Abstract
Purpose
The rapid expansion of the use of continuous critical care electroencephalogram (cEEG) monitoring and resulting multicenter research studies through the Critical Care EEG Monitoring Research Consortium has created the need for a collaborative data sharing mechanism and repository. The authors describe the development of a research database incorporating the American Clinical Neurophysiology Society standardized terminology for critical care EEG monitoring. The database includes flexible report generation tools that allow for daily clinical use.
Methods
Key clinical and research variables were incorporated into a Microsoft Access database. To assess its utility for multicenter research data collection, the authors performed a 21-center feasibility study in which each center entered data from 12 consecutive intensive care unit monitoring patients. To assess its utility as a clinical report generating tool, three large volume centers used it to generate daily clinical critical care EEG reports.
Results
A total of 280 subjects were enrolled in the multicenter feasibility study. The duration of recording (median, 25.5 hours) varied significantly between the centers. The incidence of seizure (17.6%), periodic/rhythmic discharges (35.7%), and interictal epileptiform discharges (11.8%) was similar to previous studies. The database was used as a clinical reporting tool by 3 centers that entered a total of 3,144 unique patients covering 6,665 recording days.
Conclusions
The Critical Care EEG Monitoring Research Consortium database has been successfully developed and implemented with a dual role as a collaborative research platform and a clinical reporting tool. It is now available for public download to be used as a clinical data repository and report generating tool.
Keywords: Continuous EEG monitoring, Research database, Clinical reporting tool
Recent advances in medicine have been driven by novel and large-scale collaborations, including major collaborative efforts in epilepsy (Perucca and O’Brien, 2015). The Critical Care EEG Monitoring Consortium (CCEMRC) was established in 2005 to promote collaborative research and to improve the quality of the clinical practice of critical care electroencephalogram (cEEG) monitoring. Creation of standardized terminology for cEEG monitoring (Hirsch et al., 2013) represented a major achievement. Our next goal is to incorporate this terminology into daily cEEG interpretation in a manner that would facilitate multicenter data collection for collaborative research. Through the support of an infrastructure grant from the American Epilepsy Society, a multicenter research and clinical database with data sharing mechanisms was created based on this standard terminology. The objectives of this project were to create a database (1) to serve as a repository of research data to facilitate multicenter studies and (2) to serve as a clinical tool that could be integrated into daily workflow for standardized reporting of cEEG monitoring results at member sites. Multicenter collaborative intensive care unit EEG collaboration is required to study relatively rare conditions and has resulted in key clinical findings in the field of cEEG (Gaspard et al., 2013a, 2013b; Kilbride et al., 2013; Schmitt et al., 2012).
Several past attempts to create standardized EEG reporting databases for clinical studies have failed to achieve multicenter adoption (Aurlien et al., 1999; Finnerup et al., 1999), possibly because of inefficient user interfaces or inability of the reporting system to meet local stylistic and regulatory requirements. However, a more recent effort, the Standardized Computer-based Organized Reporting of EEG (Beniczky et al., 2013a), seems to have gained substantial adoption in Europe. The CCEMRC database was specifically designed to collect detailed descriptions of EEG findings in a standardized format while providing flexibility to accommodate a large variety of prospective and retrospective studies. Our objectives were to create a database that could serve both as a repository of research data to facilitate multicenter studies and as a clinical tool that could be integrated into daily workflow for standardized reporting of cEEG monitoring results at member centers.
In this report, we summarize the major collaborative research and clinical reporting features of this database and present the results of a collaborative study designed to validate this database as a multicenter collaborative research tool among adult and pediatric CCEMRC member centers.
METHODS
Database Description
The database user interface and data structure were conceived in 2010 by a subgroup of CCEMRC members. A file server approach was implemented on a Microsoft (Redmond, WA) Access database platform by a single professional database programmer with >20 years of experience. This approach was chosen to allow self-installation at individual centers without requiring professional information technology services or back-end database servers, thus minimizing effort and cost. Successful installations onto a shared network drive or a desktop virtualization environment (e.g., Citrix) have also been achieved. The database consists of a separate interface file (containing front-end user interface code) and a database file (containing data tables) for improved performance and security. The interface file can be upgraded easily to accommodate new features without affecting the underlying data elements. It has gone through extensive development and testing, and frequent updates since its release in February 2013. The database consists of 741 data elements organized across 15 tables, many of which are self-populated. It incorporates all elements of the standardized cEEG terminology (Hirsch et al., 2013), and also some EEG patterns described more recently such as brief potentially ictal rhythmic discharges (B[I]RDs) (Yoo et al., 2014). Categories for morphological description of electrographic seizures are based on the modified Young criteria (Chong and Hirsch, 2005; Young et al., 1996) in addition to commonly observed patterns in electroclinical syndromes (fast activity >13 Hz, electrosuppression), but are not intended to be comprehensive as the definition of seizures can be difficult (Walker and Kovac, 2015). Although the presence/absence of status epilepticus is recorded, no definite criteria are defined within the database. In case of suspected nonconvulsive status epilepticus, the user is encouraged to use the Salzburg Consensus Criteria (Beniczky et al., 2013b), which is coded into an algorithm (Trinka and Leitinger, 2015).
