Abstract
The conception and deployment of cost effective Picture Archiving and Communication Systems (PACS) is a concern for small to medium medical imaging facilities, research environments, and developing countries’ healthcare institutions. Financial constraints and the specificity of these scenarios contribute to a low adoption rate of PACS in those environments. Furthermore, with the advent of ubiquitous computing and new initiatives to improve healthcare information technologies and data sharing, such as IHE and XDS-i, a PACS must adapt quickly to changes. This paper describes Dicoogle, a software framework that enables developers and researchers to quickly prototype and deploy new functionality taking advantage of the embedded Digital Imaging and Communications in Medicine (DICOM) services. This full-fledged implementation of a PACS archive is very amenable to extension due to its plugin-based architecture and out-of-the-box functionality, which enables the exploration of large DICOM datasets and associated metadata. These characteristics make the proposed solution very interesting for prototyping, experimentation, and bridging functionality with deployed applications. Besides being an advanced mechanism for data discovery and retrieval based on DICOM object indexing, it enables the detection of inconsistencies in an institution’s data and processes. Several use cases have benefited from this approach such as radiation dosage monitoring, Content-Based Image Retrieval (CBIR), and the use of the framework as support for classes targeting software engineering for clinical contexts.
Keywords: PACS, Digital Imaging and Communications in Medicine (DICOM), PACS implementation, PACS integration, PACS service, Radiation dose, Software design
Background
Over the last two decades, medical imaging has evolved to become a very valuable tool not only in clinical practice but also in research. Nowadays, it is fundamental for providing quality diagnosis and supporting practitioners’ decision-making [1, 2]. Picture Archiving and Communication Systems (PACS), originally designed as a tool to provide convenient means of storage and access to medical imaging data for the radiology department, have evolved into hospital-wide systems. Besides radiology, many other clinical areas have adopted PACS in their daily routines, such as cardiology [3], dentistry, and pathology [4]. To cope with this diversity, distinct approaches have been followed regarding PACS implementation in clinical and research institutions. Solutions range from simple models, used in small laboratories, to enterprise grade platforms, typically integrated with other healthcare information systems. The requirements and workflows for those integrated solutions may vary widely, however.
Besides the common usage of a PACS as a mere image repository, continuous technological evolution has led to the emergence of other applications ranging from teleradiology to computer-assisted diagnosis (CAD), content-based image retrieval (CBIR), and multidimensional imaging analysis. New perspectives in both data storage and distribution have also been brought forward due to the emergence of new paradigms, such as GRID, peer-to-peer, and cloud computing. The application of these technologies is likely to further improve PACS-based applications, increasing the service availability, reliability, and speed, hence enhancing overall productivity and improving patient care.
However, due to the need for market research and extensive clinical validation, the development of commercial systems lags behind the ever increasing demand for advanced processing tools. Furthermore, there is limited room for exploration of new avenues with a reasonable turnaround time that could allow users to rapidly evaluate new features [5]. It also becomes hard to justify cater to niche use cases, as the profit potential is low.
Besides the lengthy timespan required when bringing a tool to market, the high cost and complex setup required by these systems also limits severely the availability of these tools [1, 6]. This is a major constraint particularly in small medical institutions and in some research centers. Cost reduction, the ability to adapt software to particular workflows and perform experiments, modifying, or creating new functionality are, however, some of the reasons why open source and free software are becoming adopted in the medical community [7].
In that regard, these are advantages provided by Dicoogle [6], an open source PACS with a plugin-based architecture. What makes Dicoogle unique is its mindset towards the exploration and application of innovative technological approaches while providing the robustness required of a PACS for its daily operations. The software framework provides a symbiotic environment where multiple concerns are separated into self-contained units orchestrated and managed by the core application. In this article, we describe its architecture and its impact on our research and into the development processes of PACS applications, with emphasis on some selected success stories.
