Abstract
In DNA from prostate tumors, methylation patterns in gene promoter regions can be a biomarker for disease progression. It remains unclear whether methylation patterns in benign prostate tissue—prior to malignant transformation—may provide similar prognostic information. To determine whether early methylation events predict prostate cancer outcomes, we evaluated histologically benign prostate specimens from 353 men who eventually developed prostate cancer and received “definitive” treatment [radical prostatectomy (58%) or radiation therapy (42%)]. Cases were drawn from a large hospital-based cohort of men with benign prostate biopsy specimens collected between 1990 and 2002. Risk of disease progression associated with methylation was estimated using time-to-event analyses. Average follow-up was over 5 years; biochemical recurrence (BCR) occurred in 91 cases (26%). In White men, methylation of the APC gene was associated with increased risk of BCR, even after adjusting for standard clinical risk factors for prostate cancer progression (adjusted hazard ratio (aHR) = 2.26; 95%CI 1.23–4.16). APC methylation was most strongly associated with a significant increased risk of BCR in White men with low prostate specific antigen at cohort entry (HR = 3.66; 95%CI 1.51–8.85). In additional stratified analyses, we found that methylation of the RARB gene significantly increased risk of BCR in African American cases who demonstrated methylation of at least one of the other four genes under study (HR = 3.80; 95%CI 1.07–13.53). These findings may have implications in the early identification of aggressive prostate cancer as well as reducing unnecessary medical procedures and emotional distress for men who present with markers of indolent disease.
Keywords: biopsy, DNA methylation, biomarkers, field cancerization, disease recurrence
Prostate cancer is the most common noncutaneous cancer in men in the United States, with the diagnoses projected to increase 40% between 2010 and 2020.1 Prostate cancer is characterized by its marked multi-focal nature—67–96% of prostatectomy specimens display more than one tumor focus.2,3 This suggests the presence of a “field defect,” whereby factors underlying carcinogenesis result in molecular changes throughout large areas of the prostate.4 Such changes are measurable in histologically normal cells and generally precede overt carcinogenesis. Studies of tumor-adjacent benign tissue have found alterations in gene expression,5,6 telomere DNA content,7 gene copy number 8 that mimic a malignant phenotype. In addition, often there is an increased prevalence of methylation in genes such as APC,9 GSTP110 and RARB2 9,10 known to be methylated in prostate cancer. The mechanisms underlying these alterations are not clearly understood, but may be explained by one or more factors including extended effects of carcinogens, lateral expansion of the tumor or influence of the tumor on the adjacent tissue. All these fall under the general rubric of field cancerization and have clinical relevance in terms of early detection and treatment of prostate cancer.
Aberrant methylation may arise from any number of insults to the entire prostate, incidental or cumulative, many resulting in transcriptional silencing and inactivation of genes that regulate critical cell processes and is considered an early landmark event in carcinogenesis.11 Epigenetic changes are thought to occur progressively in prostate cancer, and DNA methylation may also be involved in the regulation of biologic pathways leading to metastasis,12,13 but it remains unclear at what stage these changes have prognostic significance.14 Panels of methylation markers are emerging that demonstrate added utility for prediction of disease progression beyond standard clinical parameters,15,16 but results vary across studies. Two recent studies both showed methylation of glutathione S-transferase pi 1 (GSTP1), a gene where methylation has long been associated with presence of prostate cancer, also may have predictive value in disease recurrence.17,18 To date, all studies that have examined gene promoter methylation as a risk factor for disease progression have assessed methylation at the time of disease diagnosis. None have tested whether methylation that occurs before disease onset has any prognostic value.
Methylation changes are evident in non-neoplastic cells adjacent to prostate tumors, even as far as 10–20 mm from the primary tumor.19 Troung et al.20 successfully used the methylation status of two genes (EVX1 and FGF1) in tumor-associated benign biopsy specimens to discriminate between men with and without prostate cancer. Globally, methylation patterns in tumor-adjacent benign tissue are remarkably similar to the nearby cancer.21,22 Methylation patterns that distinguish tumor from normal prostate tissue, but that can be found in tumor adjacent benign tissue, may also have utility in predicting prostate cancer relapse.21 We found both methylation of either the APC or RARB gene in benign prostate increased risk for subsequent prostate cancer;23 other studies have shown that methylation of APC 24,25 and RARB 26 may predict biochemical recurrence (BCR) of cancer.
The only published study of gene methylation in nontumor tissue and prostate cancer progression found that, after adjusting for Gleason score, methylation of both APC and GSTP1 in tumor-adjacent benign tissue was associated with a 2.4-fold increased risk of death from prostate cancer.27 Notably, methylation of these genes in tumor-adjacent benign tissue correlated with methylation status in tumor. We sought to determine whether aberrant gene promoter methylation that occurs prior to the onset of clinically detectable prostate cancer would predict BCR in prostate cancer cases that received definitive treatment. Using data from a previously conducted matched case-control study nested within a large historical cohort,23 we asked whether gene promoter methylation in prediagnostic benign tissue was associated with BCR, and whether associations between methylation status and BCR were race-specific.
Methods
Study sample
After obtaining approval from the Henry Ford Health System Institutional Review Board, a historical cohort of 6,692 men with a benign prostate specimen collected by needle core biopsy or transurethral resection of the prostate (TURP) between January 1990 and December 2002 was identified. Eligibility criteria included a recorded prostate specific antigen (PSA) level within a year of cohort entry and no history of a previous prostate cancer diagnosis. “Date of cohort entry” was defined as the date of the initial benign prostate specimen collection; “date of case diagnosis” was the date of first cancer-positive prostate tissue histology. Only cases diagnosed with prostate cancer one or more years from date of cohort entry were eligible for the study.