The database is designed to create 1 report per monitoring day, with an optional final summary at the end of the monitoring session. Common elements can easily be carried forward onto subsequent days. The main screen allows for patients to be searched or ordered by name or medical record number. To facilitate the workflow of clinical cEEG reporting, studies can also be filtered by the attending physician or fellow name, allowing for sequential approval by a fellow and attending physician.
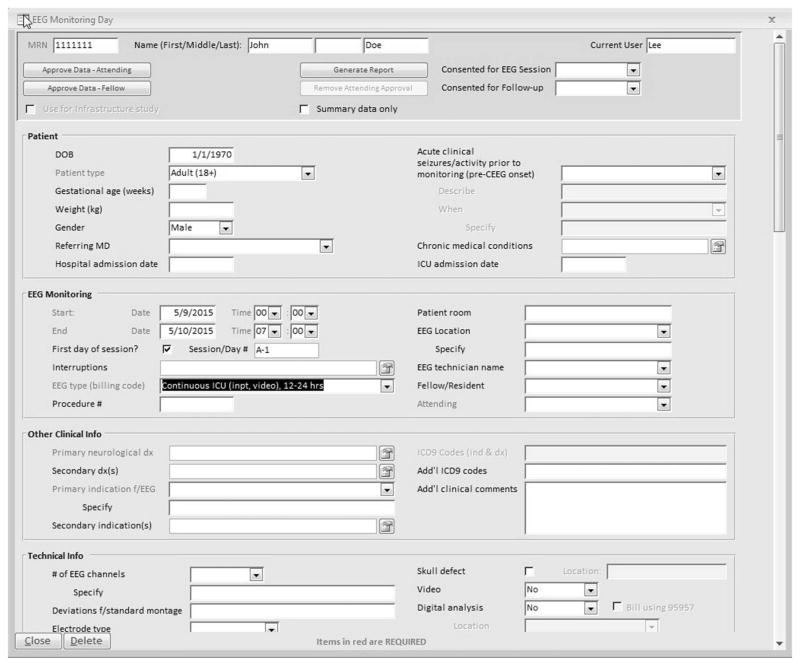
The main patient data entry screen initially consists of four subsections that can be completed by a technologist: (1) Patient (demographics and admission dates); (2) Monitoring date/time/ location, and assignment of technologist, fellow, and attending physician; (3) Other clinical information including neurological diagnosis and indication for monitoring; and (4) Technical details regarding montage and type of electrodes used (Fig. 1). Within a specific day, the record may consist of several monitoring epochs if there are significant EEG changes from baseline (e.g., start of burst suppression). Generally, 1 recording day consists of a single epoch.
FIG. 1.
Database user interface: demographics, clinical, and technical information entry screen.