Picture Archiving and Communication System
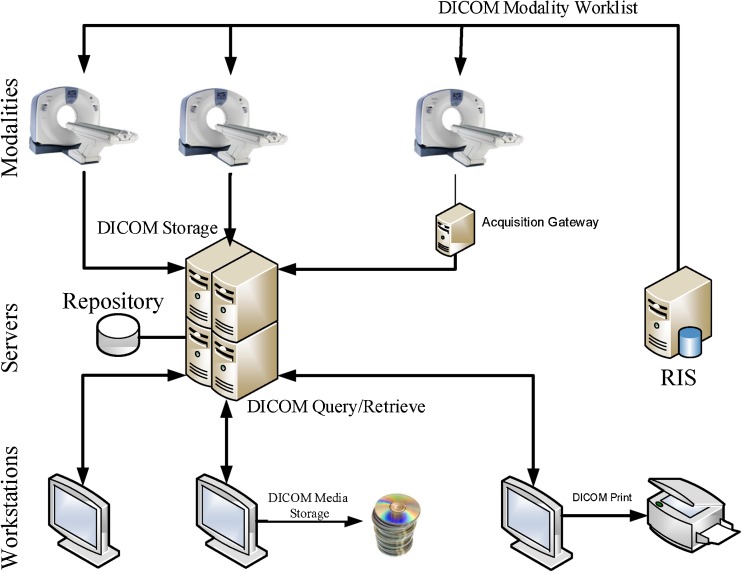
The move towards digital support for radiological images gave rise to a set of challenges that led to several implementations of what are commonly designated by the umbrella term of PACS (see Fig. 1). This concept is the embodiment of distinct hardware and software technologies comprising medical imaging and data acquisition equipment, subsequent storage devices, and display subsystems, all of which are integrated by digital networks and end-user software [1]. Such systems are designed to cope with the high storage needs and transmission requirements of an institution’s digital medical data.
Fig. 1.
A common workflow for DICOM based PACS
The implementation of a PACS provides an overall boost of productivity to an institution and decreases its operational costs [8]. It also creates an excellent opportunity for empowering healthcare practitioners with the capacity to work remotely or perform telemedicine, providing collaborative work environments and facilitating the access and sharing of multimedia information.
PACS have become one of the most valuable tools supporting the medical profession in both decision-making and treatment procedures [1]. It is estimated that over 1 EB of data pertaining to medical procedures will be produced in 2016 [9]. Due to the increasing demand for more flexible PACS solutions, researchers are actively exploring state-of-the-art paradigms and technologies such as distributed and heterogeneous computing grids [10, 11], cloud computing [12], peer-to-peer networks [6], and knowledge extraction using indexing engines [13].
The core element of PACS is, typically, a central archive server that stores images acquired by the modalities, along with complementary information about patients and studies. The archive data can be queried by a multitude of equipment, such as CAD systems and display workstations. In Fig. 1, a common disposition for the various PACS components is depicted.
Protocols for Data Sharing in a PACS Environment
The evolution of information and communication technologies creates new opportunities but also imposes new challenges for PACS. For instance, professionals are demanding new mobile Web-based access platforms to not only enhance enterprise mobility, but also to promote Internet-based scenarios for remote diagnostic and cooperative work [3, 14]. PACS rely heavily on a set of standard definitions denominated as Digital Imaging and Communications in Medicine (DICOM) [15], which, by itself, represents a major contribution to the exchange of structured medical imaging data.
The DICOM Version 3 is a well-established standard in the medical field that provides guidelines for communication and data structures in these scenarios. The first versions of this protocol were created circa 1983 by the National Electrical Manufacturers Association (NEMA), when point-to-point connections were the norm and PACS infrastructure was characterized by being a semi-static organization. This protocol stands now as one of the key pieces involved in medical imaging systems used by practically every acquisition gateway.
DICOM is an extensible object-oriented protocol and defines several normalized services [16] that enable the communication between different PACS components. For instance, it allows modalities to directly send acquired images to the PACS archive, and their later retrieval by the practitioner’s workstations for diagnostic purposes. Of great importance is the fact that it defines how medical imaging data and respective metadata is to be stored, retrieved, and transmitted, thus allowing interoperability between different modalities and applications. In DICOM, images are associated with a single patient, their correspondent series and study, as well as their respective metadata concerning patient demographics, clinical staff, equipment, radiation dosages, and other useful information. The data is organized hierarchically following DICOM Information Model (DIM) and respective levels (patient, study, series, and object).
The networking specification of DICOM comprises an application layer protocol that uses TCP/IP to move data through the network and an addressing mechanism based on application entity (AE) [2]. Another DICOM part is Web Access to DICOM Persistent Objects (WADO) [17]. WADO is an attempt to bridge web technologies and the DICOM infrastructure by allowing RESTful HTTP access to persistent objects at the image level. The lack of standard query and storage mechanisms in the original WADO has led to several extensions to the base architecture [18, 19], and recently, to the adoption by NEMA of the WADO using RESTful Services (WADO-RS), Store Over the Web (STOW-RS), and Query based on ID for DICOM Objects (QIDO-RS) as addends to the original protocol. These extensions allow, respectively, for the storage and query of DICOM objects via HTTP. They are also useful for cross-enterprise access to medical images since HTTP is often allowed to traverse firewalls.
