Abstract
The proteasome refers to a collection of complexes centered on the 20S proteasome core particle, a complex of 28 subunits that houses proteolytic sites in its hollow interior. Proteasomes are found in eukaryotes, archaea, and some eubacteria, and their activity is critical for many cellular pathways. Important advances include inhibitor binding studies and the structure of the immunoproteasome, whose specificity is altered by incorporation of inducible catalytic subunits. The inherent repression of the 20S CP is relieved by the ATP-independent activators, 11S and Blm10/PA200, whose structures reveal principles of proteasome mechanism. The structure of the ATP-dependent 19S regulatory particle, which mediates degradation of polyubiquitylated proteins, is being revealed by a combination of crystal or NMR structures of individual subunits and electron microscopy reconstruction of the intact complex. Other recent structural advances inform about mechanisms of assembly and the role of conformational changes in the functional cycle.
Keywords: macromolecule assembly, X-ray crystallography, electron microscopy, nuclear magnetic resonance, proteasome structure, conformational changes
INTRODUCTION
This review summarizes advances made in understanding structural aspects of the proteasome, which is a protease found in eukaryotes, archaea, and some bacteria, and is of critical importance for many facets of cellular metabolism because it performs most of the regulated protein turnover in the eukaryotic cytosol and nucleus. The proteasome exists as a collection of complexes that are centered on the 20S proteasome core particle (20S CP), a ~700kDa complex of 28 protein subunits. Since the first 20S CP structure was determined in 1995 (51), considerable progress has been made in understanding proteasome mechanism, including an accelerating rate of advances in structural biology that includes several important papers published in the past year.
Here we provide an overview of the current state of proteasome structural biology. We start with the 20S CP and, of the many publications on proteasome inhibitor complexes, highlight two notable recent advances, a difference in available conformational changes that may allow development of novel therapeutics for the treatment of TB, and understanding of how the inducible subunits of the immunoproteasome favor production of ligands for MHC-I molecules. This is followed by a discussion of the activators that relieve the inherently repressed 20S CP structure, including the ATP-independent activators, 11S and Blm10/PA200, whose biological function is unclear, but for which structural studies have provided insight to biochemical mechanisms of proteasome binding and activation. The other class of 20S CP activators is ATP-dependent, and includes the 19S regulatory particle (19S RP) of eukaryotes, which includes a core of six ATPases that unfold and translocate substrates to mediate most of the regulated proteolysis in the eukaryotic cytosol and nucleus. Archaea and some eubacteria encode the simpler ATP-dependent activators PAN, ARC and Mpa, which are relatively simple homohexameric homologs of the 19S RP ATPases that lack the additional non-ATPase subunits of the 19S RP. The complete 19S RP and its complex with the 20S CP, known as the 26S proteasome, is a topic for which especially exciting advances have been obtained recently in the form of reconstructions by electron microscopy (EM) that have revealed the relative location of all 19 subunits of the 19S RP. The final topic reviewed here is structural insights being revealed on the processes of assembly of the 20S CP and the 19S RP, and of their association to form the 26S proteasome. An emerging theme that runs throughout is that understanding of mechanism will require insights into the conformational changes that occur during many facets of proteasome function.
20S CORE PARTICLE (Figure 1)
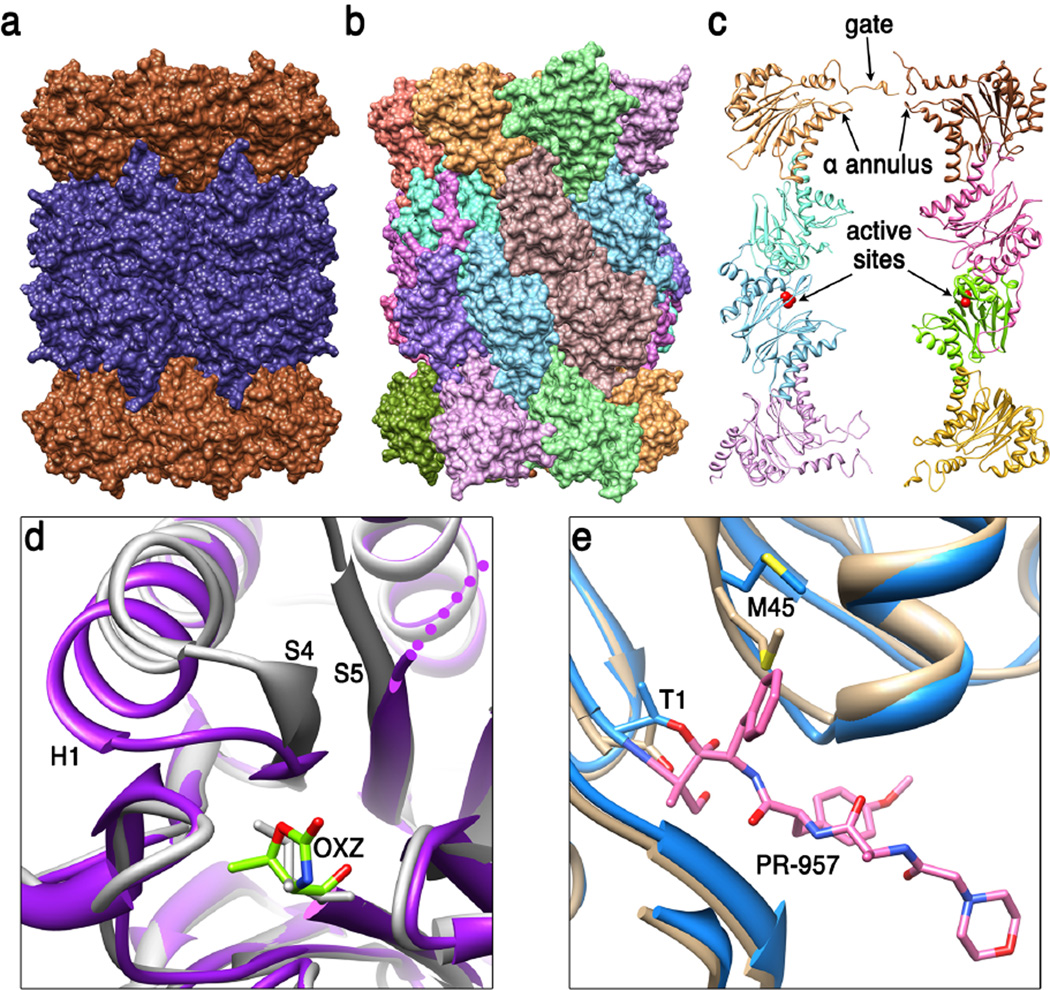
Figure 1. 20S proteasome core particle (20S CP).
(a) Side view of the archaeal T. acidophilum 20S CP (51) (pdb code 1pma). End rings comprise seven identical α subunits (brown) and the two middle rings comprise seven identical β subunits (dark blue). (b) Side view of the eukaryotic S. cerevisiae 20S CP (24) (pdb code 1ryp). Each of the seven different α subunits and seven different β subunits occupies a unique position within their respective rings. The whole structure has two-fold symmetry relating the top and bottom halves of the structure to each other, with the 2-fold axis in the horizontal plane, a little to the right of center in this view. (c) Cutaway view showing internal features. The S. cerevisiae 20S CP is shown in ribbon representation with just eight subunits displayed in order to reveal the hollow interior. Labeled features include residues that contribute to the asymmetric closed gate structure, loops that contribute to the α annulus, and the active sites of β1 and β5 in the lower β ring (only the β1, β2, and β5 subunits have active sites in eukaryotic proteasomes). (d) Conformational changes at the activate site of M. tuberculosis 20S CP that are induced upon binding of inhibitor suggest the possibility of developing a specific therapeutic (49). The loop connecting S4 and H1 of the β subunit moves from the unbound conformation (white, pdb code 2fhg) to cover OXZ, the inhibitor oxazolidin-2-one ring on Thr1 in the stabilized complex (purple, pdb code 3h6f). (e) Comparison of mouse liver constitutive and inducible β5 S1 binding pocket (30). Met45 adopts the sky blue conformation (pdb code 3unf) when bound to the PR-957 inhibitor (pink), which binds with a large hydrophobic group in the S1 pocket. Met45 also adopts this conformation in the unbound immunoproteasome but adopts the tan conformation in the unbound constitutive proteasome (pdb code 3une). This requirement for repositioning Met45 explains why immunoproteasomes prefer to cleave substrate after large hydrophobic side chains.
Determination of a crystal structure of the 20S CP from the archaeon T. acidophilum was a major landmark achievement that revealed a cylindrical structure of four rings, with seven α subunits in each of the two end rings and seven β subunits in each of the two central rings (51). The catalytic centers were localized to the central chamber, and biochemical and structural studies of inhibitor complexes further revealed essential elements of the N-terminal nucleophile catalytic mechanism (75). Whereas archaea and eubacteria typically encode a single α and a single β subunitto assemble a seven-fold symmetric 20S CP, eukaryotes encode seven distinct α subunits (α1–7) and seven distinct β subunits (β1–7), that occupy unique positions to assemble a pseudo seven-fold symmetric 20S CP, as revealed by a crystal structure of the 20S CP from the yeast S. cerevisiae (24). This structure and associated inhibitor complexes also showed how distinctive S1 pockets define the specificity of the three catalytically active β1, β2, and β5 subunits of eukaryotes, which possess caspase, trypsin, and chymotrypsin-like activities, respectively. The subsequent crystal structure of the bovine 20S proteasome indicated that all eukaryotic 20S proteasomes have closely similar structures (90).
