Abstract
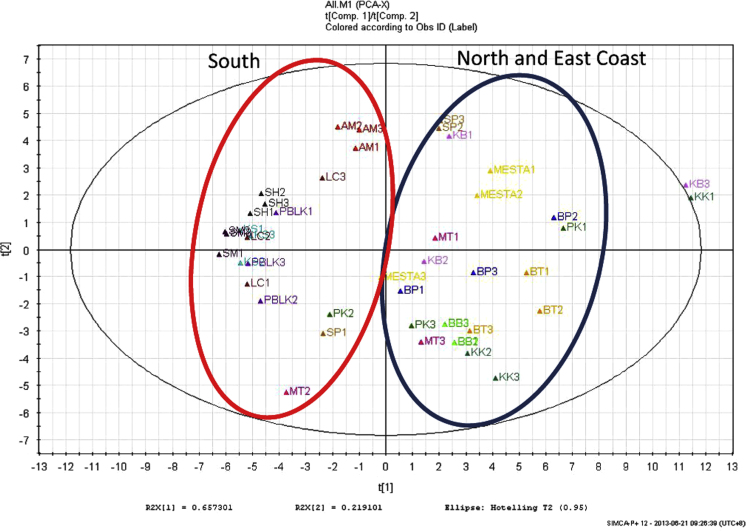
In this dataset, we distinguish 15 accessions of Garcinia mangostana from Peninsular Malaysia using Fourier transform-infrared spectroscopy coupled with chemometric analysis. We found that the position and intensity of characteristic peaks at 3600–3100 cm−1 in IR spectra allowed discrimination of G. mangostana from different locations. Further principal component analysis (PCA) of all the accessions suggests the two main clusters were formed: samples from Johor, Melaka, and Negeri Sembilan (South) were clustered together in one group while samples from Perak, Kedah, Penang, Selangor, Kelantan, and Terengganu (North and East Coast) were in another clustered group.
Keywords: Apomictic, Mangosteen, Fourier Transformed-Infrared, Peninsular Malaysia
Specifications Table
| Subject area | Biology |
|---|---|
| More specific subject area | Plant Sciences |
| Type of data | Figure; Table |
| How data was acquired | Fourier Transform-Infrared spectroscopy (Perkin-Elmer Frontier TM with a spectrum software version 10.3) |
| Data format | Analyzed |
| Experimental factors | Leaf of Garcinia mangostana from 15 different locations throughout Peninsular Malaysia were analysed using Fourier Transform-Infrared (FTIR) spectroscopy coupled with chemometric analysis. |
| Experimental features | Due to its reproductive manner, Garcinia mangostana trees are essentially clonal, FTIR coupled with chemometric analysis was used to primarily discriminate and to identify functional groups or chemical bonds in several accessions of Garcinia mangostana in Peninsular Malaysia. This approach is the first fingerprint identification for this apomictic clone plant. |
| Data source location | Peninsular Malaysia |
| Data accessibility | The data is available with this article. |
1. Value of the data
-
•
Fourier transform-infrared (FTIR) is a fast, effective and non-destructive procedure to provide unique fingerprints without any sample pretreatment [1], [2].
-
•
As an obligate apomictic plant, the genetic diversity of Garcinia mangostana is relatively narrow [3], [4]. FTIR spectroscopic data in combination with multivariate statistical analysis were performed to discriminate G. mangostana in Peninsular Malaysia.
-
•
FTIR and multivariate analysis are able to separate G. mangostana in Peninsular Malaysia into two clusters.