PACS Challenges and Trends
The PACS concept started to be broadly accepted two decades ago. The advantages provided by these systems are well understood by the medical community, and the initial hurdles related with display, storage, and transmission technology have largely been overcome. For small institutions, the question of using a PACS is commonly no longer phrased in terms of “if,” but in terms of “when” or “how.” For large medical institutions, the initial investment in a PACS can be considerable, reaching many million dollars [9]. Moreover, they are but a single piece of healthcare IT infrastructure, and integration with other healthcare systems such as the Radiology Information Systems (RIS) and Hospital Information Systems (HIS) was also an important issue in the last decade [4]. Far from stagnating, PACS have followed the ever-evolving context of healthcare industry.
A distinct issue faced by healthcare IT is how to share data between distinct institutions, efficiently and routinely. This issue is not merely technological. Medical data exchange between different institutions poses legal and ethical challenges [20]; hence, institutions are reluctant to exchange sensitive data such as non-anonymous medical images. Moreover, there are also other issues such as data ownership, integrity and the licensure, accreditation, and liability of the practitioners and institutions. To overcome such issues, the Integrating Healthcare Enterprise (IHE) defines integration profiles based on existing protocols [21]. Among those profiles, one stands out, the Cross-Enterprise Document Sharing (XDS) [22]. Particularly, XDS for imaging (XDS-i) [23] is a content profile that takes into consideration the specifics of the medical imaging field, including the PACS in the XDS workflow. One of the main challenges is to facilitate cross-institutional data transfer in a transparent, efficient manner, while maintaining the stringent privacy and confidentiality requirements of patient data [24].
A related issue to cross-institutional document sharing is teleradiology which, in 2003, was reportedly used by 70 % of American medical institutions [25]. Today’s teleradiology systems provide a multitude of services for experts, allowing them to request second opinions from colleagues, to work remotely from home, or to provide radiology services to rural areas where no radiologist is present [26]. The IHE profiles are, however, not yet widely deployed and hence not powering the current teleradiology solutions. And, while a reality for several years now, its existence has not yet been freed from all controversies. So, if 15 years ago the major issues were image quality, transmission speed, and image compression, nowadays the focus is on clinical governance, legal issues, and quality assessment. Another issue to overcome is the general inability to seamlessly integrate teleradiology systems with other healthcare information systems [26, 27]. Most solutions proposed in literature rely on custom software deployed under very particular conditions.
Dicoogle
Dicoogle is an open source PACS. Its implementation started in 2007 and has since been tested on multiple contexts. Besides supporting standard DICOM semantics, Dicoogle leverages peer-to-peer communication models and document-based indexing techniques within a DICOM network, diverging from the common architecture employing relational databases [13]. This replacement has resulted in a paradigm change since it has enabled much more information to be extracted from medical imaging repositories contrasting heavily with the standard DICOM query and retrieve services. In turn, its P2P features have allowed Dicoogle to be used as part of an inexpensive federated network of repositories.
Recently, the project has matured into a catalyst for research and development of new PACS subsystems and applications. With the project growing organically, a number of forks and branches were created providing distinct functionality. Dealing with multiple implementations [28–32] quickly became unattainable and cumbersome to scale. The merge of branched code with the stable branch has proven not to be free of issues and led to minor and sometimes major duplication of code having slightly different interfaces, to the need to adapt existing functionality to changes introduced elsewhere in the code, or even caused the introduction of race conditions. This is a common problem faced by many software developers, and in our view, it stated the need for a more decoupled and modular architecture and a more integrated development culture. Given the very active research effort in PACS systems, we felt there is a need for an environment where an idea may quickly be prototyped, tested, and validated. On the other hand, the addition of a CBIR module supported by a separated indexing engine [30] and plans to leverage cloud-based data storage required us to re-evaluate our previous assumptions (such as the internal interfaces to query and indexing based on Lucene’s API). This led us to refactor the application’s architecture, with the goal of streamlining and easing third-party development, a process that benefited from the field expertise obtained from previous iterations and deployments of Dicoogle.