Recent advances in inhibitor development
A large variety of inhibitor complex crystal structures have been studied, in large part because 20S CP inhibition is an established approach for cancer therapy, with the inhibitor bortezomib currently approved for the treatment of relapsed multiple myeloma and mantle cell lymphoma (31). Recently, crystal structures have been reported for the mouse liver 20S proteasome and immunoproteasome, a variant in which the three constitutive catalytic subunits are substituted by inducible counterparts that are upregulated in response to T-cell signaling (30). These structures explain the basis for the change in specificity, which largely occur through changes in the S1 pocket, and also explain why the PR-957 inhibitor preferentially binds the β5i subunit. These findings give impetus to efforts to develop specific inhibitors that might be efficacious in the treatment of disorders in which immunoproteasomes are upregulated, such as some autoimmune disorders, neurodegenerative diseases, and cancers. Structural studies are also guiding efforts to develop inhibitors against the proteasome of pathogens, such as M. tuberculosis, which causes tuberculosis. Interestingly, binding of oxathiazole-2-one inhibitors was shown to induce a conformational change that explains why these compounds show specificity for M. tuberculosis proteasome, whereas the equivalent conformational change is not accommodated in eukaryotic proteasomes (49).
Gating
The entrance route for substrates through an axial pore in the α subunits was indicated by EM visualization of gold-labeled substrate bound to the T. acidophilum 20S CP (96), while the crystal structure of the same 20S CP showed that the pore comprises a 13Å-diameter constriction called the α annulus that limits entry to proteins that are unstructured (51). Passage through this pore is further impeded by disordered polypeptide corresponding to the first 12 residues of the seven α subunits (4; 19). In contrast, the eukaryotic 20S CP adopts a precisely closed conformation (24). Bacterial proteasomes also appear to adopt an ordered closed gate, although the structure is striking different from that of eukaryotic proteasomes (47). Despite their different mechanisms of gate closure, it seems likely that fully activated proteasomes will all adopt the same seven-fold symmetric fully open conformation (82).
Insights from NMR
Although most of the structural data on the 20S CP has been obtained by X-ray crystallography, NMR studies by the Kay group have made a number of notable contributions. These are remarkable achievements given the very large molecular weight, and were made possible by the development of methyl-transverse relaxation optimized spectroscopy using deuterated protein and selectively labeled amino acid methyl groups (on either methionine or isoleucine, leucine, and valine) (35). These studies were performed on the T. acidophilum 20S CP, which offers the advantage of providing a number of more tractable subassemblies, including a monomeric α subunit, a heptameric α ring, and a double α ring of 14 subunits, which provided a clearer view of many of the processes studied. This allowed quantification of properties such as internal dynamics of specific labeled residues and activator binding (79). Insight to the mechanism of gate closure by the flexible N-termini of archaeal proteasomes was provided by determining that on average two of the chains pass through the α annulus to the proteasome interior, thereby plugging the passage needed for protein substrates (64). Using three model substrate proteins, this approach also demonstrated that the interior surface of the proteasome stabilizes an unstructured conformation of translocated substrates, thereby inhibiting refolding of stable protein domains inside the proteasome (67). NMR methods have also guided new approaches to developing proteasome inhibitors by demonstrating that inhibition can be achieved by binding in the vicinity of the interface between α and β subunits in a manner that is independent of binding to the active sites (80).
ATP-INDEPENDENT ACTIVATORS (Figure 2)
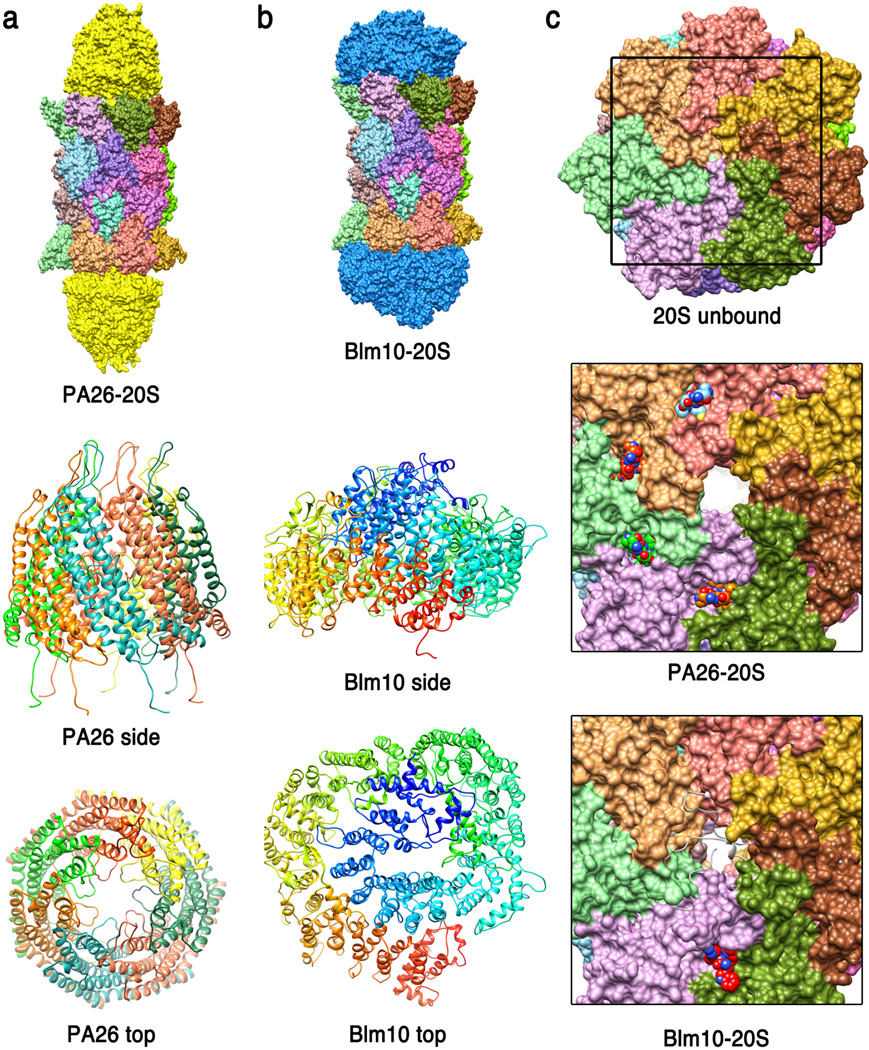
Figure 2. ATP-independent activators.
(a) Top - crystal structure of the T. brucei PA26 heptamer (yellow) in complex with S. cerevisiae 20S CP (18) (pdb code 1z7q). Middle - side view of PA26 ribbon representation with each of the seven identical subunits in a different color. Bottom - PA26 top view. Loops from an insertion in helix 3 project into the middle of the channel where they would impede transit of a potential substrate. (b) Top - crystal structure of the S. cerevisiae Blm10-20S CP complex (68) (pdb code 1vsy). Middle - side view of Blm10, rainbow colored from N-terminus (blue) to C-terminus (red). Bottom - top view. (c) Top - top surface of S. cerevisiae 20S CP in the unbound closed conformation. Middle - closer view (corresponding to frame of top panel) showing the open conformation induced by PA26 and the four ordered PA26 C-termini visible in this structure. Bottom -top surface of S. cerevisiae 20S CP from the Blm10 complex structure. The gate appears open, although not so extensively as with PA26, and the space is largely filled with disordered residues, which are indicated as white ribbons.
11S activators
The 11S activators, as illustrated by a crystal structure of the human PA28α/REGα homolog, are toroidal heptamers that present seven-fold symmetric arrays of proteasome-binding C-terminal residues and internal activation loop residues on one surface (37). The 20 Å pore through this heptamer was initially suggestive of a substrate entry channel, although it was subsequently found that this channel is occluded in the distantly related PA26 homolog of Trypanosome brucei (18). Crystal structures of PA26 in complex with the S. cerevisiae (19; 97) and T. acipophilum (18) 20S CP have revealed that the activator C-termini bind in pockets between proteasome α subunits while the activation loops reposition the 20S CP Pro17 turn to trigger formation of a seven-fold symmetric open gate conformation. Biochemical assays of mutant T. acidophilum 20S CP and the PAN activator have indicated that the ATP-dependent activators, such as the 19S RP a use a similar mechanism of binding through subunit C-termini (18) and induce a similar open gate conformation (19).
Blm10/PA200
Consistent with EM reconstructions (34; 73), the crystal structure of a proteasome-Blm10 complex revealed a very different architecture from the 11S activators, with the single-chain ~250kDa activator wrapping around the end of the proteasome α subunits like a turban (68). Curiously, Blm10 induces a disordered 20S CP gate conformation, and only limited access is apparent to the dome-like structure formed by Blm10 over the proteasome entrance pore, which is consistent with the relatively low level of peptidase stimulation by Blm10 compared to PA26 (34). The crystal structure did reveal that the one C-terminus of Blm10 binds between the 20S proteasome α5 and α6 subunits, with the C-terminal three residues overlapping closely with the C-termini of PA26 and forming the same main-chain hydrogen bonds and salt bridge to the pocket lysine of α6. This does not result in complete gate opening because other α subunits are not fully repositioned and because conserved Blm10 residues impede the fully open conformation, but it does provide an attractive model for the mechanism of binding of the ATP-dependent activators, which also appear to utilize a salt bridge between the activator C-terminal carboxylate and the pocket lysine (18) and, like Blm10 (12; 68), displays a functionally important penultimate tyrosine (or phenylalanine) (77). In this model, the ATP-dependent activators reposition the proteasome α5 Pro17 turn to the same open position seen in the PA26 complexes, albeit through quite different interactions. This model has been supported by two studies of crystal structures of PA26 mutants in complex with archaeal 20S proteasome (81; 101), albeit with some differences in interpretation, and an EM reconstruction of PAN C-terminal peptides in complex with 20S proteasome (61).