2. Data
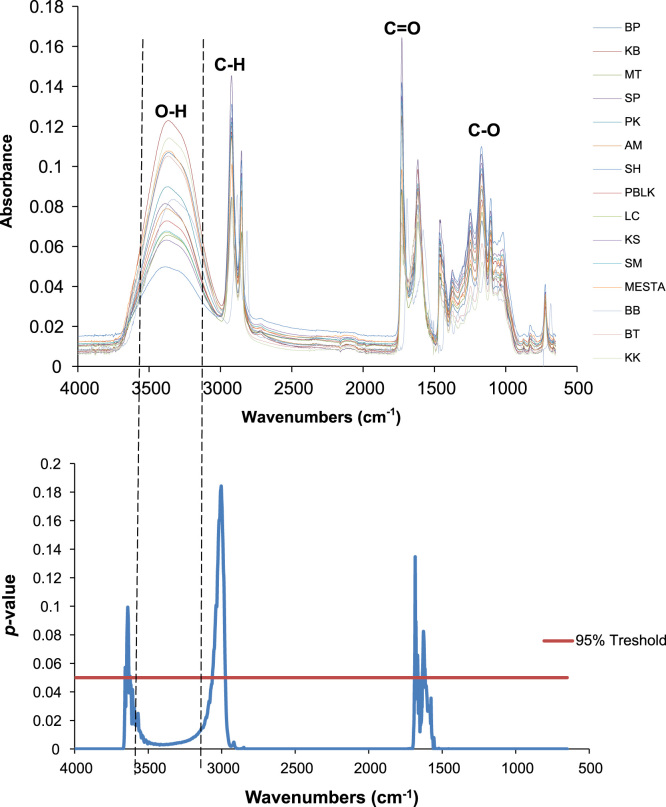
FTIR spectra (4000–650 cm−1) identified four major functional groups (O–H, C–H, C=O, and C–O) in the leaves of G. mangostana (Fig. 1) from 15 different sample locations in Peninsular Malaysia (Table 1). Principal component analysis (PCA) revealed two major clustering groups: samples from Johor, Melaka, and Negeri Sembilan (South) were clustered together in one group while samples from Perak, Kedah, Penang, Selangor, Kelantan, and Terengganu (North and East Coast) were in another clustered group (Fig. 2).
Fig. 1.
Four major functional groups present in leaves of Garcinia mangostana and their p-value. BP: Kg. Sungai Rusa, Balik Pulau; KB: Kampung Belukar, Tumpat; MT: MARDI Terengganu; SP: Kg. Tandap Batu, Sungai Petani; PK:Pengkalan Kubor, Kelantan; AM: Kg. Air Melintang Kota; SH: Kg. Senama Hilir, Rembau; PBLK: Pengkalan Balak, Alor Gajah; LC: Lubuk Cina, Melaka; KS: Kg. Solok, Tangkak; SM: Sungai Mati, Muar; MESTA: Rumah Tumbuhan, UKM; BB: Bukit Besi; BT: MARDI Bukit Tangga; KK: MARDI Kuala Kangsar.
Table 1.
15 different locations for sampling of G. Mangostana.
| Locations | Latitude (N) | Longitude (E) |
|---|---|---|
| Bukit Besi, Terengganu | N: 4° 56׳ 51.2" | E: 103° 10׳ 22.5" |
| MARDI Terengganu | N: 4° 57׳ 14.3" | E: 103° 10׳ 8.7" |
| Pengkalan Kubor, Kelantan | N: 6° 10׳ 36.3" | E: 102° 07׳ 28.5" |
| Kg. Belukar, Tumpat | N: 6° 12׳ 16.9" | E: 102° 06׳ 31.9" |
| Rumah Tumbuhan, UKM | N: 2° 55׳ 13" | E: 101° 47׳ 2" |
| Kg. Sungai Rusa, Balik Pulau | N: 5° 20׳ 43.0" | E: 100° 13׳ 45.9" |
| Kg. Tandop Batu, Sg. Petani | N: 5° 43׳ 19.9" | E: 100° 24׳ 54.1" |
| MARDI Bukit Tangga | N: 6° 29׳ 7.2" | E: 100° 28׳ 58.6" |
| MARDI Kuala Kangsar | N: 4° 45׳ 51.1" | E: 100° 54׳ 21.8" |
| Kg. Senama Hilir, Rembau | N: 2° 34׳ 21.0" | E: 102° 05׳ 49.6" |
| Kg. Air Melintang Kota | N: 2° 30׳ 6.4" | E: 102° 06׳ 39.4" |
| Lubok Cina, Melaka | N: 2° 27׳ 40.3" | E: 102° 04׳ 4.6" |
| Pengkalan Balak, Alor Gajah | N: 2° 22׳ 54.5" | E: 102° 13׳ 5.7" |
| Kg. Solok, Tangkak | N: 2° 15׳ 33.4" | E: 102° 32׳ 36.8" |
| Sg. Mati, Muar | N: 2° 07׳ 36.0" | E: 102° 33׳ 27.6" |
Fig. 2.
PCA analysis showed two major clustered groups: South, and North and East Coast region.