The Dicoogle framework enables the development of plugins that modify or enhance its core functionality. Plugin communication, services, and lifetime are all managed by the core application. A plugin-based approach maximizes the decoupling between components, enabling orthogonal features to be developed separately and deployed easily. Custom builds are done by packing only the desired plugins, which minimizes the area of impact of experimental components.
There are innumerous scenarios in healthcare institutions and academia where clinical practitioners are working together with computer scientists to develop innovative solutions to improve the quality of services provided in medical imaging field. However, the researching and developing of new software pieces, for instance for CAD or image analysis, is very difficult to perform in traditional PACS-DICOM solutions. By facilitating the access to a fully fledged and extensible PACS server, Dicoogle permits us to bridge the gap between state-of-the-art investigation and integration with DICOM networks, a complex and time-spending task. By leveraging its DICOM and Web capabilities, Dicoogle has also been used to bridge PACS with the cloud, third-party applications, and as a learning platform in the academia.
Methods
Dicoogle Framework
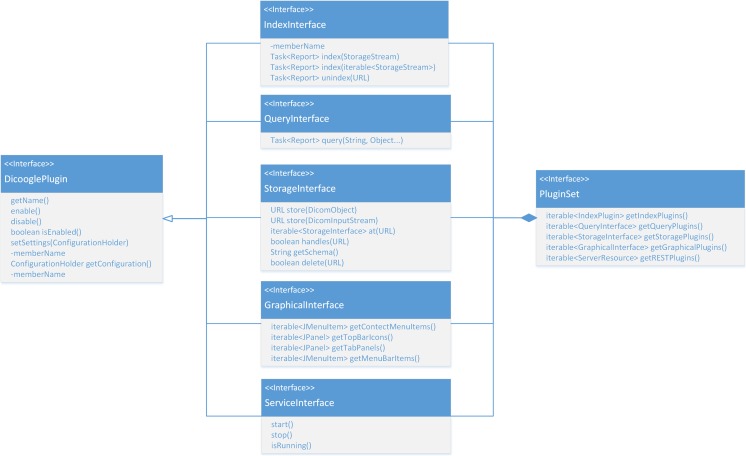
Our analysis of DICOM and PACS usage and trends coupled with the field experience obtained from previous iterations of the Dicoogle project have led us to separate and streamline the functionalities provided into multiple categories: storage, indexing, query, service, and presentation (as shown in Fig. 2). Each category is associated with a particular interface, the implementation of which is loaded at runtime, as a plugin. It is up to the core application to orchestrate the operations provided by the various plugins.
Fig. 2.
Class diagram of the SDK’s main interfaces
Although the various plugin categories embody an orthogonal semantic context often, in practice, there is a strong degree of intermingling that needs to be explored in order to provide a fast and robust solution. For instance, an indexing plugin using Lucene as backend will likely have a counterpart in terms of a query plugin accessing the same database. As such, the entry point for the plugin framework is a class representing a set of plugins, PluginSet. This data structure aggregates plugins from multiple categories into a functionally consistent unit simplifying both development and deployment.
The plugin’s life cycle is managed by the Dicoogle core allowing each module to be enabled or disabled per user request. During the application startup, the plugin directory is scanned and identified plugins are loaded according to their configuration file.
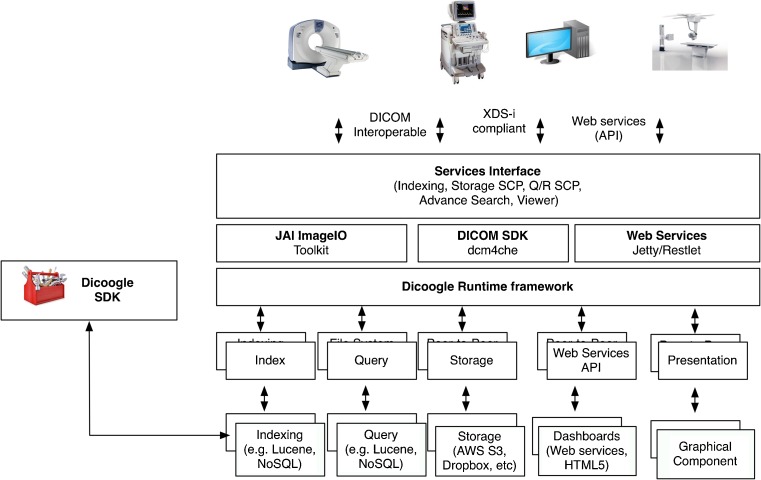
From the application’s core point of view, the various plugins within a set are independent of each other, accessed only via the respective interfaces. However, internally, complex plugins may need to have their state shared. These types of dependencies are easily solved inside the PluginSet constructor which has the opportunity to properly instantiate its child plugins and share any data structure required to achieve communication between its internal components. From the external user point of view, functionality is accessible through the DICOM protocol, the exported web services, an RMI interface, or the Web frontend, depending on purpose and configuration (as show in Fig. 3).