We identified 617 potentially eligible cases diagnosed with prostate cancer prior to July 2007.28 Of these 617 cases, we measured methylation levels for at least one gene for 554 cases. For the cases with methylation data, we then reviewed medical chart follow-up data to determine which cases had both definitive primary treatment (radical prostatectomy or external beam radiation therapy) and a minimum of two or more PSA tests after treatment, resulting in an analytic sample of 353 cases. The percentage of missing methylation data or positive methylation for any of the genes under study did not differ significantly between excluded and included cases. For these 353 cases, 58% underwent radical prostatectomy, while 42% received radiation. For surgical patients, BCR was defined as a postsurgery undetectable PSA followed by two consecutive detectable (>0.2 ng/ml) rising PSA levels four weeks or more postsurgery.29 For radiation-treated patients, we used the Phoenix criteria (PSA increase ≥2 ng/ml above nadir).30
Clinical and demographic data were abstracted from case medical records from five years before the date of cohort entry through the date of diagnosis. Data on all PSA tests were recorded and the PSA test value immediately prior to the initial benign biopsy was used as the PSA level at time of cohort entry. The PSA test value closest to, but preceding, the date of diagnosis, was considered the PSA level at time of diagnosis. All surgical specimens were reviewed for the absence of malignancy and presence of inflammation by a single urological pathologist (ONK) blinded to outcomes 28 (Fig. 1). Inflammation in biopsy specimens was classified by grade (mild, moderate, severe), extent (focal, multifocal, diffuse) and compartmental location (stromal, periglandular, glandular).31 For the purposes of analysis, we dichotomized inflammation as present or absent where inflammation of any grade or extent and in any location was considered positive.
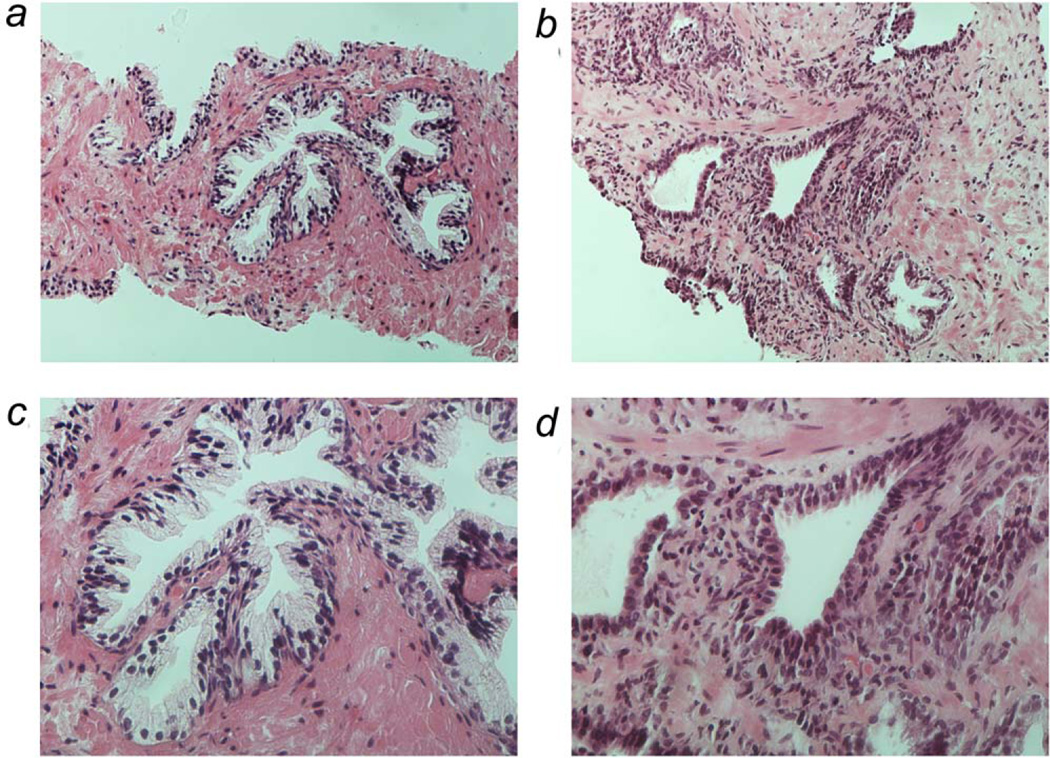
Figure 1.
Representative histology of benign prostate specimens used in study. Photographs taken at 20 × (panel A and B) and 40 × (panel C and D) of two different prostate specimens. Panels A and C depict normal prostate glandular architecture of a biopsy specimen of a 61 year old African American man. Panels on B and D depict prostate glands of biopsy specimen of a 79 year old white man with significant atrophy and inflammation. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
DNA isolation and methylation-specific PCR
Methylation studies were conducted using DNA from benign prostate specimens acquired by needle biopsy or TURP. Five-micron sections of paraffin-embedded prostate tissues were processed as previously described.23 Nested methylation-specific PCR was used to detect methylation in ~200 base pair promoter regions of the RARB, APC, CTNND2, RASSF1 and MGMT genes. Stage I primers were multiplexed to maximize DNA recovered from tissue (<200 ng for most biopsies). Because Stage I primers recognize bisulfite-modified templates but do not discriminate between methylated and unmethylated alleles, the PCR product from Stage I was diluted and then subjected to a second PCR (Stage II) with primers specific for a methylated template located around the transcriptional start site, where methylation—if present—is strongly correlated with gene silencing. Visualization of a gel band from stage II primer PCR product was considered positive methylation. Absence of gel band from stage II primer PCR product, but visualization of gel band from a stage I primer PCR product was considered negative methylation. Absence of both stage I and II PCR product bands was considered missing data. To ensure that only methylated alleles were amplified, Stage II PCR reactions were performed at annealing temperatures greater than the melting temperature of the Stage I primers. Sensitivity was approximately 1 in 500 even with the nested methylation-specific PCR approach, due to tissue degradation from formalin fixation and storage in paraffin.
Statistical analysis
χ2 tests and two sample t-tests were used to compare differences in demographic, clinical characteristics and methylation status with respect to BCR. Additional analyses that considered time to BCR included log-rank tests and Cox proportional hazard models that analyzed the difference between DNA methylation categories with respect to risk of BCR. Time to BCR was defined as the duration between the date of surgery or radiation and the second PSA test that defined the recurrence event or censored at the last postoperative PSA test for men that did not recur. Models were adjusted for tumor stage, advanced tumor grade, PSA level at cancer diagnosis, treatment type and age at cancer diagnosis. Stratified models were compared using a Cox proportional hazard model that included interaction terms with the stratified variable.