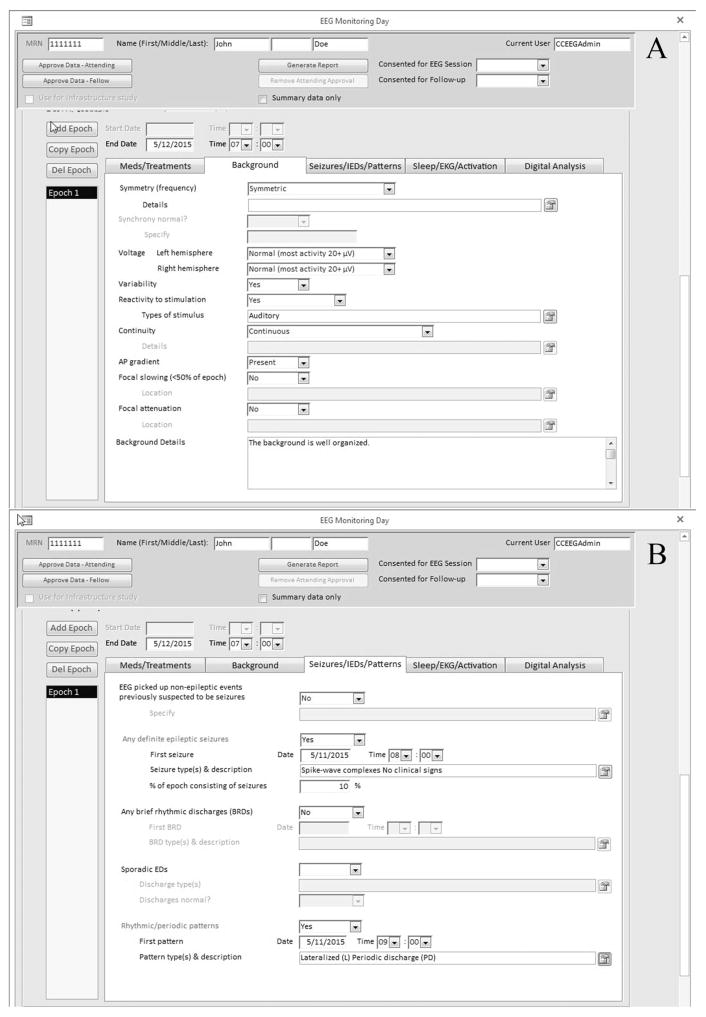
Within each recording day or epoch, there are further descriptions of medication, treatments, EEG background, seizures, sporadic interictal epileptiform discharges, and periodic or rhythmic patterns; additional information such as sleep, electrocardiogram, and activation procedures can be described and also digital analysis (quantitative EEG) (Fig. 2) by clicking the appropriate tab. Daily and end-of-recording summaries consisting of an impression and clinical correlations are entered as free text. Although the database encourages selection of preset values for data consistency, there are free text entry fields to describe the EEG background and seizure description, if needed. Finally, the database allows for assignment of reports to a specific technologist, clinical fellow, and attending neurophysiologist, and also dual attestation by the clinical fellow and attending physician with customizable wording and workflow requirements.
FIG. 2.
Database user interface: EEG results entry screen: (A) background; (B) seizures, interictal discharges, rhythmic/periodic patterns, others.
To achieve its primary function as a research repository for the CCEMRC, secure data sharing capabilities were implemented. From within the database interface file, data can be deidentified, encrypted, and transferred through the Internet to a central data repository, provided that interinstitutional data sharing agreements and local institutional review board approvals are in place. The database has Structured Query Language capabilities accessible both locally and from the central data repository to easily search for records satisfying specific criteria, facilitating local and multicenter research and quality improvement initiatives. More detailed data analysis can be performed by downloading either portions or the entire database as data tables that can be imported into analysis software including Matlab (The MathWorks, Natick, MA) and R (The R Foundation, Vienna, Austria). If a patient is part of a study, the interface file can be modified to highlight fields that are required to be completed for that particular study.
A second major function of the database is the ability to function as a clinical reporting tool. Particular attention was paid to form design and workflow integration to promote ease-of-use. The database can accommodate data entry from technologists, clinical fellows, and attending clinical neurophysiologists, each of whom can be tracked separately for incomplete reports. A large number of fields may be autogenerated with institution-specific preferences. Report generation can be optimized to comply with the local regulatory requirements, such as report headers and attestation requirements. The output is formatted as a text file that can be printed or copied into the patient’s electronic medical record.
Multicenter Collaborative Study
To assess the feasibility of data collection across a large number of institutions, members of the CCEMRC were invited to enter and submit data on 12 or more consecutive adult and/or pediatric subjects (aged >28 days) who underwent ≥12 hours of continuous EEG monitoring in intensive care units at their institution during the year 2014. Subjects who underwent elective EEG monitoring for epilepsy evaluation were excluded. All participating centers had completed data sharing agreements in place and obtained institutional review board approval.
Data quality was ensured by limiting EEG data entry to consortium members who had completed a training module and certification examination on the appropriate use of the American Clinical Neurophysiology Society (ACNS) standardized cEEG terminology (Gaspard et al., 2014). A minimum of 26 data fields were requested for each subject. The following variables of interest were analyzed on a per-subject basis: duration of cEEG recording, frequency of seizures or status epilepticus, frequency of rhythmic or periodic patterns, and characterization of epileptiform discharges, if present.
RESULTS
Multicenter Collaborative Study
All 21 participating CCEMRC centers were able to install the database, enter data locally, and upload deidentified data to the central repository, contributing a total of 280 subjects of which 41.8% were female and 23% were under age 18 (Table 1). Of the 6,048 requested data points, 99.98% were completed by participating centers.
TABLE 1.