Fig. 3.
Dicoogle framework—full stack architecture
In order to facilitate plugin development by third parties, Dicoogle provides an Software Development Kit (SDK). By using this SDK, a plugin may be created and distributed as a .jar. If it requires external dependencies (for instance, the jfreechar library used for data visualization for the CBIR validation plugin), they may be also included in the jar.
The SDK provides common functionalities to unify and ease the development of various plugins. For instance, a settings management system and a logging interface are both supported by Dicoogle and exposed to the developer. The SDK further provides a message-based inter-plugin communication infrastructure and exposes an interface to the application task manager allowing plugins to dispatch asynchronous tasks without having to deal with boilerplate code. Convenience methods to handle DICOM objects and access indexed data are also provided.
Storage Plugins
A crucial part of any PACS is the persistency of medical image studies. A standard Dicoogle deployment relies on the local file-system as the backend of choice to store persistent DICOM objects. However, with the coming of age of cloud storage technologies, peer-to-peer distribution mechanisms and grid networks, a distinct set of storage policies comes into play. Data may not, and needs not, in all cases to be locally available to the system. The distinct types of storage policies are fertile ground for investigation and need to be normalized in the context of an application if we are to provide storage functionality in a consistent manner regardless of the underlying technology.
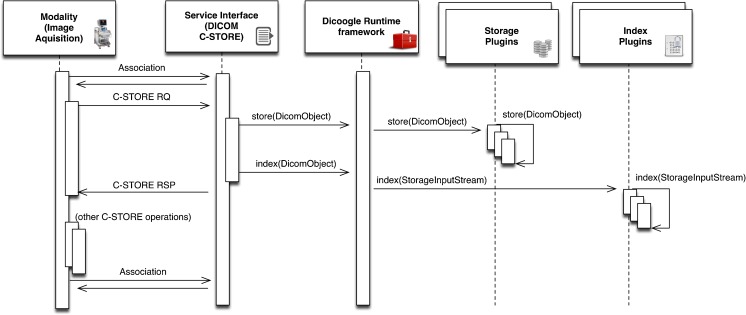
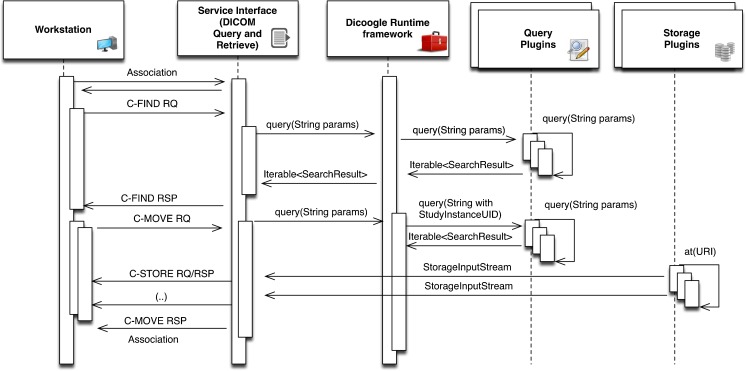
In Dicoogle, we need access to DICOM data when responding to services and when extracting information for fast querying and indexing (see Figs. 4 and 5). Hence, we need to unambiguously identify each file so that a relation may be established between separate indexed data and the actual physical location of the DICOM object. Moreover, given the wide array of potential technologies that may be employed for storage, we do not particularly care where the files are stored, as long as we can retrieve them on demand.
Fig. 4.
DICOM C-STORE operation—the sequence diagram explaining the interaction with the plugins
Fig. 5.
DICOM query and retrieve of medical images
The solution adopted by Dicoogle employs storage plugins to provide the persistency mechanisms, and universal resource locators (URL) to uniquely identify a resource, i.e., DICOM file, and the means to retrieve it, i.e., a storage plugin that knows how to decode and retrieve the specified data. For instance: “cloud:///dicomsrv1/file1.dcm“or “file:///var/repo/1.2.12345.dcm.” A storage Plugin must implement a store method that takes either a DICOM object or stream and return a URI for the stored resource.