Biological function of the ATP-independent activators
Although the Blm10 and PA26 complex structures provide a wealth of biochemical insight, they do not clarify the rather confused understanding of biological function for either of these activators (63). For example, a large literature implicates some 11S homologs in the production of ligands for MHC-I molecules, although a mechanism for this process is not securely established and many species that express an 11S homolog do not encode MHC-I (76). One of the 11S homologs, PA28γ/REGγ, is reported to promote the degradation of some natively unstructured transcription factors (9; 48). There is even more confusion for Blm10/PA200, where there almost seems to be as many proposed biological functions as there are publications (72). One attractive possibility is that the 11S and Blm10/PA200 activators function in the context of hybrid proteasomes, in which different classes of activator, including the ATP-dependent 19S activator, bind to opposite ends of the same 20S proteasome.
ATP-DEPENDENT ACTIVATORS (Figure 3)
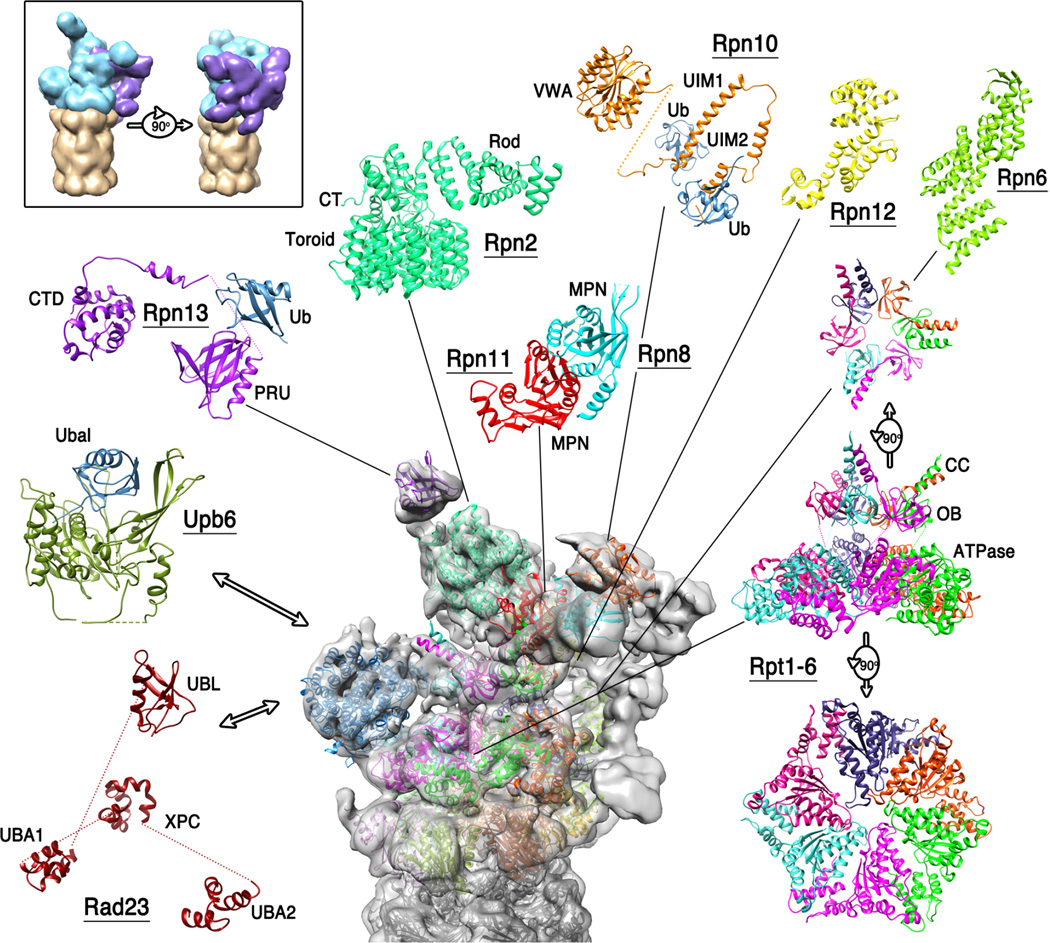
Figure 3. 19S regulatory particle/26S proteasome.
Top left, boxed, two views of a cartoon depiction of the 26S proteasome EM structures showing the 20S CP (tan), base (cyan), and lid (purple). A charge density map of the S. cerevisiae 26S proteasome reconstruction (43) is shown centrally. Atomic models for individual protein subunits whose structures are known at atomic resolution have been positioned following the analyses of references (43) and (3), and are shown around the periphery in an expanded view. Also included are Rad23, Ubp6/USP14, and the C-terminal domain of human Rpn13, which are not part of the reconstructed complex but illustrate how additional structural components contribute to proteasome function. See box for details.
26S proteasome
In contrast to the 11S and Blm10/PA200 activators, the biological function of the ATP-dependent 19S RP is well established to be the selection, conditioning, and delivery of substrates for proteolysis, especially those modified by conjugation to a polyubiquitin chain (17). Complexes of the 19S RP with the 20S CP are known as the 26S proteasome, and include assemblies with a 19S RP on one or both ends of the 20S CP, as well as hybrid complexes with 11S or Blm10 activators on the opposite end of a 20S CP from the 19S RP. The extraordinarily complex 19S RP comprises 19 stoichiometric subunits. Numerous substoichiometric or transient proteasome interacting proteins have also been described, but with a few exceptions will not be discussed here. The assembly can be described in terms of lid and base components (21). The base comprises the six ATPases (Rpt1–6), the two largest (~100kDa) subunits Rpn1 and Rpn2, and the ubiquitin receptors Rpn10 and Rpn13. The lid comprises nine subunits (Rpn3, 5–9, 11, 12, and 15), just one of which, the deubiquitylase Rpn11, displays enzyme activity. Although the 19S RP and 26S proteasome present daunting challenges, they are yielding to structural studies at the level of EM reconstructions of the assembled complex and NMR and x-ray crystal structures of individual domains and subunits.
ATPase subunits of the 19S regulatory particle
The Rpt subunits are members of the classical family of AAA ATPases (16). Rpt1–6, form a heterohexameric ring at the heart of the eukaryotic 19S RP, while the homologous PAN and ARC/Mpa activators of archaea and eubacteria are homohexamers that form functional proteasome activators in the absence of additional subunits. These ATPases comprise an N-terminal coiled-coil (CC) domain, a central oligonucleotide/oligosaccharide binding (OB) domain, and a C-terminal AAA ATPase cassette. Crystal structures of the OB domain and portions of the CC domain of archaeal and eubacterial homologs revealed a symmetry mismatch between the six-fold rotational symmetry of the OB domain ring and a trimer of dimers formed by the CC domains that is accommodated by formation of a cis proline conformation in three of the six subunits (13; 95; 102; 103). The sequence requirements of this interaction guided cross-linking experiments that defined the order of the unique ATPase subunits in the ring of the 19S activator to be Rpt1-Rpt2-Rpt6-Rpt3-Rpt4-Rpt5 (89), in agreement with an earlier EM study (20).
The three coiled-coils projecting at the N-terminal face of the ATPase hexamer resemble chaperones such as profilin and can promote protein unfolding (13), an activity that likely conditions substrates prior to their entrance through the central 13-Å diameter ring of OB domains. Moreover, the eubacterial Mpa coiled-coils directly bind the Pup tag of conjugates targeted for degradation by pupylation (94). The need for substrate to reach from the distal side of the OB pore to the pore loops of the ATPase cassette, the structural features that engage and actively translocate substrate in an ATP-dependent manner, explains why substrates displaying a 30–40 residue segment of unstructured polypeptide are efficiently hydrolyzed whereas proteins lacking disordered segments are protected from proteasomal degradation (33; 59; 85). The separation of initial recognition and substrate engagement further explains why the ubiquitin tag can be on a separate subunit of a complex from the subunit that displays an unstructured segment and is degraded (58). Because the unstructured initiation sequence can be on either N or C-terminus of the substrate, it seems that the ATPases can translocate protein chains in either direction (59), and the finding that proteolysis can start from flexible loops that are removed from either terminus indicates that more than two chains can pass through the channel at the same time (50; 62). The finding that some domains within substrate proteins can escape degradation is explained by the requirement that continued translocation can only occur if the translocating sequence engages efficiently with the ATPase pore loops and the domain entering the ATPase conduit does not strongly resist unfolding (88).