Results
Demographic and clinical characteristics of our analytic sample are shown in Table 1. Among the 353 cases, 91 (25.8%) recurred during the course of follow-up, with BCR events occurring between 2.3 and 167.6 months of follow-up with a median recurrence time of 22.5 months. Cases that did not recur were followed between 1 month and 19 years, with a median follow-up of 75.5 months. As expected, recurrent cases had higher PSA levels both at cohort entry and diagnosis as well as higher PSA velocity prior to diagnosis. Recurrent cases were also more likely to demonstrate higher Gleason grade (8–10) tumors than nonrecurrent cases.
Table 1.
Demographic and clinical characteristics of prostate cancer cases by recurrence status
| Variable | Response | Nonrecurrent cases (N = 262) |
Recurrent cases (N = 91) |
p values |
|---|---|---|---|---|
| Race | White | 152 (58%) | 54 (59%) | 0.825 |
| African American | 110 (42%) | 37 (41%) | ||
| Age at cohort entry | (n = 262) 63.7 ± 6.8 | (n = 91) 64.5 ± 6.8 | 0.325 | |
| Age at diagnosis | (n = 262) 68.3 ± 7.0 | (n = 91) 68.8 ± 7.4 | 0.515 | |
| Median Date of Cohort Entry | 07/11/1995 | 09/15/1994 | 0.424 | |
| Median Time until Dx in years | 3.84 | 3.78 | 0.607 | |
| Median time until recur- rence/last follow-up (months) | 75.48 | 22.54 | <.001 | |
| Type of benign specimen | Biopsy | 247 (94%) | 85 (93%) | 0.763 |
| Transurethral Resection of the Prostate | 15 (6%) | 6 (7%) | ||
| PSA level at cohort entry (ng/ml) | (n = 262) 6.4 ± 3.6 | (n = 91) 7.8 ± 5.8 | 0.033 | |
| PSA level at diagnosis (ng/ml) | (n = 260) 9.2 ± 7.3 | (n = 91) 17.4 ±28.9 | 0.009 | |
| PSA Velocity (ng/ml per year) | (n = 257) 1.0 ± 1.9 | (n = 90) 1.6 ±4.0 | 0.154 | |
| Number of PSA test prior to diagnosis | (n = 262) 9.2 ± 5.5 | (n = 91) 9.0 ±5.1 | 0.712 | |
| Inflammation Present | 147 (57%) | 48 (53%) | 0.508 | |
| High Grade Prostatic Intra- epithelial Neoplasia present | 29 (11%) | 5 (5%) | 0.114 | |
| Type of treatment | Prostatectomy | 153 (58%) | 53 (58%) | 0.979 |
| Radiation | 109 (42%) | 38 (42%) | ||
| Tumor Stage | 1 | 72 (27%) | 24 (26%) | 0.337 |
| 2 | 168 (64%) | 54 (59%) | ||
| 3 | 21 (8%) | 13 (14%) | ||
| 4 | 1 (0%) | 0 (0%) | ||
| Gleason Grade | <6 | 136 (54%) | 27 (31%) | <.001 |
| 7 (3 + 4) | 53 (21%) | 18 (21%) | ||
| 7 (4 + 3) | 29 (11%) | 12 (14%) | ||
| 8–10 | 36 (14%) | 30 (34%) |
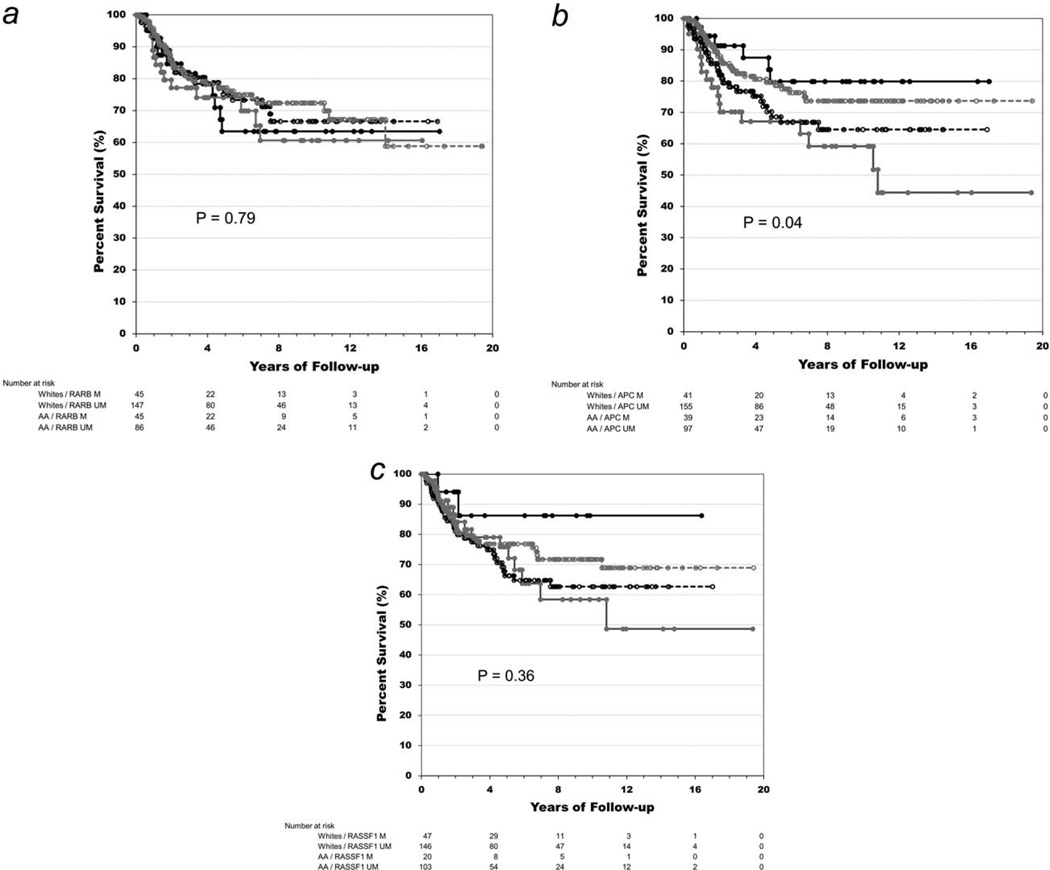
Of the five genes studied, RARB, APC and CCND2 had slightly higher percentages of positive methylation among recurrent cases, but methylation status was not significantly different for any of the five genes between recurrent and nonrecurrent cases (Table 2). When we stratified by race, we found APC methylation was significantly higher among Whites with BCR (p = 0.006) and RASSF1 methylation was marginally higher in nonBCR African Americans (20% vs. 6%; p = 0.06). Using time-to-BCR as the outcome variable, we next estimated crude and adjusted hazard ratios for the three genes with the highest methylation percentage (RARB, APC and RASSF1) in both the full and race-stratified samples (Table 3). APC methylation demonstrated the strongest association with BCR in Whites in both crude (HR = 2.07; 95%CI 1.15–3.74) and adjusted analyses (Hazard Ratio (HR) = 2.26; 95% confidence interval (CI) = 1.23–4.16). In Figure 2b, the race-stratified survival curves for APC methylation show a clear separation by methylation status (overall log rank p values = 0.04) with the survival curves for White cases with and without APC methylation significantly different from each other (p = 0.03). In addition, when we found a race x APC methylation interaction term to be statistically significant (p = 0.01). There were no significant associations between methylation status and BCR for any of the other genes, in either the full or race-stratified samples.