Characteristics of Subjects and cEEG Recordings (n = 280)
| Characteristics | Values |
|---|---|
| Subject characteristics | |
| Female, % | 41.8 |
| Age, median (IQR), years | 52.5 (20.4–65.8) |
| Children (age <18), % | 23 |
| cEEG recording characteristics | |
| Duration of recording, median (IQR), hours | 25.5 (19.0–46.2) |
| Days recorded with video, % | 72.1 |
| Days recorded with quantitative EEG analysis, % | 57.7 |
| Prevalence of cEEG abnormalities, % | |
| Seizures | 17.6 |
| Rhythmic or periodic patterns | 35.7 |
cEEG, critical care electroencephalogram; IQR, interquartile range.
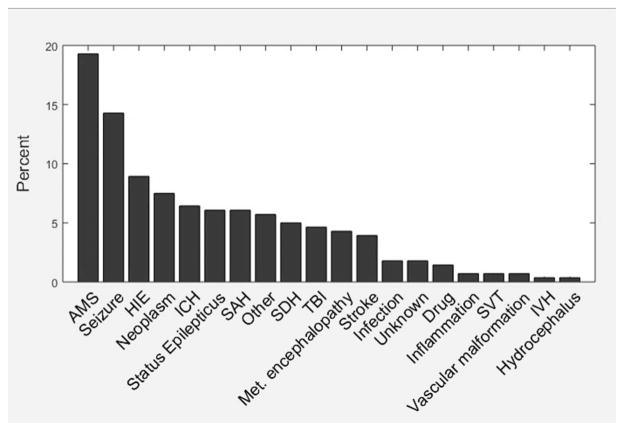
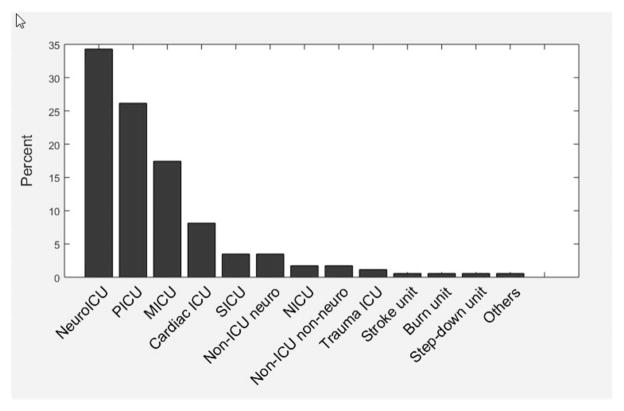
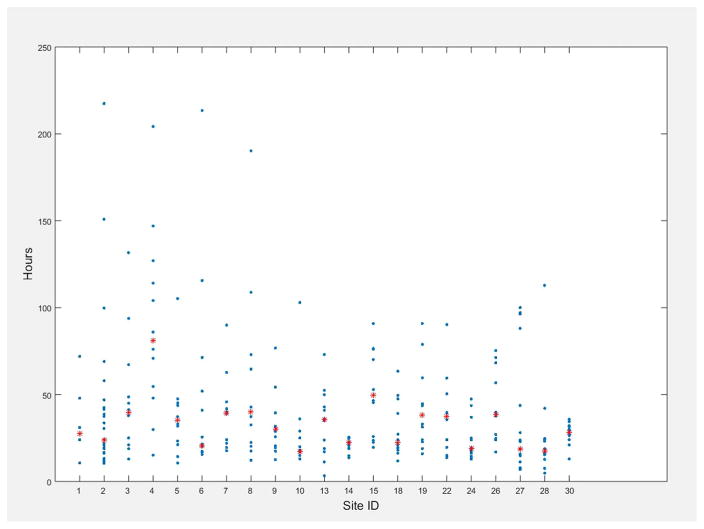
The primary indication for continuous EEG monitoring is shown in Fig. 3. The most common primary indication was alteration in mental status (19.3%) followed by the detection of electrographic seizures (14.3%). Approximately, one third (34.3%) of the recordings occurred in a dedicated neurological intensive care unit (Fig. 4). The median duration of EEG recording was 25.5 hours (interquartile range, 19.0–46.2). The duration of EEG recording varied significantly by site but not by indication (Fig. 5, two-factor analysis of variance, main effect for site F(20,240) = 1.91, P = 0.01; main effect for indication F(19,240) = 0.8, P = 0.71; insufficient data to calculate interaction). There was no significant correlation between subject age and duration of monitoring (r2 = 0.534; P = 0.37).
FIG. 3.
Indication for critical care EEG monitoring in 280 consecutive patients from 21 participating Critical Care EEG Monitoring Research Consortium centers. AMS, altered mental status; HIE, hypoxic ischemic encephalopathy; ICH, intracranial hemorrhage; IVH, intraventricular hemorrhage; Met. Encephalopathy, metabolic encephalopathy; SAH, subarachnoid hemorrhage; SDH, subdural hemorrhage; SVT, sinus venous thrombosis; TBI, traumatic brain injury.