Non-ATPase subunits of the 19S regulatory particle
The two largest 19S subunits, Rpn1 and Rpn2, share low sequence identity but display similar three-dimensional structures, and they each bind at least one ubiquitin receptor and a deubiquitylating enzyme. A crystal structure of S. cerevisiae Rpn2 revealed a central domain composed of 11 proteasome/cyclosome (PC) repeats in which the inner and outer PC helices form a closed ring that is filled by two additional helices (27). Projecting from one face of this central domain are an N-terminal rod-like domain of 17 stacked helices and a globular C-terminal domain comprising β structure. Negative stain EM analysis of purified Rpn1 indicates that it shares this architecture, with some reorientation of the rod domain. This study also found that the C-terminal 20 residues of Rpn2 are unstructured and mediate binding to the Rpn13 subunit.
Earlier work had shown that Rpn13 comprises an N-terminal domain that binds ubiquitin and is termed the pleckstrin-like receptor for ubiquitin (PRU) domain (32; 74). In most species, this domain is followed by an unstructured linker (~150 residues in human) and a helical C-terminal domain (10) that provides the primary binding module for the Uch37/Uch-L5 deubiquitylating enzyme, (25; 60; 99), which likely functions to edit inappropriately or inadequately ubiquitylated conjugates and to disassemble free ubiquitin chains (42). Crystal structures of Uch37 shows that it comprises a catalytic domain that closely resembles structures of other UCH enzymes, followed by a C-terminal helical segment that includes the Rpn13-binding epitope (8). Interestingly, Uch37 is activated by association with Rpn13 (60; 99), and its specificity is altered by association with the 19S activator (41).
Rpn1 is also the binding module for the shuttle ubiquitin receptors Rad23 and Dsk2, and the deubiquitylating enzyme Ubp6/USP14 (14; 46; 66). These proteins all bind through their N-terminal Ubl domains with micromolar binding affinity, and the Ubp6 catalytic domain provides an additional interaction that results in nanomolar affinity for the full-length protein. This is consistent with the respective roles of Rad23 and Dsk2 as transiently associating shuttle receptors, and Ubp6 as an integral 19S RP subunit. Interestingly, a recent report concluded that the three Ubl domains preferentially bind to different regions of Rpn1 (66).
Ubp6/USP14 employs the same cysteine protease mechanism as UCH37 but belongs to the distinct Ubp structural class (29). It is of special interest because its inhibition enhances degradation of some proteasome substrates implicated in neurodegenerative disease (45). As with the case of Uch37 binding to Rpn13, Ubp6 is activated by association with Rpn1 (46), and Ubp6 also seems to modify 19S RP structure because its binding delays proteolysis by a mechanism that is independent of its catalytic activity (26). Another example of functionally important conformational change is provided by the shuttle receptors, which likely adopt an autoinhibited conformation that is opened to release their Ubl domains for proteasome association upon binding of ubiquitylated substrate to the shuttle’s UBA domains (23).
EM reconstructions of the 26S proteasome
The overall architecture of the 19S RP has been revealed in recent flurry of EM reconstructions of 26S proteasomes from S. cerevisiae, S. pombe, and human (3; 7; 11; 43; 44; 56; 69). Two of the highest resolution reconstructions, which were both performed on the S. cerevisiae complex, used different approaches to assign all of the subunits to regions of the reconstructed map. One study coexpressed the nine lid subunits in E. coli, which allowed the lid structure to be determined separately and for the N and C-termini of specific subunits to be localized by expressing fusions with MBP (43). The alternative approach of incorporating crosslinking data and computational methods of map fitting has provided a similar model at ~7Å resolution (3).
A provocative observation from the 7.4 Å resolution reconstruction is that the two 19S RP complexes bound to one 20S CP are not identical to each other (3). Significant differences are indicated, although at the current time, only the more precisely defined RP structure has been discussed in detail. It is not apparent how conformational changes might propagate through the 20S CP in order to provide communication between the two 19S RP binding surfaces, which would presumably be a requirement for asymmetry to be an inherent property of fully assembled complexes. The potential of allosteric communication between two ends of a 26S proteasome complex and between the 20S CP proteolytic sites and the 19S RP is therefore an interesting but currently unresolved possibility. Another possibility is that a fraction of the double-capped 26S proteasomes analyzed had a defect in one of the RP, such as a partly assembled/disassembled conformation, or perhaps even an alternative binding partner, such as Blm10. Thus, the alignment procedure would have favored superimposing the most clearly defined 19S RP at one end of the reconstruction, with all of the less clearly defined 19S RP at the opposite end, where the inclusion of noise would yield apparent structural differences. Resolving this issue and understanding the possibility of allostery between two ends of the 26S proteasome will be an important challenge as the structural studies are pushed to higher resolution.
A surprise from these studies is that the lid sits on the side of the 19S RP, rather than on the top as had been generally imagined. Rpn3, 5, 6, 7, 9, and 12 associate in a horseshoe-like configuration through their PCI modules, while their N-terminal solenoid domains radiate widely. This allows Rpn6, and to a lesser extent Rpn5, to contact the C-termini of 20S α2 and α1, respectively, and so presumably contribute to overall stability of the 26S complex. This is consistent with a very recent report that increased expression of Rpn6 confers resistance to proteotoxic stress and increases longevity in C. elegans, perhaps because increased Rpn6 promotes stability of active 26S proteasome complex (92). Rpn8 and Rpn11 dimerize through their MPN domains, and their C-terminal helices associate with the C-terminal helices of the six PCI-containing lid subunits in a bundle arrangement (3). This places the Rpn11 deubiquitylase over the mouth of the ATPases, and superposition with a structure of the homologous AMSH enzyme bound with diubiquitin (71) supports the model that Rpn11 removes ubiquitin as substrate enters the ATPase channel.
The Rpt1-6 ATPases form a hexameric ring in which the N-terminal domains project up to contact other 19S RP subunits, and the ATPase cassettes lie close to the 20S CP α subunits. The C-termini of Rpt2, Rpt3 and Rpt5, which are the ATPase subunits that display C-terminal HbYX motifs, dock at the α3/α4, α1/α2, and α5/α6 pockets, respectively, consistent with findings from site-directed crosslinking (87). The details of these interactions are not currently resolved, but presumably resemble the structures seen earlier for the ATP-independent activators.
Another major surprise is that the pore region of the ATPase subunits assemble into a spiral staircase-like arrangement, with the lowest and highest subunits, Rpt2 and Rpt3, respectively, separated by Rpt6 in an intermediate position (3; 43). It is generally thought that hexameric ATPase unfoldases, including the proteasome, function in a mixed nucleotide state with ATP or ADP bound to some subunits while other subunits are unbound (22; 28; 78). Beautiful structures of analogous nucleic acid helicases provide models for how propagation of a wave of conformational changes, driven by ATP binding, hydrolysis, and release around the ring are coupled to translocation of the bound substrate (15; 86). The homohexameric nucleic acid helicase structures revealed a spiral configuration, analogous to that of the proteasome Rpt subunits, presumably because they were complexes with substrate, which induces asymmetry, and because crystallization selected just one of the six orientations that represent propagation of the spiral staircase conformation around the ring. It is not clear, however, how to reconcile this attractive “wave” model of the helicases with the proteasome reconstructions because, unlike the constraints of a crystal lattice, the 26S proteasome EM reconstructions are not expected to favor one particular arrangement of the propagating ATPase spiral, and the multiple staircase configurations would presumably appear as an averaged/blurred map with an apparently more circular arrangement of ATPase density. Thus, understanding the mechanistic implications of the defined spiral conformation observed for the Rpt subunits presents a challenging and enticing problem for future studies.
It is striking that the ubiquitin receptor subunits Rpn10 and Rpn13 are located at the distal end of the activator from the 20S proteasome interface. Similarly, the ubiquitin shuttle receptors are likely to be bound distant from the entrance to the ATPase hexamer. This arrangement is consistent with the model that ubiquitin binding promotes degradation by increasing the affinity of tagged substrate, without playing a more direct role in the processes of unfolding or translocation. Nevertheless, important functional questions remain, including the possibility of coordination between different ubiquitin binding sites, the mechanistic basis for preference of binding polyubiquitin rather than monoubiquitin, and the possibility of coupling between ubiquitin binding and substrate processing by the ATPases (57).
The location of deubiquitylating enzymes within the 19S RP is of mechanistic relevance. As discussed above, Rpn11 is poised to remove ubiquitin as substrate enters the ATPase channel. Interestingly, the substantial conformational differences seen between the isolated lid and the 26S proteasome may serve to maintain Rpn11 in an inactive state until assembly is completed, with a possible trigger for the conformational change being association of Rpn5 with the 20S CP (43). The more peripheral locations inferred for UCH37 and Ubp6/USP14 is consistent with their likely roles in editing. The EM reconstructions suggest that Rpn1-Upb6 may have some mobility within the 19S RP, and Uch37 is likely to enjoy considerable conformational freedom due to the flexible ~150 residue linker between the Rpn13 N-terminal PRU domain that binds Rpn2 and the C-terminal domain that binds Uch37. This flexibility may allow Uch37 and Ubp6 to efficiently disassemble polyubiquitin chains that might otherwise clog the 26S proteasome. Ubp6 also provides an additional example of the complexity of proteasome regulation and the importance of further studies to understand conformational changes because its binding is reported to regulate proteasome activity independently of its catalytic activity (26).