Table 2.
Number and percent of prostate cancer cases with positive methylation by race and recurrence status
| Total sample |
Whites |
African Americans |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | n1 | Nonrecurrent cases |
Recurrent cases |
p values |
n1 | Nonrecurrent cases |
Recurrent cases |
p values |
n1 | Nonrecurrent cases |
Recurrent cases |
p values |
| RARB | 323 | 64 (27%) | 26 (31%) | 0.463 | 192 | 31 (22%) | 14 (28%) | 0.376 | 131 | 33 (34%) | 12 (35%) | 0.893 |
| APC | 332 | 57 (23%) | 23 (28%) | 0.335 | 196 | 24 (16%) | 17 (35%) | 0.006 | 136 | 33 (32%) | 6 (18%) | 0.126 |
| RASSF1 | 316 | 50 (21%) | 17 (20%) | 0.852 | 193 | 32 (22%) | 15 (30%) | 0.280 | 123 | 18 (20%) | 2 (6%) | 0.063 |
| CCND2 | 314 | 19 (8%) | 12 (15%) | 0.083 | 187 | 9 (7%) | 7 (14%) | 0.095 | 127 | 10 (11%) | 5 (16%) | 0.440 |
| MGMT | 307 | 18 (8%) | 5 (6%) | 0.486 | 182 | 9 (7%) | 3 (6%) | 0.777 | 125 | 9 (10%) | 2 (6%) | 0.482 |
Number of cases with methylation data for each study gene.
Table 3.
Crude and adjusted recurrence risk estimates for methylation of the RARB, APC and RASSF1 genes
| Model | Gene | n | n events | Hazard ratio | 95% confidence interval | p values |
|---|---|---|---|---|---|---|
| Total sample | ||||||
| Crude | RARB | 323 | 84 | 1.26 | 0.79–2.00 | 0.33 |
| APC | 332 | 82 | 1.21 | 0.75–1.96 | 0.43 | |
| RASSF1 | 316 | 83 | 0.98 | 0.57–1.67 | 0.93 | |
| Adjusted1 | RARB | 306 | 83 | 1.23 | 0.77–1.98 | 0.39 |
| APC | 315 | 81 | 1.21 | 0.73–2.00 | 0.46 | |
| RASSF1 | 299 | 82 | 1.11 | 0.65–1.91 | 0.71 | |
| Whites | ||||||
| Crude | RARB | 192 | 50 | 1.34 | 0.72–2.48 | 0.35 |
| APC | 196 | 49 | 2.07 | 1.15–3.74 | 0.02 | |
| RASSF1 | 193 | 50 | 1.36 | 0.74–2.49 | 0.32 | |
| Adjusted1 | RARB | 179 | 50 | 1.29 | 0.69–2.41 | 0.43 |
| APC | 183 | 49 | 2.26 | 1.23–4.16 | 0.01 | |
| RASSF1 | 180 | 50 | 1.14 | 0.60–2.18 | 0.68 | |
| African Americans | ||||||
| Crude | RARB | 131 | 34 | 1.17 | 0.58–2.37 | 0.65 |
| APC | 136 | 33 | 0.51 | 0.21–1.23 | 0.13 | |
| RASSF1 | 123 | 33 | 0.36 | 0.09–1.51 | 0.16 | |
| Adjusted1 | RARB | 127 | 33 | 1.17 | 0.56–2.47 | 0.68 |
| APC | 132 | 32 | 0.40 | 0.15–1.06 | 0.07 | |
| RASSF1 | 119 | 32 | 0.52 | 0.12–2.21 | 0.37 |
Adjusted for age, tumor stage and grade, PSA level at diagnosis, treatment type (radical prostatectomy or radiation).
Figure 2.
Kaplan-Meier survival plots for methylation-race subgroups. The curves show separate analyses of the methylation status of the RARB (a), APC (b) and RASSF1 (c) genes in benign prostate specimen. Prostate cancer disease-free survival is depicted for cases with a methylated—M (solid lines) or unmethylated—UM (broken lines) gene stratified by race: White (grey lines) and African American—AA (black lines) patients.
Because gene promoter methylation may potentially interact with other factors that presage a malignant environment such as an increased PSA level or presence of inflammation, we examined RARB and APC methylation associations stratified on these factors as well as the methylation of the other genes under study (Table 4). The latter was a crude indicator of possible field cancerization in the benign specimen in which methylation was being measured. For RARB methylation, the greatest differences across risk strata were observed for African American cases. RARB methylation conferred a significant increased risk for BCR in cases where another gene was methylated (“high methylation index”) and when inflammation was absent in the benign prostate specimen. Absence of prostatic inflammation had a similar positive effect on RARB methylation-associated prostate cancer risk of progression across racial groups, with a two-fold increased risk of BCR associated with the combination of positive RARB methylation and absence of histologic prostatic inflammation in the full sample (OR = 2.38; 95%CI 1.22–4.65).
