization of cGMP-dependent protein kinase Ibeta is mediated by an extensive amino-terminal leucine zipper motif, and dimerization modulates enzyme function. J Biol Chem. 2003;278:50070–9. doi: 10.1074/jbc.M306796200. [DOI] [PubMed] [Google Scholar]
- 29.Lander HM, Jacovina AT, Davis RJ, Tauras JM. Differential activation of mitogen-activated protein kinases by nitric oxide-related species. J Biol Chem. 1996;271:19705–9. doi: 10.1074/jbc.271.33.19705. [DOI] [PubMed] [Google Scholar]
- 30.Lang P, et al. Protein kinase A phosphorylation of RhoA mediates the morphological and functional effects of cyclic AMP in cytotoxic lymphocytes. EMBO J. 1996;15:510–9. [PMC free article] [PubMed] [Google Scholar]
- 31.Trifonov V, Pasqualucci L, Tiacci E, Falini B, Rabadan R. SAVI: a statistical algorithm for variant frequency identification. BMC Syst Biol. 2013;7(Suppl 2):S2. doi: 10.1186/1752-0509-7-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pasqualucci L, et al. Genetics of follicular lymphoma transformation. Cell Rep. 2014;6:130–40. doi: 10.1016/j.celrep.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magi A, et al. EXCAVATOR: detecting copy number variants from whole-exome sequencing data. Genome Biol. 2013;14:R120. doi: 10.1186/gb-2013-14-10-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1
Suplementary Table 2. Exome Sequencing parameters
Supplementary Table 3: CNV in SS
Supplementary Table 4: CNV in CTCL samples
Supplementary Table 5: Number of SNV per sample
Supplementary Table 6: Somatic SNVs in Sezary Syndrome samples
Supplementary Table 7: Somatic SNVs in CTCL samples
Supplementary Table 8: Sequence of primers used in the study