PROTEASOME ASSEMBLY (Figure 4)
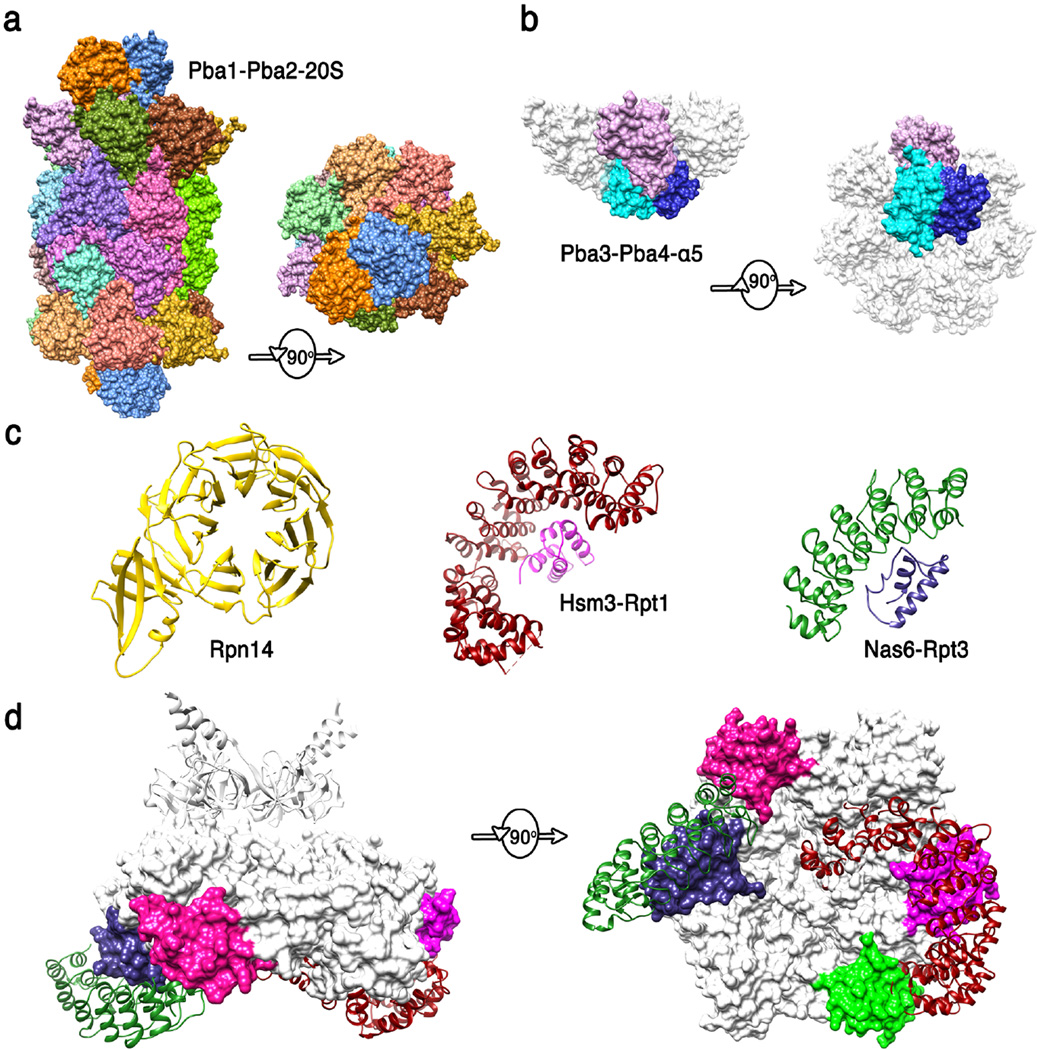
Figure 4. Structures of proteasome chaperones.
(a) Pba1-Pba2 (orange and blue) structure from (83) (pdb code 4g4s). Side and top views are shown of the complex with the 20S CP. The contacts seen in this structure are presumably maintained from the earliest stages of α-ring assembly to maturation of the 20S CP. (b) Pba3-Pba4 (shades of blue) structure from (100) (pdb code 2z5c). Side and bottom views are shown of this complex with α5, with the other α subunits modeled in white based on their structure in the mature 20S CP. This structure explains why Pba3-Pba4 are lost as β subunits are added to the assembling 20S CP. (c) Structures of 19S RP chaperones. Rpn14 (36) (pdb code 3acp), Hsm3 complex with C-terminal domain of Rpt1 (1) (pdb code 4a3v), and Nas6/gankyrin complex with the C-terminal domain of Rpt3 (54) (pdb code 2dzn). (d) Side and bottom model of Hsm3 and Nas6 docked onto the Rpt hexamer model. Substantial steric clashes would occur with the 20S CP (not shown) in the 26S proteasome, and minor steric clashes are suggested with Rpt subunits, consistent with the mechanisms that the 19S RP chaperones modulate interactions between ATPase subcomplexes and with the 20S CP. The C-terminal domains of Rpt5 and Rpt6 that bind Nas2 and Rpn14 are colored green and pink, respectively.
Assembly chaperones of the 20S core particle
In most species, 20S CP assembly proceeds with formation of a ring of α subunits followed by addition of β subunits to form half proteasomes, which dimerize to form the 20S CP, with a final maturation step coupled to cleavage of the β subunit pro-peptides (53). Assembly is promoted by chaperones, including the heterodimer Pba1-Pba2/Poc1-Poc2/PAC1-PAC2, which associates with the assembling 20S CP from the earliest stages of α ring formation to completion of the mature 20S CP. Although biochemical studies indicate that archaeal 20S proteasomes do not require assembly factors, the archaeal proteins PbaA and PbaB are thought to function analogously to the eukaryotic Pba1-Pba2 (39). The structure of a complex between Pba1-Pba2 and the 20S CP demonstrates that Pba1-Pba2 directly contacts α4, α5, α6, and α7, and that it binds through its C-terminal residues using very similar principles to those observed for PA26 and Blm10, although Pba1-Pba2 is not itself a proteasome activator (83). Binding of Pba1-Pba2 does not substantially alter 20S CP structure, suggesting that binding may promote assembly by stabilizing the correct relative positions of α sub units.
Although it is unrelated to Pba1-Pba2, the Pba3-Pba4/Dmp3-Dmp4/PAC3-PAC4 heterodimer also chaperones early stages of 20S CP assembly. The crystal structure of Pba3-Pba4/Dmp3-Dmp4 in complex with α5 demonstrates that binding occurs on the face opposite from that contacted by Pba1-Pba2, which explains why Pba3-Pba4 dissociates as β subunits are added following assembly of the α ring (100). The interaction with Pba3-Pba4 is important for promoting the appropriate association of α subunits, especially α3 and α4; α3 is notable in being non-essential in yeast, with α4 able to substitute in the case of α3 deficiency (38)
The final stages of associating two half proteasomes is promoted by Ump1, which is degraded upon proteasome assembly (52). Although structural data are not available for Ump1 interactions, structural insights to the final stages of maturation have been provided by the crystal structure a mutant Rhodococcus proteasome that retains its pro-peptides. This reveals that the pro-peptide contacts two adjacent α subunits, thereby promoting assembly (40). Similarly, the structure of another mutant proteasome guides models of the detailed structural requirements for maturation (98).
Assembly chaperones of the 19S regulatory particle
Assembly of the 19S RP base complex is facilitated by four chaperones, Hsm3/S5b, Nas2/p27, Rpn14/PAAF1, and Nas6/gankyrin (55). The leading model holds that assembly proceeds via formation of three subcomplexes that each contains two of the ATPases, one or two chaperones, and in some cases one or more Rpn subunits: Hsm3-Rpt1-Rpt2-Rpn2; Nas6-Rpn14-Rpt3-Rpt6-Rpn2-Rpn13; Nas2-Rpt4-Rpt5. Formation of the base is followed by addition of the lid to form the 19S RP, which then associates with the 20S CP to form the 26S proteasome. Interestingly, the base chaperones each bind to the C-terminal domain of a specific Rpt ATPase (Hsm3-Rpt1; Nas2-Rpt5; Nas6-Rpt3; Rpn14-Rpt6). Despite this functional similarity, the four base chaperones adopt quite different structures, as indicated by the sequence prediction of a PDZ domain for Nas2, and crystal structures that show Nas6 comprises ankyrin repeats (54), Rpn14 forms a WD40 propeller (36), and Hsm3 comprises HEAT repeats (84). The mechanism of binding to Rpt C-termini was revealed for Hsm3 and Nas6, whose structures were determined as complexes with the Rpt1 and Rpt3 C-terminal domains, respectively. Docking of these complex structures onto the EM model of the 26S proteasome indicates that binding of Hsm3 and Nas6 is incompatible with the assembled structure due to clashes with the 20S CP. This modeling also suggests that Hsm3 may clash with Rpt5, although this apparent overlap may be relieved by relatively modest conformational changes. The modeling also suggests that binding of Nas6 is incompatible with the positions of Rpn5 and Rpn6 in the 26S proteasome, which may indicate that Nas6 may regulate association of base and lid, although conformational changes in Rpn5 and 6 have been noted in the isolated lid (43). Due to the location of the Rpt C-terminal domains, it is likely that binding of Nas2 and Rpn14 are also incompatible with 19S RP-20S CP association, and clashes between Hsm3 and Nas2 also seem possible. Thus, despite the uncertain nature of this simple modeling, the current structures are consistent with roles for the base chaperones in regulating interactions between specific Rpt subunits, between base and lid, and between the 19S RP and the 20SCP.