Table 4.
Stratified recurrence risk estimates for methylation of the RARB and APC genes
| Study sample/stratification variable |
n | n events | Hazard ratio |
95% confidence interval |
p values | n | n events | Hazard ratio | 95% confidence interval |
p values | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total Sample | RARB gene | APC gene | |||||||||
| Methylation Index1 | High | 145 | 43 | 1.23 | 0.66–2.29 | 0.52 | 157 | 42 | 1.03 | 0.54–1.95 | 0.93 |
| Low | 175 | 41 | 1.33 | 0.61–2.88 | 0.48 | 172 | 40 | 1.19 | 0.52–2.70 | 0.68 | |
| PSA at cohort entry2 | High | 149 | 49 | 1.42 | 0.75–2.69 | 0.28 | 152 | 48 | 0.77 | 0.37–1.60 | 0.48 |
| Low | 172 | 35 | 1.36 | 0.66–2.81 | 0.41 | 178 | 34 | 1.81 | 0.89–3.68 | 0.10 | |
| Inflammation | Present | 180 | 45 | 0.84 | 0.42–1.68 | 0.62 | 184 | 41 | 1.34 | 0.67–2.71 | 0.41 |
| Absent | 138 | 39 | 2.38 | 1.22–4.65 | 0.01 | 143 | 41 | 0.97 | 0.49–1.95 | 0.94 | |
| Whites | |||||||||||
| Methylation Index1 | High | 83 | 30 | 0.76 | 0.34–1.70 | 0.50 | 87 | 26 | 0.90 | 0.38–2.12 | 0.81 |
| Low | 108 | 20 | 2.24 | 0.79–6.40 | 0.13 | 108 | 23 | 3.28 | 1.33–8.11 | 0.01 | |
| PSA at cohort entry2 | High | 88 | 27 | 1.07 | 0.43–2.63 | 0.89 | 89 | 27 | 0.80 | 0.30–2.10 | 0.65 |
| Low | 103 | 23 | 1.78 | 0.71–4.50 | 0.22 | 106 | 22 | 3.66 | 1.51–8.85 | 0.004 | |
| Inflammation | Present | 109 | 27 | 1.05 | 0.42–2.66 | 0.91 | 110 | 24 | 3.24 | 1.38–7.60 | 0.007 |
| Absent | 79 | 23 | 1.96 | 0.76–5.05 | 0.16 | 82 | 25 | 1.01 | 0.40–2.55 | 0.99 | |
| African Americans | |||||||||||
| Methylation Index1 | High | 62 | 13 | 3.80 | 1.07–13.53 | 0.04 | 70 | 16 | 1.02 | 0.36–2.94 | 0.96 |
| Low | 67 | 21 | 0.91 | 0.29–2.85 | 0.87 | 64 | 17 | Could not estimate | |||
| PSA at cohort entry2 | High | 61 | 22 | 1.85 | 0.66–5.19 | 0.25 | 63 | 21 | 0.55 | 0.16–1.98 | 0.36 |
| Low | 69 | 12 | 1.13 | 0.34–3.82 | 0.84 | 72 | 12 | 0.45 | 0.11–1.78 | 0.25 | |
| Inflammation | Present | 71 | 18 | 0.84 | 0.29–2.43 | 0.75 | 74 | 17 | 0.30 | 0.07–1.33 | 0.11 |
| Absent | 59 | 16 | 3.35 | 0.98–11.41 | 0.05 | 61 | 16 | 0.48 | 0.13–1.81 | 0.28 | |
High (RARB gene): methylation of APC, RASSF1, MGMT or CCND2; High (APC gene): methylation of RARB, RASSF1, MGMT or CCND2; low: none of the 4 other genes methylated.
Stratified at median.
For methylation of APC, the greatest differences across risk strata were observed for White men. APC methylation conferred a significant increased risk for BCR when no other gene was methylated (“low methylation index”), PSA was low at time of cohort entry, or when inflammation was present in the benign prostate specimen. Risk estimates for APC methylation were significantly different only between strata for the high and low PSA risk subsets (p = 0.02). In African Americans, APC methylation tended to be associated with decreased risk of BCR, however, none of the strata-specific estimates were significant. Stratified analyses of RASSF1 methylation did not reveal any significant associations for either the full or race-stratified samples (data not shown).
Discussion
In our earlier study,23 we demonstrated that gene promoter methylation may be an indicator of field cancerization—a “field” indicative of clonal expansion and spreading of cells with genetic alterations that can lead to cancer initiation and potentially progression32—in the prostate. We now report results from the first study to examine methylation in preneoplastic prostate tissue and its association with later disease progression. We found a suggestive association between APC methylation and BCR in White men; the association remained after adjustment for standard clinical disease progression risk factors (HR = 2.26; 95%CI 1.23–4.16). While in our earlier report, it was mainly in African American men where methylation of APC and to a greater extent, RARB, increased risk of prostate cancer, APC did increase risk of high grade disease two-fold in White men.23 In the present study, we found the hazard ratio for BCR associated with APC methylation was increased in White men and further accentuated in certain clinical substrata. These results support our hypothesis that aberrant DNA methylation—detected before the tumor becomes morphologically evident—is an early event in carcinogenesis that may drive cancer progression.