The converse operation must also be supported. Given an URL, the underling resources must be delivered. This is achieved by defining the method “at(URL).” URLs may refer to collections of resources; accordingly, this method returns an iterator into a set of StorageInputStream objects allowing access to the raw DICOM data. Specializations of this class are used to provide data prefetching and caching in a transparent manner to the core.
Indexing and Query Plugins
Besides acting as a PACS, a goal of Dicoogle is to provide to a practitioner access to as much information as it might be required for decision support or statistical analysis. To do so, we initially relied on the extraction of metadata present in the objects of DICOM repositories and its respective indexation in a Lucene database [13]. This proved a successful approach, validated on the field, where it provided insights efficiency and service quality [28] and on the radiology dosage variation [33]. Our foray into new strategies for information retrieval, such as CBIR, required us to come up with new abstractions that, while allowing us to maintain all previous functionality, are more amenable to extension.
Indexing plugins are the components responsible for organizing data in a format that allows quick access to the stored information, while query plugins mediate access to that information. This solution harmonizes the processes of extracting, storing, and retrieving information, hence allowing the exploration of other data representation and information retrieval mechanisms, such as query-by-example, and study of the scalability and performance of distinct databases [34]. Due to the variety of data that can be indexed (textual, visual, hierarchical), the indexing policy and information extraction mechanisms are left entirely to the indexing plugin, which is free to contain additional dependencies to specific databases or libraries not part of the core and to define its own internal data representation. The indexer interface requires as input a StorageInputStream which has the effect of decoupling the indexing plugins from the DICOM storage policy. We could have the same effect by passing a URL; however, this approach does not require the extra step to retrieve the URL, an operation that would have to be repeated through each index plugin.
The output of a set of operations is a report data structure with a status, successful or not, and some additional information. However, instead of directly returning a report, the plugin must return a task. The reasons are twofold. Firstly, while the indexing process is fairly quick when handling the textual component of a DICOM file, the same is not true when extracting image features (for instance, in CBIR plugins or any kind of image analysis). Hence, we need to execute that code asynchronously. We could do that by creating tasks inside Dicoogle’s core, however, that would leave us no guarantee that the method is thread safe. Requesting an explicit task does not offer that guarantee as well; however, it gives the programmer a very strong hint that the code will execute asynchronously and not at the place of call. Each index method is scheduled for execution on new data as it arrives, independently of its source, which may be a DICOM service or a user-initiated action.
Query plugins are the natural counterpart to the indexing plugins and provide the means for information to be retrieved. They provide the bridge from an index’s internal data representation into an object understood by Dicoogle’s frontend. Typically, for each index plugin, we have at least a query plugin that knows how to leverage the indexed data to provide answers quickly to a user’s query.
Services
Dicoogle supports some of the most important DICOM services, such as C-STORE, C-FIND and C-MOVE, and WADO, and has some degree of support for other services such as XDS-i [24], with plans to integrate QIDO-RS and STOW-RS.
The DICOM storage and retrieve actions are fully supported through the C-STORE command (also used in C-MOVE operations). The DICOM C-STORE operations are typically called when a modality (or any other entity) produces an image or study and sends it to the PACS archive. In this process, as shown in Fig. 3, C-STORE operations will invoke functionality provided by the storage and indexing plugins.
The indexed information can be accessed programmatically using the SDK API, which has a query language similar to Lucene, or through instantiated graphical plugins. Queries are automatically performed whenever any DICOM service requests them, such as DICOM query/retrieve (Fig. 5). This is fully interoperable with workstations that support the DICOM standard. Results returned by query plugins can be retrieved or forwarded using the storage plugins.
REST Plugins
The usage of Dicoogle in clinical environments highlighted the necessity of sharing information with external applications that do not understand DICOM, and how cumbersome it is to rely on the DICOM protocol to share data with mobile applications and transverse firewalls. Nowadays, one of the best ways to share information is through the use of web services. Motivated by the good results obtained on a previous work [19], we decided to place emphasis on the REST paradigm. This methodology has gained wide acceptance around the web as a simple, resource-oriented alternative to SOAP and its companion the Web Services Description Language (WSDL). The inherent simplicity of accessing resources is the main reason it was chosen to drive the interfaces. Another advantage of using it instead of SOAP is the fact that it is simple to transmit binary data where SOAP would require an inefficient base 64 encoding and posterior decoding. These are likely the same reasons that have led NEMA to base the new revisions of WADO (WADO-RS, STOW-RS, and QIDO-RS) on RESTful web services. Dicoogle’s WADO implementation is based on REST plugins which are being used to drive zero footprint DICOM visualization tools [35], and mobile applications [32].