FIG. 4.
Location of critical care EEG monitoring among participating Critical Care EEG Monitoring Research Consortium centers. MICU, medical intensive care unit; NICU, neonatal intensive care unit; PICU, pediatric intensive care unit; SICU, surgical intensive care unit.
FIG. 5.
Duration of EEG monitoring among participating Critical Care EEG Monitoring Research Consortium centers (numbers assigned at random). Red stars represent the median duration of monitoring.
Clinical seizures before their cEEG study were noted in 79 subjects (28.2%) of whom 20 (25.3%) subsequently had seizures during the cEEG study. Clinical or electrographic seizures during cEEG recording were identified in a total of 37 subjects (17.6%) and present in the recordings from 15 of 21 sites. Among subjects who experienced seizures, 10 subjects (27.0%) had generalized seizures (tonic, clonic, tonic–clonic, or myoclonic), 3 subjects (8.1%) had focal seizures, 1 subject (2.7%) had subtle seizures, 4 subjects (10.8%) had seizures not otherwise specified, and 19 (51.4%) had nonconvulsive seizures; status epilepticus was present in 14 subjects (37.8%). Rhythmic or periodic patterns were seen in 75 subjects (35.7%) in the recordings from 16 sites, ranging from 16.7% to 81.3% of subjects at those sites. Brief rhythmic discharges were present in 17 subjects (8.1%) and present in the recordings from 8 sites.
Epileptiform discharges were noted in 33 subjects (11.8%). Among subjects with epileptiform discharges, they were classified as lateralized (67%), generalized (16%), multifocal (13%), and bilateral independent (4%) discharges. Discharge frequency was abundant in 4.2% (greater than 1 per 10 seconds), frequent in 31.9% (greater than 1/minute but less than 1/10 seconds), occasional in 48.9% (greater than 1/hour but less than 1/minute), and rare in 14.9% (less than 1/hour).
Clinical Reporting Tool
The database has thus far been successfully implemented as a clinical reporting tool at three consortium sites (Brigham and Women’s Hospital, Emory University, Yale University). At each institution, the clinical fellow was required to take and pass the 37-item certification test at the start of the academic year. A total of 3,144 clinical patient entries have been uploaded to the central repository as of August 5, 2015 (Brigham and Women’s Hospital [BWH]: 1,374; Emory 1,138; Yale 632), totaling 6,665 recording days. The database was sufficiently flexible to account for differences in reporting requirements (Yale and Emory required daily reporting and BWH required a single report covering the entire recording period) and regulatory concerns, such as institutional differences in attestation statements.
DISCUSSION
We describe the creation and implementation of the CCEMRC database with the dual role as a multicenter research data repository and a daily clinical reporting tool. The software has undergone several rounds of testing and is now available as a stable release.
Critical Care EEG Monitoring Research Consortium Database as a Collaborative Research Tool
We have demonstrated the potential for the CCEMRC database to be used as a collaborative research tool across 21 CCEMRC centers. All participating centers were able to install the database locally, enter at least 12 subjects, and securely transmit deidentified data to the central repository for analysis. This has yielded standardized high-quality data on critically ill patients undergoing cEEG monitoring across a large number of CCEMRC member centers, which will serve as valuable preliminary data to inform the design of future prospective studies. Our finding that seizures were present in 17.6% of patients and rhythmic/periodic discharges were present in 35.7% is consistent with previous reports (Claassen et al., 2004).
The CCEMRC database represents an ideal platform for future collaborative studies because it allows for dynamic addition of new studies, each with their own specific required and optional data elements. Furthermore, the database may be easily cross-indexed with another database housing non-EEG data elements such as more detailed clinical or neuroimaging data. Currently, two CCEMRC studies are using the CCEMRC database to collect EEG data, which is then cross-indexed or interfaced with other databases such as the REDCap (Research Electronic Data Capture) (Harris et al., 2009) database system.
Critical Care EEG Monitoring Research Consortium Database as a Clinical Tool
When institutions commit to provide a cEEG monitoring service, large volumes of data need to be collected, analyzed, and stored. The CCEMRC database presents several advantages for use as a daily clinical cEEG reporting tool. By incorporating the latest ACNS standardized terminology for cEEG monitoring, the database facilitates standardization of reporting. This structure is particularly useful as a teaching tool for fellows in training and for clinicians who less frequently participate in cEEG monitoring and may not be as familiar with the terminology. Furthermore, because cEEG monitoring studies may extend for several days and may require interpretation by multiple physicians, the database helps standardize reporting, potentially improving reporting consistency across multiple readers. The workflow tools are particularly robust, allowing technologists, fellows, and attendings to edit and authenticate the report in stages. In practice, the database has proven itself to function as a daily clinical reporting tool at three institutions performing a high volume of cEEG monitoring.