Because we have previously found that inflammation in benign prostate is inversely associated with subsequent risk of prostate cancer,28 we performed analyses stratified on inflammation status. Methylation of APC and RARB demonstrated divergent results. Methylation of APC was associated with a three-fold increased risk of BCR in White men with inflammation in their benign prostate specimens, but not in White men without inflammation. This is consistent with reports in other cancers. Methylation levels of both APC and RARB are increased in inflammatory versus noninflammatory breast cancer.33 In glioblastoma, methylation of RARB and RASSF1 are positively associated with expression of IL-6, a marker of chronic inflammation.34 In APC (Min/+) mutant mice that spontaneously develop prostate intraepithelial neoplasia and microinvasive carcinoma with increasing age, development of subsequent prostate cancer occurs in an inflammation-dependent manner following infection with Helicobacter hepaticus.35
Conversely, RARB methylation in benign prostate was associated with a two-fold increased risk of BCR in the absence of inflammation with this increased risk observed mainly in African American men where positive RARB methylation and absence of inflammation in benign prostate was associated with a three-fold increased the risk of BCR. Racial differences are known to exist in prostate tumor immunobiology,36 particularly in genes related to inflammation,37 which may in part explain this finding. RARB methylation likely occurs early in prostate carcinogenesis,38 and the majority of studies investigating methylation as a marker of prostate cancer progression have found RARB methylation to be associated with an increased risk of BCR after adjusting for other known risk factors.25,26,39–42
Only one study to date has examined whether methylation markers in a prostate cancer field are predictive of disease outcome. Richiardi et al. showed that methylation of both the APC and GSTP1 genes in tumor-adjacent benign tissue were associated with a 2.4-fold increased risk in death from prostate cancer after adjusting for Gleason score.27 Methylation of both genes in tumor-adjacent benign tissue correlated strongly with methylation status in tumor. The results of this study suggest that the molecular changes indicative of field cancerization in prostate cancer are also linked to how the disease will progress in the malignant state. Although the authors note that they focused on methylation of only two genes—GSTP1 and APC—because of their involvement in crucial pivotal cell control pathways, it is likely that additional prognostic information could be obtained from other genes. Richiardi et al. also found increasing risk for prostate cancer mortality when both GSTP1 and APC were methylated, compared with when only one was methylated.27 This result is consistent with our finding of RARB methylation being associated with a greater increased risk for BCR in African Americans when APC, RASSF1, CTND2 or MGMT is methylated. However, it conflicts with our finding that BCR risk associated with APC methylation is highest in White men with a low methylation index. One explanation for latter finding could be that GSTP1 and APC work in complementary carcinogenesis pathways, whereas APC and the other four genes we studied (RARB, RASSF1, CTNND2, MGMT) work in antagonistic pathways. These results collectively suggest that promoter gene methylation is not an independent event, and should be considered in the context of gene networks and biological pathways.
While all study prostate tissue specimens were systematically reviewed by a pathologist to confirm absence of malignancy, the nature of our study design did not allow us to exclude men with small undectable tumor lesions at time of cohort entry. We chose the cutoff of one year for inclusion of incident cases based on earlier unpublished work that showed the cumulative incidence distribution of prostate cancer in men with a benign prostate specimen in our cohort formed a left skewed distribution with the majority of cases diagnosed within three months of the time of benign prostate specimen collection with a steady decline of incident cases thereafter. Since the slope in this decline leveled off at one year, and because a year would also generally encompass the time of any re-evaluation of suspicious biopsy findings,43 we chose one year of follow-up as the cutoff for study eligibility, which should eliminate most of the men with prostate cancers that were “missed” at time of cohort entry.
DNA methylation patterns may differ by race in both healthy and diseased tissue.44,45 In benign prostate, African Americans appear to have a higher baseline level of RARB methylation compared with White men.23,46 However, if racial differences in RARB methylation exist in benign prostate, these differences apparently disappear when the prostate becomes malignant.23,46,47 Interestingly, RARB methylation does not appear to be related to clinical parameters of disease in African American prostate cancer cases.23,46 If high levels of RARB methylation are a precursor for prostate cancer, then the elevated RARB methylation observed in histologically benign prostate tissue from African American men may suggest a higher baseline risk for this racial group, consistent with epidemiologic observations.48,49 In the current study, we found that RARB methylation was associated with disease outcome only in certain subgroups of African American patients, while APC methylation increased risk for disease progression only in White patients. These results are different than what we previously reported as far as the race-specific associations of methylation of these genes with prostate cancer risk.23 Biologic pathways involved in early development of cancer versus later cancer progression are likely different so congruence between risk estimates of gene methylation for disease occurrence versus progression would not necessarily be expected. For example, cigarette smoking is not considered a strong risk factor for prostate cancer risk, but has been shown to increase risk for poor outcome.50
In summary, we show for the first time that promoter gene methylation in the earliest stages of prostate carcinogenesis may drive later disease outcomes. If an aggressive disease phenotype is epigenetically “programmed” in the earliest stages of disease—before cellular morphological changes that distinguish malignancy are apparent—this has important implications in the prevention and treatment of clinically significant prostate cancer. Moreover, if early epigenetic markers of aggressive disease vary by race, this will affect the development and implementation of future molecular-based diagnostic tests. Our results are novel, and need to be replicated in independent ethnically diverse patient cohorts, especially where more definitive disease outcome data, such as development of metastatic disease and death due to prostate cancer, are available
What’s new?
Methylation patterns hold clues to the risk of disease progression in men with prostate cancer. Whether risk of malignant transformation from benign prostatic disease can be similarly deduced is unknown. Here, methylation patterns in gene promoter regions were analyzed in benign tissue samples collected from men who later developed prostate cancer. In white men only, methylation of a known tumor suppressor gene in premalignant benign prostate was associated with an increased risk for subsequent disease progression. The results suggest that race-specific methylation panels could play a role in the early detection of aggressive prostate cancer.