IMPLICATIONS FOR FUTURE STUDIES
Recent years have seen remarkable progress in proteasome structural biology. Detailed structures are available for the 20S CP including numerous complexes with active site inhibitors, two ATP-independent activator complexes, several isolated 19S RP subunits, and several 20S CP and 19S RP chaperone complexes. Moreover, EM reconstructions coupled with high-resolution structures of individual subunits is providing valuable models of the 26 proteasome. Major goals for future structural studies include pushing models of the 26S proteasome to higher resolution and providing structural information on additional proteasome complexes, such as the numerous proteins reported to interact substoichiometrically with the proteasome (5; 91), and the recently reported functional association, at least in archaea, of the 20S CP with Cdc48 (2).
It is apparent that conformational changes are an important component of proteasome function. This is most apparent for the 19S RP ATPases, which drive substrate unfolding and translocate substrate into the 20S CP. Understanding how these Rpt subunits move during ATP binding and hydrolysis, and if the pore regions remain in the spiral stair configuration seen in the EM reconstructions or undergoes a wave of conformational changes analogous to those proposed for the rho and E1 helicases, is a high priority. The functional importance of movement is also apparent for the ubiquitin receptors and for the associated deubiquitylating enzymes, and it will be important to understand how binding and conformational changes are coordinated and how they function to regulate proteasome activity. Finally, changes in association are explicit in the processes of proteasome assembly, and one exciting possibility for future functional studies is that these might be regulated events that are of physiological importance.
SUMMARY POINTS.
Gate/repression
The proteolytic sites of 20S proteasome core particles are sequestered in a hollow structure that promotes protein unfolding, and are accessed via gates through the α subunits that are closed by different mechanisms in eukaryotes, archaea, and eubacteria.
Proteasome inhibitors offer therapeutic potential, with recent advances including structures that explain the basis for a specific M. tuberculosis 20S CP inhibitor and the increased preference for hydrophobic P1 residues in the immunoproteasome
Mechanisms of binding and activation by the ATP-independent activators are now understood in structural detail, although their biological roles are less clear. The principles of binding and open gate structure seem to apply broadly, including to the 19S RP
Structures of many of the 19S RP subunits have been determined at atomic resolution either directly or on the basis of homology modeling.
Electron microscopy has recently produced models of the 26S proteasome at ~7Å resolution. Especially important insights include the overall arrangement of base and lid subcomplexes, the location of ubiquitin receptors and deubiquitylating enzymes, and the arrangement of the Rpt ATPase subunits.
Proteasome assembly follows a highly regulated pathway that is guided by molecular chaperones that promote some specific subunit interactions and appear to inhibit other interactions until the appropriate binding partners are assembled.
There is considerable scope for future structural studies including a need for higher resolution structures of the 26S proteasome, understanding the importance of numerous implied conformational changes and other dynamic processes such as binding/release of substoichiometric binding partners, and the potential role of additional activators such as Cdc48.
Box. Structures 19S RP subunits and associated proteins shown in Figure 3.
Rad23
Structures from (93) (pdb code 1qze). This shuttle receptor comprises four folded domains that are connected by flexible linkers. The UBA domains bind ubiquitin, or in its absence can bind its own Ubl domain. The Ubl domain binds Rpn1, which is shown in blue in the central panel but not in an expanded view because it is a homology model based on the structure of Rpn2.
Ubpβ
Structure of this ubiquitin aldehyde (Ubal) complex from (29) (pdb code 2ayo). Ubp6 is a deubiquitylating enzyme that binds Rpn1 through its Ubl domain, whose structure has not yet been determined.
Rpn13
Structures from (10; 74) (pdb codes 2z59 and 2kqz). Rpn13 binds a flexible sequence at the C-terminus of Rpn2 through its N-terminal PRU domain, which also binds ubiquitin. In most species, although not S. cerevisiae, the PRU domain is followed by a flexible linker and a helical C-terminal domain that binds the deubiquitylating enzyme Uch37.
Rpn2
Structure from (27) (pdb code 4ady). Rpn2 is the second largest 19S RP subunit after Rpn1, and also provides a homology model for Rpn1. These proteins comprise a helical toroid domain from which a helical N-terminal rod domain and a mostly β C-terminal domain project on one side. The C-terminus of the ordered structure, from which the Rpn13 binding site projects, is labeled CT.
Rpn6 and Rpn12
Structures from (6; 56) (pdb codes 3txm and 4b0z). These proteins closely resemble each other and serve as homology models for Rpn3, Rpn5, Rpn7, and Rpn9.
Rpn8 and Rpn11
The model of this heterodimer follows the analysis of reference (3) and the crystal structure of Rpn8/MOV34 (70) (pdb code 2o95). Rpn11 is the enzyme that removes ubiquitin from substrates as they are translocated by the ATPases. Rpn8 shares sequence similarity with Rpn11 but lacks active site residues.
Rpn10
The N-terminal VWA domain (65) (pdb code 2×5n) is followed by a flexible segment that includes one (in yeast) or two UIM domains as seen in this structure of a human S5a construct in complex with diubiquitin (104) (pdb code 2kde).
Rpt1-6
The Rpt subunits form a hexamer that is modeled in the side view of the central panel. This is based on the structure of the N-terminal CC-OB hexamer (102) (pdb code 3h43), which is shown from the top in the upper panel, and the structure of a monomeric PAN ATPase cassette (102) (pdb code 3h4m), which is docked into a hexamer based on the EM map and viewed from the bottom in the lower panel.
GLOSSARY AND ACRONYMS
- Proteasome
refers to a variety of complexes of the 20S core particle that can be bound on one or both ends by activators
- 20S core particle (20S CP)
a 28-subunit protease that houses proteolytic sites in a central chamber
- Immunoproteasome
a 20S CP variant of higher eukaryotes in which the three constitutive catalytic subunits are replaced by inducible counterparts
- Proteasome activator
proteins or protein complexes that stimulate 20S CP peptidase activity in biochemical studies by inducing an open conformation of the entrance/exit gate
- 11S
a family of heptameric ATP-independent proteasome activators that include PA26 from T. brucei and the α, β, γ homologs of PA28/REG in higher eukaryotes
- Blm10
an ATP-independent proteasome activator that was named for the mistaken belief that it confers resistance to bleomycin. Blm10 is the yeast homolog of Proteasome Activator 200 (PA200)
- 19S regulatory particle (19S RP)
an ATP-dependent proteasome activator that comprises 19 subunits, including six ATPases. Unlike the 11S and Blm10 activators, it has the well-defined biological role of recognizing polyubiquitylated proteins, unfolding them, and translocating them into the 20S CP for degradation
- 26S proteasome
complexes of the 20S CP with one or two 19S RP
- Hybrid proteasomes
complexes of the 20S CP with a 19S RP on one end and another activator such as 11S or PA200 on the other end
- Ubiquitin
an 8.5 kDa protein that is covalently attached to other proteins to modify their properties, including their degradation by the 26S proteasome
- Polyubiquitylation
ubiquitin can itself be ubiquitylated, which can give rise to polyubiquitylation. Polyubiquitylation through Lys48 is typically associated with degradation by the 26S proteasome
- Ubiquitylation
the conjugation of ubiquitin to target proteins is accomplished by an E1, E2, E3 enzyme cascade that includes many different E2/E3 enzymes that determine the specificity of ubiquitylation. Although not discussed in this review, ubiquitylation enzymes have been reported to bind substoichiometrically with the 26S proteasome
- Deubiquitylation
three deubiquitylases, which belong to different structural families, are tightly associated with the 26S. Rpn11/POH1 removes ubiquitin attached to substrate as it is translocated by the Rpt ATPases. Ubp6/USP14 and Uch37 are more peripherally located and edit inadequately ubiquitylated substrates and disassembly unattached polyubiquitin chains
- Proteasome/cyclosome (PC) repeat
a 35–40 amino acid residue motif that folds into two helices. The largest 19S RC subunits, Rpn1 and Rpn2, each contain 11 PC repeats, which fold into a toroidal domain
- Pup
prokaryotic ubiquitin-like protein, a 7 kDa protein that is natively unstructured and covalently conjugated to other proteins to target them for degradation in a manner analogous to ubiquitylation, with the N-terminal sequence of Pup binding to the N-terminal domain of the Mpa ATPase
- PAN/ARC/Mpa
homohexameric ATPases of archaea (PAN) or eubacteria (ARC/Mpa) that function analogously to the 19S RP
- Shuttle receptors
proteins, typified by Rad23, that can bind ubiquitin through C-terminal UBA domains and associate transiently with Rpn1 and perhaps other subunits of the 19S RP through an N-terminal Ubl domain, with the Ubl and UBA domains contacting each other in the absence of a binding partner
- Chaperones
specific molecular chaperones appear to promote 20S CP and 19S RP assembly by promoting some appropriate subunit contacts while delaying other interactions
- Cdc48
a hexameric ATPase, known as p97 in higher eukaryotes, that is implicated in numerous biological processes, including interactions with ubiquitin. A recent report indicates that Cdc48 serves as a proteasome activator in archaea. It will be of interest to determine if Cdc48/p97 functions as an alternative ATP-dependent activator to the 19S RP in other species
REFERENCES
- 1.Barrault MB, Richet N, Godard C, Murciano B, Le Tallec B, et al. Dual functions of the Hsm3 protein in chaperoning and scaffolding regulatory particle subunits during the proteasome assembly. Proc Natl Acad Sci U S A. 2012;109:E1001–E1010. doi: 10.1073/pnas.1116538109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barthelme D, Sauer RT. Identification of the Cdc48*20S proteasome as an ancient AAA+ proteolytic machine. Science. 2012;337:843–846. doi: 10.1126/science.1224352. Reports an exciting recent discovery that an archaeal Cdc48 functions as a proteasome activator. Eukaryotic Cdc48/p97 conserves the distinctive C-terminal tripeptide of established proteasome activators, and it will be of considerable interest to see if Cdc48/p97 functions as a proteasome activator in other species.