To facilitate the development of new web services, an API, based on RESTlet, is provided. The web services are enabled, configured and orchestrated by Dicoogle’s control panel leaving to the application only the semantics of the service. Plugins loaded on demand and web services are no exception.
Having the functionality to view indexed DICOM files as RESTful resources, we can make them available on demand by performing a standard HTTP GET on the plugin-defined URL. That is the basis of our WADO implementation. Using HTTP content type negotiation mechanisms, besides various representations for DICOM files (DCM, JPEG, PNG for image types, JSON or XML for the metadata), we can also return the results of a query plugin, some of which perform complex analysis to the indexed dataset.
Inter-plugin Communication
By design, a plugin has no direct access to any of Dicoogle’s methods or classes outside the Platform SDK package. Direct schemes of communication using object references are disabled for plugins except when mediated by Dicoogle via a proxy object. This also means that a plugin cannot directly execute any of another plugin’s methods which forces the plugins to be decoupled and insures that we have no illegal accesses to plugins disabled by the user. There are situations, however, where a plugin must have access to data provided by other plugin. Such an example is the CBIR validation plugin which must execute CBIR queries and at the same time access DICOM data for result validation. While we could conceivably merge the validation functionality with CBIR plugin, in the general case, we do require that plugins may communicate with each other, meaning we must be able to handle dependencies. We handle them by allowing each plugin to specify a unique name, which is then used to resolve any dependency during runtime. Plugins whose dependencies have not been satisfied will be disabled.
For a plugin which dependencies have been successfully resolved, we provide three inter-plugin communication mechanisms.
Interface request. If a plugin depends on another, it can use a proxy object to request a reference to a plugin interface by specifying the plugin name. Having the interface, it can call the methods exposed in its own thread. This mechanism makes the assumption of thread safety in the called plugin.
Message Passing. This is a non-blocking one-way communication mechanism. A Plugin is instantiated with a message queue that acts as a message pool. Thread safety is guaranteed by Dicoogle, which routes the communication, places the messages in the message pool, and notifies the receiver. A message contains the sender’s identification, the message tag, and a payload. These messages can be broadcast or routed to a specific plugin (using its name or placing them using methods exposed on the Plugin interface). Furthermore, Dicoogle core also issues some messages on its own, for instance when an indexing operation is requested or a query is to be performed.
REST services. If a plugin exposes a RESTful webservice, another one can use it to request data. This has the advantage of allowing to check if a plugin is currently instantiated and allows for very high decoupling at the expense of having to parse or handle the returned data.
User Interface and Graphical Plugins
Dicoogle has three modes of interface with the user. It can operate as a stand-alone command line application where its indexing, analysis, and querying functionality can be scripted. For end-users, we provide a graphical user interface (GUI) based on remote method invocation (RMI) that allows the PACS server and data engine to run on a machine and the graphical frontend on another. The standard RMI user interface sports the basic mechanisms for plugin control, configuration, and data visualization. Graphical plugins further extend Dicoogle’s functionality in a predefined manner. We provide several hooks that may be used to attach custom graphical components. In Fig. 6, it is shown the interface with the placeholders for the graphical components highlighted.
Fig. 6.
RMI user interface. The areas highlighted in blue are extensible through graphical plugins. The main window is a tabbed interface. Its main tab, displaying a tree of results, also has a context-specific menu which can be extended
Each plugin may request a panel which is provided to it by the application and of which the plugin has unique ownership. That panel will be placed as part of a tab panel available from the main interface where it will be used for configuration or to cope with use cases not handled by the core application’s user interface, for instance, CBIR’s query-by-example.
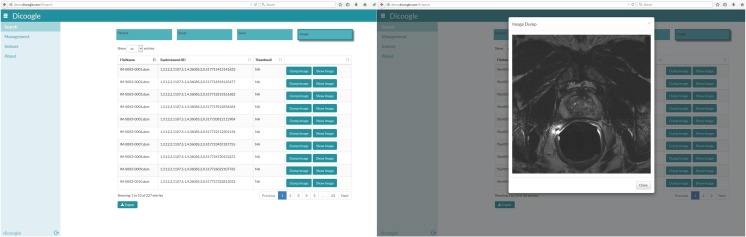
Recently, web technologies have become ubiquitous due to their ability to drive zero-install applications that run on any browser and the fact that the protocols are often allowed to transverse firewalls. The service-oriented architecture of Dicoogle lends itself nicely to the development of web-interfaces as shown in Fig. 7. The web interface allows a user to index various repositories, perform queries, and control the operations performed by Dicoogle.