References
- 1.Mariotto AB, Yabroff KR, Shao Y, et al. Projections of the cost of cancer care in the United States: 2010–2020. J Natl Cancer Inst. 2011;103:117–128. doi: 10.1093/jnci/djq495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arora R, Koch MO, Eble JN, et al. Heterogeneity of Gleason grade in multifocal adenocarcinoma of the prostate. Cancer. 2004;100:2362–2366. doi: 10.1002/cncr.20243. [DOI] [PubMed] [Google Scholar]
- 3.Douglas TH, McLeod DG, Mostofi FK, et al. Prostate-specific antigen-detected prostate cancer (stage T1c): An analysis of whole-mount prostatectomy specimens. Prostate. 1997;32:59–64. doi: 10.1002/(sici)1097-0045(19970615)32:1<59::aid-pros8>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 4.Nonn L, Ananthanarayanan V, Gann PH. Evidence for field cancerization of the prostate. Prostate. 2009;69:1470–1479. doi: 10.1002/pros.20983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Risk MC, Knudsen BS, Coleman I, et al. Differential gene expression in benign prostate epithelium of men with and without prostate cancer: Evidence for a prostate cancer field effect. Clin Cancer Res. 2010;16:5414–5423. doi: 10.1158/1078-0432.CCR-10-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandran UR, Dhir R, Ma C, et al. Differences in gene expression in prostate cancer, normal appearing prostate tissue adjacent to cancer and prostate tissue from cancer free organ donors. BMC Cancer. 2005;5:45. doi: 10.1186/1471-2407-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fordyce CA, Heaphy CM, Joste NE, et al. Association between cancer-free survival and telomere DNA content in prostate tumors. J Urol. 2005;173:610–614. doi: 10.1097/01.ju.0000143195.49685.ce. [DOI] [PubMed] [Google Scholar]
- 8.Yu YP, Song C, Tseng G, et al. Genome abnormalities precede prostate cancer and predict clinical relapse. Am J Pathol. 2012;180:2240–2248. doi: 10.1016/j.ajpath.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehrotra J, Varde S, Wang H, et al. Quantitative, spatial resolution of the epigenetic field effect in prostate cancer. Prostate. 2008;68:152–160. doi: 10.1002/pros.20675. [DOI] [PubMed] [Google Scholar]
- 10.Hanson JA, Gillespie JW, Grover A, et al. Gene promoter methylation in prostate tumor-associated stromal cells. J Natl Cancer Inst. 2006;98:255–261. doi: 10.1093/jnci/djj051. [DOI] [PubMed] [Google Scholar]
- 11.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 12.Aryee MJ, Liu W, Engelmann JC, et al. DNA methylation alterations exhibit intraindividual stability and interindividual heterogeneity in prostate cancer metastases. Sci Transl Med. 2013;5:169ral0. doi: 10.1126/scitranslmed.3005211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Shang Y. Epigenetic control of epithelial-to-mesenchymal transition and cancer metastasis. Exp Cell Res. 2013;319:160–169. doi: 10.1016/j.yexcr.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 14.Gurel B, Iwata T, Koh CM, et al. Molecular alterations in prostate cancer as diagnostic, prognostic, and therapeutic targets. Adv Anat Pathol. 2008;15:319–331. doi: 10.1097/PAP.0b013e31818a5c19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashour N, Angulo JC, Andres G, et al. A DNA hypermethylation profile reveals new potential biomarkers for prostate cancer diagnosis and prognosis. Prostate. 2014;74:1171–1182. doi: 10.1002/pros.22833. [DOI] [PubMed] [Google Scholar]
- 16.Stott-Miller M, Zhao S, Wright JL, et al. Validation study of genes with hypermethylated promoter regions associated with prostate cancer recurrence. Cancer Epidemiol Biomarkers Prev. 2014;23:1331–1339. doi: 10.1158/1055-9965.EPI-13-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maldonado L, Brait M, Loyo M, et al. GSTP1 promoter methylation is associated with recurrence in early stage prostate cancer. J Urol. 2014;192:1542–1548. doi: 10.1016/j.juro.2014.04.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Litovkin K, Van Eynde A, Joniau S, et al. DNA methylation-guided prediction of clinical failure in high-risk prostate cancer. PLoS One. 2015;10:e0130651. doi: 10.1371/journal.pone.0130651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steiner I, Jung K, Schatz P, et al. Gene promoter methylation and its potential relevance in early prostate cancer diagnosis. Pathobiology. 2010;77:260–266. doi: 10.1159/000318017. [DOI] [PubMed] [Google Scholar]
- 20.Truong M, Yang B, Livermore A, et al. Using the epigenetic field defect to detect prostate cancer in biopsy-negative patients. J Urol. 2013;189:2335–2341. doi: 10.1016/j.juro.2012.11.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo JH, Ding Y, Chen R, et al. Genome-wide methylation analysis of prostate tissues reveals global methylation patterns of prostate cancer. Am J Pathol. 2013;182:2028–2036. doi: 10.1016/j.ajpath.2013.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim JH, Dhanasekaran SM, Prensner JR, et al. Deep sequencing reveals distinct patterns of DNA methylation in prostate cancer. Genome Res. 2011;21:1028–1041. doi: 10.1101/gr.119347.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang D, Kryvenko ON, Mitrache N, et al. Methylation of the RARB gene increases prostate cancer risk in black Americans. J Urol. 2013;190:317–324. doi: 10.1016/j.juro.2013.01.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richiardi L, Fiano V, Vizzini L, et al. Promoter methylation in APC, RUNX3, and GSTP1 and mortality in prostate cancer patients. J Clin Oncol. 2009;27:3161–3168. doi: 10.1200/JCO.2008.18.2485. [DOI] [PubMed] [Google Scholar]
- 25.Ellinger J, Bastian PJ, Jurgan T, et al. CpG island hypermethylation at multiple gene sites in diagnosis and prognosis of prostate cancer. Urology. 2008;71:161–167. doi: 10.1016/j.urology.2007.09.056. [DOI] [PubMed] [Google Scholar]
- 26.Moritz R, Ellinger J, Nuhn P, et al. DNA hypermethylation as a predictor of PSA recurrence in patients with low- and intermediate-grade prostate cancer. Anticancer Res. 2013;33:5249–5254. [PubMed] [Google Scholar]