- 3. Beck F, Unverdorben P, Bohn S, Schweitzer A, Pfeifer G, et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1213333109. Currently, the highest resolution EM reconstruction of the 26S proteasome. Involved development of methods for map fitting and the integration of multiple sources of information.
- 4.Benaroudj N, Zwickl P, Seemuller E, Baumeister W, Goldberg AL. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol Cell. 2003;11:69–78. doi: 10.1016/s1097-2765(02)00775-x. [DOI] [PubMed] [Google Scholar]
- 5.Besche HC, Haas W, Gygi SP, Goldberg AL. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry. 2009;48:2538–2549. doi: 10.1021/bi802198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boehringer J, Riedinger C, Paraskevopoulos K, Johnson EO, Lowe ED, et al. Structural and functional characterisation of Rpn12 identifies residues required for Rpn10 proteasome incorporation. Biochem J. 2012 doi: 10.1042/BJ20120542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bohn S, Beck F, Sakata E, Walzthoeni T, Beck M, et al. From the Cover: Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc Natl Acad Sci U S A. 2010;107:20992–20997. doi: 10.1073/pnas.1015530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgie SE, Bingman CA, Soni AB, Phillips GN., Jr Structural characterization of human Uch37. Proteins. 2011 doi: 10.1002/prot.23147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Barton LF, Chi Y, Clurman BE, Roberts JM. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol Cell. 2007;26:843–852. doi: 10.1016/j.molcel.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X, Lee BH, Finley D, Walters KJ. Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein hRpn2. Mol Cell. 2010;38:404–415. doi: 10.1016/j.molcel.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.da Fonseca PC, He J, Morris EP. Molecular model of the human 26S proteasome. Mol Cell. 2012;46:54–66. doi: 10.1016/j.molcel.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 12.Dange T, Smith D, Noy T, Rommel PC, Jurzitza L, et al. Blm10 protein promotes proteasomal substrate turnover by an active gating mechanism. J Biol Chem. 2011;286:42830–42839. doi: 10.1074/jbc.M111.300178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Djuranovic S, Hartmann MD, Habeck M, Ursinus A, Zwickl P, et al. Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal ATPases. Mol Cell. 2009;34:580–590. doi: 10.1016/j.molcel.2009.04.030. One of papers that established the hexameric structure of the proteasomal ATPase N-terminal domains. Also established that the coiled-coils promote protein unfolding.
- 14.Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 15.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 16.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 17.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 19.Forster A, Whitby FG, Hill CP. The pore of activated 20S proteasomes has an ordered 7-fold symmetric conformation. EMBO J. 2003;22:4356–4364. doi: 10.1093/emboj/cdg436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forster F, Lasker K, Beck F, Nickell S, Sali A, Baumeister W. An atomic model AAA-ATPase/20S core particle sub-complex of the 26S proteasome. Biochem Biophys Res Commun. 2009;388:228–233. doi: 10.1016/j.bbrc.2009.07.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, et al. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 22.Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goh AM, Walters KJ, Elsasser S, Verma R, Deshaies RJ, et al. Components of the ubiquitin-proteasome pathway compete for surfaces on Rad23 family proteins. BMC Biochem. 2008;9:4. doi: 10.1186/1471-2091-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 25.Hamazaki J, Iemura S, Natsume T, Yashiroda H, Tanaka K, Murata S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 2006;25:4524–4536. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 27.He J, Kulkarni K, da Fonseca PC, Krutauz D, Glickman MH, et al. The structure of the 26S proteasome subunit Rpn2 reveals its PC repeat domain as a closed toroid of two concentric alpha-helical rings. Structure. 2012;20:513–521. doi: 10.1016/j.str.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 28.Horwitz AA, Navon A, Groll M, Smith DM, Reis C, Goldberg AL. ATP-induced structural transitions in PAN, the proteasome-regulatory ATPase complex in Archaea. J Biol Chem. 2007;282:22921–22929. doi: 10.1074/jbc.M702846200. [DOI] [PubMed] [Google Scholar]
- 29.Hu M, Li P, Song L, Jeffrey PD, Chenova TA, et al. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huber EM, Basler M, Schwab R, Heinemeyer W, Kirk CJ, et al. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell. 2012;148:727–738. doi: 10.1016/j.cell.2011.12.030. Structures of mouse liver consititutive and immunoproteasome 20S CP were determined alone and in complex with an inhibitor. These explained how specificity of the β1, and β5 subunits is modified to promote cleavage after hydrophobic side chains by the immunoproteasome.
- 31.Huber EM, Groll M. Inhibitors for the Immuno- and Constitutive Proteasome: Current and Future Trends in Drug Development. Angew Chem Int Ed Engl. 2012 doi: 10.1002/anie.201201616. [DOI] [PubMed] [Google Scholar]
- 32.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inobe T, Fishbain S, Prakash S, Matouschek A. Defining the geometry of the two-component proteasome degron. Nat Chem Biol. 2011;7:161–167. doi: 10.1038/nchembio.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwanczyk J, Sadre-Bazzaz K, Ferrell K, Kondrashkina E, Formosa T, et al. Structure of the Blm10-20 S proteasome complex by cryo-electron microscopy. Insights into the mechanism of activation of mature yeast proteasomes. J Mol Biol. 2006;363:648–659. doi: 10.1016/j.jmb.2006.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kay LE. Solution NMR spectroscopy of supra-molecular systems, why bother? Amethyl-TROSYview. J Magn Reson. 2011;210:159–170. doi: 10.1016/j.jmr.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 36.Kim S, Saeki Y, Fukunaga K, Suzuki A, Takagi K, et al. Crystal structure of yeast rpn14, a chaperone of the 19 S regulatory particle of the proteasome. J Biol Chem. 2010;285:15159–15166. doi: 10.1074/jbc.M110.104042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knowlton JR, Johnston SC, Whitby FG, Realini C, Zhang Z, et al. Structure of the proteasome activator REGalpha (PA28alpha) Nature. 1997;390:639–643. doi: 10.1038/37670. [DOI] [PubMed] [Google Scholar]
- 38.Kusmierczyk AR, Kunjappu MJ, Funakoshi M, Hochstrasser M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat Struct Mol Biol. 2008;15:237–244. doi: 10.1038/nsmb.1389. [DOI] [PubMed] [Google Scholar]
- 39.Kusmierczyk AR, Kunjappu MJ, Kim RY, Hochstrasser M. A conserved 20S proteasome assembly factor requires a C-terminal HbYX motif for proteasomal precursor binding. Nat Struct Mol Biol. 2011;18:622–629. doi: 10.1038/nsmb.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwon YD, Nagy I, Adams PD, Baumeister W, Jap BK. Crystal structures of the Rhodococcus proteasome with and without its pro-peptides: implications for the role of the pro-peptide in proteasome assembly. J Mol Biol. 2004;335:233–245. doi: 10.1016/j.jmb.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 41.Lam YA, DeMartino GN, Pickart CM, Cohen RE. Specificity of the ubiquitin isopeptidase in the PA700 regulatory complex of 26 S proteasomes. J Biol Chem. 1997;272:28438–28446. doi: 10.1074/jbc.272.45.28438. [DOI] [PubMed] [Google Scholar]
- 42.Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 43. Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. One of the highest resolution EM reconstructions of the 26S proteasome. The lid subcomplex was purified and reconstructed separately following coexpression in E. coli. This allowed localization of the subunit termini by expression of MBP fusions.
- 44.Lasker K, Forster F, Bohn S, Walzthoeni T, Villa E, et al. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci U S A. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. Inhibition of the Ubp6/USP14 proteasomal deubiquitylase enhances degradation of proteins prone to aggregation, and motivates efforts to develop novel therapeutics.
- 46.Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, et al. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 47.Li D, Li H, Wang T, Pan H, Lin G. Structural basis for the assembly and gate closure mechanisms of the Mycobacterium tuberculosis 20S proteasome. EMBO J. 2010;29:2037–2047. doi: 10.1038/emboj.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, Amazit L, Long W, Lonard DM, Monaco JJ, O’Malley BW. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Mol Cell. 2007;26:831–842. doi: 10.1016/j.molcel.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 49. Lin G, Li D, de Carvalho LP, Deng H, Tao H, et al. Inhibitors selective for mycobacterial versus human proteasomes. Nature. 2009;461:621–626. doi: 10.1038/nature08357. Crystal structures of the M. tuberculosis 20S CP unbound and inhibited show how a conformational change that has not need seen in the 20S CP from other species protects the inhibited complex.