Several barriers to widespread adoption of the database as a clinical reporting tool have been identified. These included (1) limitations imposed by local electronic medical record systems that prohibit the generated EEG report from being copied into the medical record, despite its capability to export formatted or unformatted text; (2) lack of approval by local institutional review boards to enter clinical data into the database because it was designed by an outside party (despite its full HIPAA compliance); (3) lack of full integration into electronic medical records, resulting in duplication of work as a result of repeated entry of demographic information; (4) database learning curve; (5) clinicians being unfamiliar or uncomfortable with the ACNS standardized terminology for cEEG monitoring; (6) dissatisfaction with template data entry or with the template generated reports. Nevertheless, the experience of the three centers using the CCEMRC database as a daily clinical reporting tool indicates that the database can be time-saving in the long term.
Growth of cEEG monitoring has been rapid (Gavvala et al., 2014; Sanchez et al., 2013), resulting in recent consensus statements about its use (Herman et al., 2015a, 2015b). At a given institution, the workload of cEEG monitoring is typically divided between multiple neurophysiologists, who often have varying interest and expertise in cEEG monitoring, with resulting variability in their willingness to adopt the new standardized ACNS standardized cEEG terminology. For example, terms such as “periodic lateralized epileptiform discharges” and “triphasic waves” are likely to persist in common use. Nevertheless, the revised ACNS terminology offers distinct advantages: EEG pattern description and clinical correlation are kept separate (Hirsch et al., 2013), and interrater reliability has been validated (Gaspard et al., 2014; Mani et al., 2012). Although some clinicians may feel constrained by the predefined elements of the database, the alternative of entering a report in a free-text format or a point-by-point short form introduces local institution-specific terminologies and has the potential to reduce interrater reliability. We believe that standardized data entry and reporting supersedes the value of potentially more readable narrative free-text report because it ensures consistency in report writing within and across different institutions. The presence of multiple locations for free-text data entry in “additional description” fields, the final impression, and the clinical correlation provides ample opportunities to complement standardized reporting with narrative reports.
CONCLUSIONS
The CCEMRC database has been successfully developed and implemented as both a collaborative research platform to collect multicenter standardized EEG data and a clinical reporting tool. The database and instruction manual are available as a free of charge download to be used as a clinical data repository and report generating tool at https://www.acns.org/research/critical-care-eeg-monitoring-research-consortium-ccemrc.
Acknowledgments
This work was supported by research infrastructure awards by the American Epilepsy Society and Epilepsy Foundation of America.
L. J. Hirsch: Research support to Yale for investigator initiated studies from UCB-Pharma, Upsher-Smith, Lundbeck, Eisai and Sunovion, consultation fees for advising from Lundbeck, Upsher-Smith, Neuropace, and GSK, honoraria for speaking from Neuropace (last 12/2014), royalties for authoring chapters for UpToDate-Neurology, chapters for Medlink-Neurology, and from Wiley for coauthoring the book “Atlas of EEG in Critical Care,” by Hirsch and Brenner, 2010; Dr. Hirsch spends about 25% of his clinically billable time implementing and interpreting critical care EEG studies.