- 27.Richiardi L, Fiano V, Grasso C, et al. Methylation of APC and GSTP1 in non-neoplastic tissue adjacent to prostate tumour and mortality from prostate cancer. PLoS One. 2013;8:e68162. doi: 10.1371/journal.pone.0068162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kryvenko ON, Jankowski M, Chitale DA, et al. Inflammation and preneoplastic lesions in benign prostate as risk factors for prostate cancer. Mod Pathol. 2012;25:1023–1032. doi: 10.1038/modpathol.2012.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freedland SJ, Sutter ME, Dorey F, et al. Defining the ideal cutpoint for determining PSA recurrence after radical prostatectomy. Prostate-specific antigen. Urology. 2003;61:365–369. doi: 10.1016/s0090-4295(02)02268-9. [DOI] [PubMed] [Google Scholar]
- 30.Roach M, III, Hanks G, Thames H, Jr, et al. Defining biochemical failure following radiotherapy with or without hormonal therapy in men with clinically localized prostate cancer: Recommendations of the RTOG-ASTRO phoenix consensus conference. Int J Radiat Oncol Biol Phys. 2006;65:965–974. doi: 10.1016/j.ijrobp.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 31.Nickel JC, True LD, Krieger JN, et al. Consensus development of a histopathological classification system for chronic prostatic inflammation. BJU Int. 2001;87:797–805. doi: 10.1046/j.1464-410x.2001.02193.x. [DOI] [PubMed] [Google Scholar]
- 32.Trujillo KAJ, Griffith AC, Bisoffi JKM. Markers of field cancerization: Proposed clinical applications in prostate biopsies. Prostate Cancer. 2012;2012:12. doi: 10.1155/2012/302894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van der Auwera I, Bovie C, Svensson C, et al. Quantitative assessment of DNA hypermethylation in the inflammatory and non-inflammatory breast cancer phenotypes. Cancer Biol Ther. 2009;8:2252–2259. doi: 10.4161/cbt.8.23.10133. [DOI] [PubMed] [Google Scholar]
- 34.Piperi C, Themistocleous MS, Papavassiliou GA, et al. High incidence of MGMT and RARbeta promoter methylation in primary glioblastomas: Association with histopathological characteristics, inflammatory mediators and clinical outcome. Mol Med. 2010;16:1–9. doi: 10.2119/molmed.2009.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poutahidis T, Cappelle K, Levkovich T, et al. Pathogenic intestinal bacteria enhance prostate cancer development via systemic activation of immune cells in mice. PLoS One. 2013;8:e73933. doi: 10.1371/journal.pone.0073933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wallace TA, Prueitt RL, Yi M, et al. Tumor immunobiological differences in prostate cancer between African–American and European–American men. Cancer Res. 2008;68:927–936. doi: 10.1158/0008-5472.CAN-07-2608. [DOI] [PubMed] [Google Scholar]
- 37.Powell IJ, Dyson G, Land S, et al. Genes associated with prostate cancer are differentially expressed in African American and European American men. Cancer Epidemiol Biomarkers Prev. 2013;22:891–897. doi: 10.1158/1055-9965.EPI-12-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Troyer DA, Lucia MS, de Bruine AP, et al. Prostate cancer detected by methylated gene markers in histopathologically cancer-negative tissues from men with subsequent positive biopsies. Cancer Epidemiol Biomarkers Prev. 2009;18:2717–2722. doi: 10.1158/1055-9965.EPI-09-0068. [DOI] [PubMed] [Google Scholar]
- 39.Henrique R, Ribeiro FR, Fonseca D, et al. High promoter methylation levels of APC predict poor prognosis in sextant biopsies from prostate cancer patients. Clin Cancer Res. 2007;13:6122–6129. doi: 10.1158/1078-0432.CCR-07-1042. [DOI] [PubMed] [Google Scholar]
- 40.Woodson K, O’Reilly KJ, Ward DE, et al. CD44 and PTGS2 methylation are independent prognostic markers for biochemical recurrence among prostate cancer patients with clinically localized disease. Epigenetics. 2006;1:183–186. doi: 10.4161/epi.1.4.3530. [DOI] [PubMed] [Google Scholar]
- 41.Rosenbaum E, Hoque MO, Cohen Y, et al. Promoter hypermethylation as an independent prognostic factor for relapse in patients with prostate cancer following radical prostatectomy. Clin Cancer Res. 2005;11:8321–8325. doi: 10.1158/1078-0432.CCR-05-1183. [DOI] [PubMed] [Google Scholar]
- 42.Vanaja DK, Ehrich M, van den BD, et al. Hypermethylation of genes for diagnosis and risk stratification of prostate cancer. Cancer Invest. 2009;27:549–560. doi: 10.1080/07357900802620794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terris MK. Strategies for repeat prostate biopsies. Curr Urology Rep. 2009;10:172–178. doi: 10.1007/s11934-009-0030-y. [DOI] [PubMed] [Google Scholar]
- 44.Adkins RM, Krushkal J, Tylavsky FA, et al. Racial differences in gene-specific DNA methylation levels are present at birth. Birth Defects Res Clin Mol Teratol. 2011;91:728–736. doi: 10.1002/bdra.20770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fraser HB, Lam LL, Neumann SM, et al. Population-specificity of human DNA methylation. Genome Biol. 2012;13:R8. doi: 10.1186/gb-2012-13-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwabi-Addo B, Wang S, Chung W, et al. Identification of differentially methylated genes in normal prostate tissues from African American and Caucasian men. Clin Cancer Res. 2010;16:3539–3547. doi: 10.1158/1078-0432.CCR-09-3342. [DOI] [PubMed] [Google Scholar]
- 47.Woodson K, Hanson J, Tangrea J. A survey of gene-specific methylation in human prostate cancer among black and white men. Cancer Lett. 2004;205:181–188. doi: 10.1016/j.canlet.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 48.Rebbeck TR, Devesa SS, Chang BL, et al. Global patterns of prostate cancer incidence, aggressiveness, and mortality in men of african descent. Prostate Cancer. 2013;2013:560857. doi: 10.1155/2013/560857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Powell IJ. Epidemiology and pathophysiology of prostate cancer in African–American men. J Urol. 2007;177:444–449. doi: 10.1016/j.juro.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 50.Moreira DM, Aronson WJ, Terris MK, et al. Cigarette smoking is associated with an increased risk of biochemical disease recurrence, metastasis, castration-resistant prostate cancer, and mortality after radical prostatectomy: Results from the SEARCH database. Cancer. 2014;120:197–204. doi: 10.1002/cncr.28423. [DOI] [PMC free article] [PubMed] [Google Scholar]