- 50.Liu CW, Corboy MJ, DeMartino GN, Thomas PJ. Endoproteolytic activity of the proteasome. Science. 2003;299:408–411. doi: 10.1126/science.1079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 52.Matias AC, Ramos PC, Dohmen RJ. Chaperone-assisted assembly of the proteasome core particle. Biochem Soc Trans. 2010;38:29–33. doi: 10.1042/BST0380029. [DOI] [PubMed] [Google Scholar]
- 53.Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- 54.Nakamura Y, Nakano K, Umehara T, Kimura M, Hayashizaki Y, et al. Structure of the oncoprotein gankyrin in complex with S6 ATPase of the 26S proteasome. Structure. 2007;15:179–189. doi: 10.1016/j.str.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 55.Park S, Tian G, Roelofs J, Finley D. Assembly manual for the proteasome regulatory particle: the first draft. Biochem Soc Trans. 2010;38:6–13. doi: 10.1042/BST0380006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pathare GR, Nagy I, Bohn S, Unverdorben P, Hubert A, et al. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc Natl Acad Sci U S A. 2012;109:149–154. doi: 10.1073/pnas.1117648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peth A, Uchiki T, Goldberg AL. ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Mol Cell. 2010;40:671–681. doi: 10.1016/j.molcel.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prakash S, Inobe T, Hatch AJ, Matouschek A. Substrate selection by the proteasome during degradation of protein complexes. Nat Chem Biol. 2009;5:29–36. doi: 10.1038/nchembio.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 60.Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006;25:5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rape M, Jentsch S. Taking a bite: proteasomal protein processing. Nat Cell Biol. 2002;4:E113–E116. doi: 10.1038/ncb0502-e113. [DOI] [PubMed] [Google Scholar]
- 63.Rechsteiner M, Hill CP. Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 2005;15:27–33. doi: 10.1016/j.tcb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 64.Religa TL, Sprangers R, Kay LE. Dynamic regulation of archaeal proteasome gate opening as studied by TROSY NMR. Science. 2010;328:98–102. doi: 10.1126/science.1184991. [DOI] [PubMed] [Google Scholar]
- 65.Riedinger C, Boehringer J, Trempe JF, Lowe ED, Brown NR, et al. Structure of Rpn10 and its interactions with polyubiquitin chains and the proteasome subunit Rpn12. J Biol Chem. 2010;285:33992–34003. doi: 10.1074/jbc.M110.134510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosenzweig R, Bronner V, Zhang D, Fushman D, Glickman MH. Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J Biol Chem. 2012;287:14659–14671. doi: 10.1074/jbc.M111.316323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ruschak AM, Religa TL, Breuer S, Witt S, Kay LE. The proteasome antechamber maintains substrates in an unfolded state. Nature. 2010;467:868–871. doi: 10.1038/nature09444. One of a series of papers that use NMR approaches to investigate proteasome structure and dynamics. This paper found that the proteasome interior favors the unfolded state of substrate proteins.
- 68.Sadre-Bazzaz K, Whitby FG, Robinson H, Formosa T, Hill CP. Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Mol Cell. 2010;37:728–735. doi: 10.1016/j.molcel.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sakata E, Bohn S, Mihalache O, Kiss P, Beck F, et al. Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proc Natl Acad Sci U S A. 2012;109:1479–1484. doi: 10.1073/pnas.1119394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanches M, Alves BS, Zanchin NI, Guimaraes BG. The crystal structure of the human Mov34 MPN domain reveals a metal-free dimer. J Mol Biol. 2007;370:846–855. doi: 10.1016/j.jmb.2007.04.084. [DOI] [PubMed] [Google Scholar]
- 71.Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, et al. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–362. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 72.Savulescu AF, Glickman MH. Proteasome activator 200: the heat is on. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.006890. R110 006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmidt M, Haas W, Crosas B, Santamaria PG, Gygi SP, et al. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat Struct Mol Biol. 2005;12:294–303. doi: 10.1038/nsmb914. [DOI] [PubMed] [Google Scholar]
- 74.Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, et al. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seemuller E, Lupas A, Stock D, Lowe J, Huber R, Baumeister W. Proteasome from Thermoplasma acidophilum: a threonine protease. Science. 1995;268:579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- 76.Sijts EJ, Kloetzel PM. The role of the proteasome in the generation of MHC class I ligands and immune responses. Cell Mol Life Sci. 2011;68:1491–1502. doi: 10.1007/s00018-011-0657-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith DM, Fraga H, Reis C, Kafri G, Goldberg AL. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell. 2011;144:526–538. doi: 10.1016/j.cell.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sprangers R, Kay LE. Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature. 2007;445:618–622. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]
- 80.Sprangers R, Li X, Mao X, Rubinstein JL, Schimmer AD, Kay LE. TROSY-based NMR evidence for a novel class of 20S proteasome inhibitors. Biochemistry. 2008;47:6727–6734. doi: 10.1021/bi8005913. [DOI] [PubMed] [Google Scholar]
- 81.Stadtmueller BM, Ferrell K, Whitby FG, Heroux A, Robinson H, et al. Structural models for interactions between the 20S proteasome and its PAN/19S activators. J Biol Chem. 2010;285:13–17. doi: 10.1074/jbc.C109.070425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stadtmueller BM, Hill CP. Proteasome activators. Mol Cell. 2011;41:8–19. doi: 10.1016/j.molcel.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stadtmueller BM, Kish-Trier E, Ferrell K, Petersen CN, Robinson H, et al. Structure of a Proteasome-Pba1-Pba2 Complex: Implications for Proteasome Assembly, Activation, and Biological Function. J Biol Chem. 2012 doi: 10.1074/jbc.M112.367003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takagi K, Kim S, Yukii H, Ueno M, Morishita R, et al. Structural basis for specific recognition of Rpt1p, an ATPase subunit of 26 S proteasome, by proteasome-dedicated chaperone Hsm3p. J Biol Chem. 2012;287:12172–12182. doi: 10.1074/jbc.M112.345876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Takeuchi J, Chen H, Coffino P. Proteasome substrate degradation requires association plus extended peptide. EMBO J. 2007;26:123–131. doi: 10.1038/sj.emboj.7601476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tian G, Park S, Lee MJ, Huck B, McAllister F, et al. An asymmetric interface between the regulatory and core particles of the proteasome. Nat Struct Mol Biol. 2011 doi: 10.1038/nsmb.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tian L, Holmgren RA, Matouschek A. A conserved processing mechanism regulates the activity of transcription factors Cubitus interruptus and NF-kappaB. Nat Struct Mol Biol. 2005;12:1045–1053. doi: 10.1038/nsmb1018. [DOI] [PubMed] [Google Scholar]
- 89.Tomko RJ, Jr, Funakoshi M, Schneider K, Wang J, Hochstrasser M. Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: implications for proteasome structure and assembly. Mol Cell. 2010;38:393–403. doi: 10.1016/j.molcel.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, et al. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure. 2002;10:609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- 91.Verma R, Chen S, Feldman R, Schieltz D, Yates J, et al. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vilchez D, Morantte I, Liu Z, Douglas PM, Merkwirth C, et al. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012 doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 93.Walters KJ, Lech PJ, Goh AM, Wang Q, Howley PM. DNA-repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a. Proc Natl Acad Sci U S A. 2003;100:12694–12699. doi: 10.1073/pnas.1634989100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wang T, Darwin KH, Li H. Binding-induced folding of prokaryotic ubiquitin-like protein on the Mycobacterium proteasomal ATPase targets substrates for degradation. Nat Struct Mol Biol. 2010;17:1352–1357. doi: 10.1038/nsmb.1918. The bacterial Pup protein functions analogously to ubiquitin to target substrate proteins to the proteasome. This paper shows how Pup, binds to the coiled-coil of Mpa, thereby positioning the substrate over the entrance to the central conduit to the ATPase pore loops and the proteasome.
- 95.Wang T, Li H, Lin G, Tang C, Li D, et al. Structural insights on the Mycobacterium tuberculosis proteasomal ATPase Mpa. Structure. 2009;17:1377–1385. doi: 10.1016/j.str.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wenzel T, Baumeister W. Conformational constraints in protein degradation by the 20S proteasome. Nat Struct Biol. 1995;2:199–204. doi: 10.1038/nsb0395-199. [DOI] [PubMed] [Google Scholar]
- 97.Whitby FG, Masters EI, Kramer L, Knowlton JR, Yao Y, et al. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- 98.Witt S, Kwon YD, Sharon M, Felderer K, Beuttler M, et al. Proteasome assembly triggers a switch required for active-site maturation. Structure. 2006;14:1179–1188. doi: 10.1016/j.str.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 99.Yao T, Song L, Xu W, DeMartino GN, Florens L, et al. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- 100.Yashiroda H, Mizushima T, Okamoto K, Kameyama T, Hayashi H, et al. Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes. Nat Struct Mol Biol. 2008;15:228–236. doi: 10.1038/nsmb.1386. [DOI] [PubMed] [Google Scholar]
- 101.Yu Y, Smith DM, Kim HM, Rodriguez V, Goldberg AL, Cheng Y. Interactions of PANs C-termini with archaeal 20S proteasome and implications for the eukaryotic proteasome-ATPase interactions. EMBO J. 2010;29:692–702. doi: 10.1038/emboj.2009.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zhang F, Hu M, Tian G, Zhang P, Finley D, et al. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009;34:473–484. doi: 10.1016/j.molcel.2009.04.021. One of papers that established the hexameric structure of the proteasomal ATPase N-terminal domains. Also reported the structure of the PAN ATPase cassette.
- 103.Zhang F, Wu Z, Zhang P, Tian G, Finley D, Shi Y. Mechanism of substrate unfolding and translocation by the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009;34:485–496. doi: 10.1016/j.molcel.2009.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang N, Wang Q, Ehlinger A, Randles L, Lary JW, et al. Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13. Mol Cell. 2009;35:280–290. doi: 10.1016/j.molcel.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]