N. S. Abend: NINDS (NIH NS076550); H. Choi: UCB-Pharma, Lundbeck, Sunovion, and Eisai (research support); K. E. Dombrowski: Sage Therapeutics (salary support); E. Fertig: UCB (research funding), Acorda Therapeutics Inc (research funding), and Marinus Pharmaceuticals (research funding); Gerard: NINDS (research funding), SAGE pharmaceuticals (site-PI), and UCB/Duke (research consultant work); C. D. Hahn: Canadian Institutes of Health Research, the Physicians’ Services Incorporated Foundation, and The Hospital for Sick Children Foundation (research support); S. Herman: Biotie, Inc (consultant), UCB Pharma, Lundbeck, Inc, National Institutes of Health (NIH)/National Institute of Neurological Disorders and Stroke (NINDS), Fidelity Biosciences Research Initiative, and Epilepsy Therapy Development Project (research support); L. J. Hirsch: Research support to Yale for investigator-initiated studies from UCB-Pharma, Upsher-Smith, Lundbeck, Eisai and Sunovion, consultation fees for advising from Lundbeck, Upsher-Smith, Neuropace, and GSK, honoraria for speaking from Natus (last 1/2014) and Neuropace (last 12/2014), royalties for authoring chapters for UpToDate-Neurology, chapters for Medlink-Neurology, and from Wiley for coauthoring the book “Atlas of EEG in Critical Care,” by Hirsch and Brenner, 2010, spends about 25% of his clinically billable time implementing and interpreting critical care EEG studies; J. L. Hopp: NETT study member, UpToDate (royalties); S. LaRoche: Demos Publishing (royalties); J. W. Lee: NINDS (research funding), Advance Medical and SleepMed/ DigiTrace (consultant/contract work), UCB (research funding), and Sunovion (research funding); T. Loddenkemper: T. Loddenkemper American Epilepsy Society, Epilepsy Foundation of America, Epilepsy Therapy Project, PCORI, Pediatric Epilepsy Research Foundation, CURE, Danny-Did Foundation, HHV-6 Foundation, Lundbeck, Eisai, and Upsher-Smith (research support); S. R. Sinha: UCB (research funding), Acorda Therapeutics Inc (research funding), Biogen (research funding), Sage Therapeutics (research funding), and Cyberonics Inc (research funding, speakers bureau); J. P. Szaflarski: Funding: NIH, Shor Foundation for Epilepsy Research, AES, EFA, FDA, State of AL General Funds, DoD, Epilepsy Study Consortium (HEP), UAB; Editorial Boards: Associate Editor, Journal of Epileptology; Associate Editor: Restorative Neurology and Neuroscience; Editorial Board Member: Folia Medica Copernicana, Journal of Medical Science; Epilepsy and Behavior; Consultant: Sage Pharmaceuticals; Biomedical Systems, Inc, Medico-legal expert; and M. B. Westover: American Brain Foundation, NIH-NINDS (1K23NS090900), and the Rappaport Foundation (research support).
We would like to thank the following CCEMRC investigators for their commitment to this database: Linda Huh, MD (British Columbia Children’s Hospital), Jessica Carpenter, MD (Children’s National Medical Center), Stephen Hantus, MD (Cleveland Clinic Epilepsy Center) Jan Claassen, MD, PhD (Columbia University Medical Center), Aatif M. Husain, MD (Duke University Medical Center), Nicolas Gaspard, MD, PhD (Hôpital Erasme, Université Libre de Bruxelles), David Gloss, MD (Geisinger Medical Center), Tennile Gofton, MD (London Health Science Centre), Sara Hocker, MD (Mayo Clinic), Jonathan Halford, MD (Medical University of South Carolina), Jennifer Jones, MD (Mission Hospital), Korwyn Williams, MD, PhD (Phoenix Children’s Hospital), Andreas Kramer, MD (University of Calgary), Brandon Foreman, MD (University of Cincinnati), Leslie Rudzinski, MD (University of Florida), Rup Sainju, MD (University of Iowa Carver School of Medicine), Ram Mani, MD (University of Medicine and Dentistry New Jersey), Sarah E. Schmitt, MD (University of Pennsylvania), Giridhar P Kalamangalam, MD, DPhil (University of Texas Health Science Center – Houston), Puneet Gupta, MD, MSE (University of Texas – Southwestern Medical Center), Mark S. Quigg, MD (University of Virginia School of Medicine), and Adam Ostendorf, MD (Washington University).