3. Experimental design, materials and methods
3.1. FTIR absorption spectra
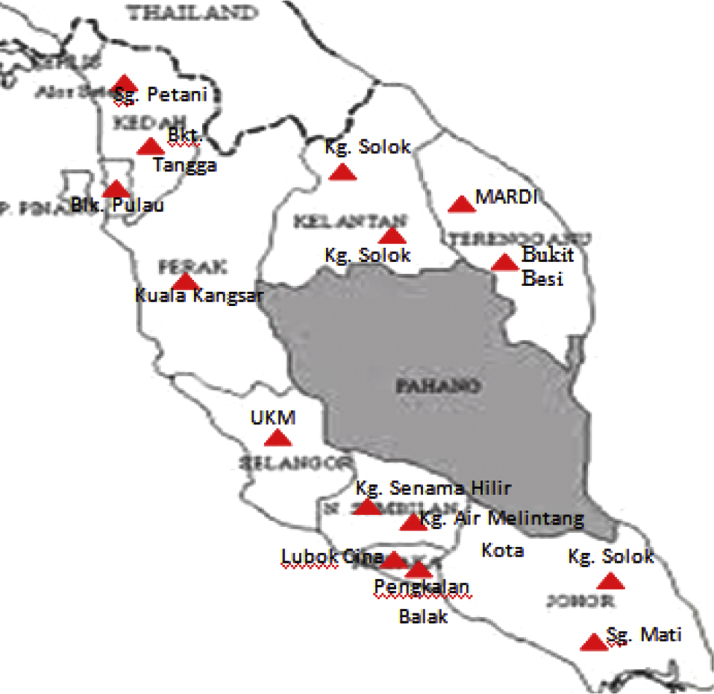
Leaves of G. mangostana from 15 different locations throughout Peninsular Malaysia (Fig. 3) were collected and the GPS location were recorded (Table 1). FTIR analysis was conducted using Perkin-Elmer FrontierTM with spectrum software version 10.3 (Perkin-Elmer, USA) for sample discrimination. Samples were placed on the top surface of the crystal and the gripper plate positioned gently on top to avoid damage to the crystal. The crystal was protected from scratches to ensure even contact with the sample and avoid undue effect on penetration depth that could confer aberrant results. IR spectra were recorded in the 4000–650 cm−1 range. All analyses were conducted with three biological replicates, each with three technical replicates, and the samples were randomly ordered to avoid bias. Data sets were baseline-corrected and area-normalized before statistical analysis.
Fig. 3.
Map of 15 different locations where accessions of G. Mangostana were collected.
3.2. Statistical analysis
Principal component analysis (PCA) was conducted using SIMCA-P software to discriminate and classify the samples. Differences between combined data of different locations were analyzed using student׳s t-test analysis in SPSS version 12.0.1 software. A value of p<0.05 was considered to be significant.
Conflict of Interest
The authors declare there is no conflict of interest.
Acknowledgments
The authors would like to thank Universiti Kebangsaan Malaysia for providing the funding (UKM-AP-KPB-18-2010).
Contributor Information
Sri A’jilah Samsir, Email: sriajilah.samsir@gmail.com.
Hamidun Bunawan, Email: hamidun.bunawan@ukm.edu.my.
Choong Chee Yen, Email: cychoong@ukm.edu.my.
Normah Mohd Noor, Email: normah@ukm.edu.my.
References
- 1.Grasel F.S., Ferrão M.F. A rapid and non-invasive method for the classification of natural tannin extracts by near-infrared spectroscopy and PLS-DA. Anal. Methods. 2016;8:644–649. [Google Scholar]
- 2.Allwood J.W., Ellis D.I., Goodacre R. Metabolomic technologies and their application to the study of plants and plant–host interactions. Physiol. Plant. 2008;132:117–135. doi: 10.1111/j.1399-3054.2007.01001.x. [DOI] [PubMed] [Google Scholar]
- 3.Yapwattanaphun C., Kobayashi S., Yonemori K., Ueda J. Hormone analysis in the locule of mangosteen fruit during apomictic seed development. Acta Hortic. 2014;1024:141–146. [Google Scholar]
- 4.Yonemori K., Nishiyama S., Yapwattanaphun C., Ueda J. Identification of plant hormones in endosperm liquid of mangosteen fruits at young developmental stages. Acta Hortic. 2014;1042:89–95. [Google Scholar]