References
- Aurlien H, Gjerde IO, Gilhus NE, et al. A new way of building a database of EEG findings. Clin Neurophysiol. 1999;110:986–995. doi: 10.1016/s1388-2457(99)00037-1. [DOI] [PubMed] [Google Scholar]
- Beniczky S, Aurlien H, Brogger JC, et al. Standardized computer-based organized reporting of EEG: SCORE. Epilepsia. 2013a;54:1112–1124. doi: 10.1111/epi.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beniczky S, Hirsch LJ, Kaplan PW, et al. Unified EEG terminology and criteria for nonconvulsive status epilepticus. Epilepsia. 2013b;54(suppl 6):28–29. doi: 10.1111/epi.12270. [DOI] [PubMed] [Google Scholar]
- Chong DJ, Hirsch LJ. Which EEG patterns warrant treatment in the critically ill? reviewing the evidence for treatment of periodic epileptiform discharges and related patterns. J Clin Neurophysiol. 2005;22:79–91. doi: 10.1097/01.wnp.0000158699.78529.af. [DOI] [PubMed] [Google Scholar]
- Claassen J, Mayer SA, Kowalski RG, et al. Detection of electrographic seizures with continuous EEG monitoring in critically ill patients. Neurology. 2004;62:1743–1748. doi: 10.1212/01.wnl.0000125184.88797.62. [DOI] [PubMed] [Google Scholar]
- Finnerup NB, Fuglsang-Frederiksen A, Rossel P, et al. A computer-based information system for epilepsy and electroencephalography. Int J Med Inform. 1999;55:127–134. doi: 10.1016/s1386-5056(99)00002-7. [DOI] [PubMed] [Google Scholar]
- Gaspard N, Foreman B, Judd LM, et al. Intravenous ketamine for the treatment of refractory status epilepticus: a retrospective multicenter study. Epilepsia. 2013a;54:1498–1503. doi: 10.1111/epi.12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspard N, Manganas L, Rampal N, et al. Similarity of lateralized rhythmic delta activity to periodic lateralized epileptiform discharges in critically ill patients. JAMA Neurol. 2013b;70:1288–1295. doi: 10.1001/jamaneurol.2013.3475. [DOI] [PubMed] [Google Scholar]
- Gaspard N, Hirsch LJ, LaRoche SM, et al. Interrater agreement for critical care EEG terminology. Epilepsia. 2014;55:1366–1373. doi: 10.1111/epi.12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavvala J, Abend N, LaRoche S, et al. Continuous EEG monitoring: a survey of neurophysiologists and neurointensivists. Epilepsia. 2014;55:1864–1871. doi: 10.1111/epi.12809. [DOI] [PubMed] [Google Scholar]
- Harris PA, Taylor R, Thielke R, et al. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman ST, Abend NS, Bleck TP, et al. Consensus statement on continuous EEG in critically ill adults and children, part I: indications. J Clin Neurophysiol. 2015a;32:87–95. doi: 10.1097/WNP.0000000000000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman ST, Abend NS, Bleck TP, et al. Consensus statement on continuous EEG in critically ill adults and children, part II: personnel, technical specifications, and clinical practice. J Clin Neurophysiol. 2015b;32:96–108. doi: 10.1097/WNP.0000000000000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch LJ, LaRoche SM, Gaspard N, et al. American clinical neurophysiology Society’s standardized critical care eeg terminology: 2012 version. J Clin Neurophysiol. 2013;30:1–27. doi: 10.1097/WNP.0b013e3182784729. [DOI] [PubMed] [Google Scholar]
- Kilbride RD, Reynolds AS, Szaflarski JP, et al. Clinical outcomes following prolonged refractory status epilepticus (PRSE) Neurocrit Care. 2013;18:374–385. doi: 10.1007/s12028-013-9823-4. [DOI] [PubMed] [Google Scholar]
- Mani R, Arif H, Hirsch LJ, et al. Interrater reliability of ICU EEG research terminology. J Clin Neurophysiol. 2012;29:203–212. doi: 10.1097/WNP.0b013e3182570f83. [DOI] [PubMed] [Google Scholar]
- Perucca P, O’Brien TJ. Epilepsy in 2014: novel and large collaborations drive advances in epilepsy. Nat Rev Neurol. 2015;11:74–76. doi: 10.1038/nrneurol.2014.255. [DOI] [PubMed] [Google Scholar]
- Sanchez SM, Carpenter J, Chapman KE, et al. Pediatric ICU EEG monitoring: current resources and practice in the United States and Canada. J Clin Neurophysiol. 2013;30:156–160. doi: 10.1097/WNP.0b013e31827eda27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt SE, Pargeon K, Frechette ES, et al. Extreme delta brush: a unique EEG pattern in adults with anti-NMDA receptor encephalitis. Neurology. 2012;79:1094–1100. doi: 10.1212/WNL.0b013e3182698cd8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinka E, Leitinger M. Which EEG patterns in coma are nonconvulsive status epilepticus? Epilepsy Behav. 2015;49:203–222. doi: 10.1016/j.yebeh.2015.05.005. [DOI] [PubMed] [Google Scholar]
- Walker MC, Kovac S. Seize the moment that is thine: how should we define seizures? Brain. 2015;138:1127–1128. doi: 10.1093/brain/awv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo JY, Rampal N, Petroff OA, et al. Brief potentially ictal rhythmic discharges in critically ill adults. JAMA Neurol. 2014;71:454–462. doi: 10.1001/jamaneurol.2013.6238. [DOI] [PubMed] [Google Scholar]
- Young GB, Jordan KG, Doig GS. An assessment of nonconvulsive seizures in the intensive care unit using continuous EEG monitoring: an investigation of variables associated with mortality. Neurology. 1996;47:83–89. doi: 10.1212/wnl.47.1.83. [DOI] [PubMed] [Google Scholar]