Fig. 7.
Dicoogle’s web-based frontend
Results
Nowadays, Dicoogle provides several key features to extract metaimaging information for retrospective assessments, which is useful for statistics, management, and reporting tasks. It can be used for wide-ranging clinical studies requiring, for example, dose metrics that are now increasingly available in DICOM persistent objects created by recent models of digital image equipment [36, 37]. By enabling multiple views over the medical data repository in a flexible and efficient way, and with the possibility of exporting data for further statistical analysis, Dicoogle allows identification of inconsistencies in data and processes. This platform can be used to audit PACS information data and contribute to the improvement of radiology department’s practices [28, 29, 33]. Dicoogle has been also extended to support CBIR, using a profile-based approach. Currently, our research group has been using the platform in several hospitals and more than 22 million of DICOM images metadata have already been indexed, corresponding to a population of 160 thousand patients, and more than 450 thousand studies. To better expose the extensibility and usefulness of Dicoogle software architecture, the next subsections will present distinct use cases supported by plugin implementations.
CBIR Engine
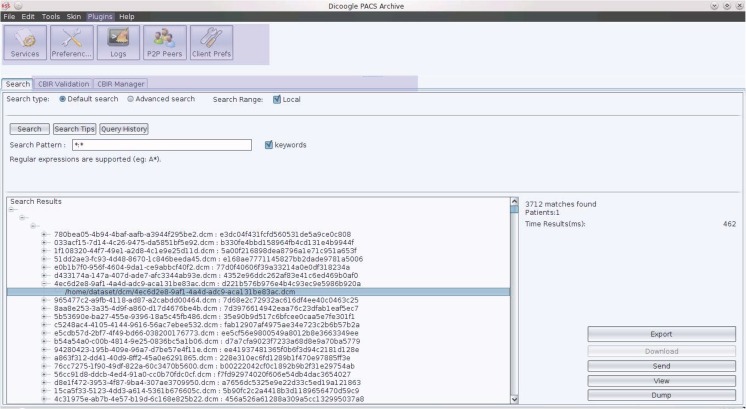
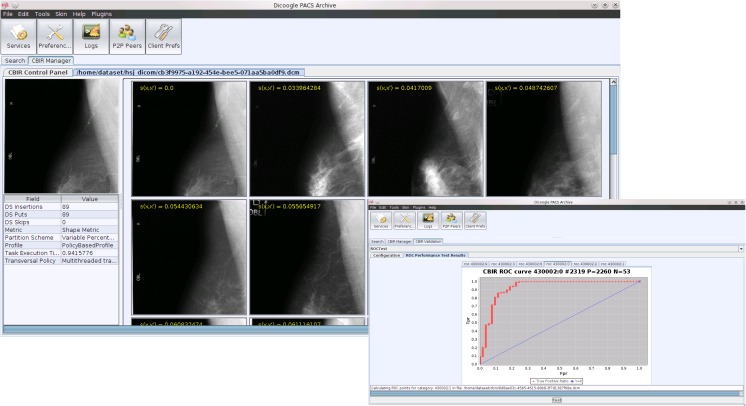
Content-based image retrieval methods have shown great promise in helping practitioners sift through the large amounts of data present in medical institutions. These methods rely on the automatic extraction of content from a source image to provide the query terms for a search. In practical terms, CBIR systems allow practitioners to use images from any study they are working on as query to the image database, hence obtaining a set of results that, in some sense, are similar to the original image. CBIR has the potential to save a significant amount of time to practitioners, enabling them to quickly move from a source image to a set of similar ones, potentially containing diagnosis reports. These reports, when compared to the original image, may strengthen the case for the diagnosis or provide the practitioner with additional insight. Given that radiologists often rely in second opinions in order to validate their diagnosis and increase their confidence levels, CBIR provides query mechanisms that are very close to the way a practitioner operates. We will be analyzing the implementation of this mechanism as an extension for Dicoogle using the plugin system mentioned before. A detailed analysis of the strategies and algorithms employed can be found in [30]. The objective of Dicoogle CBIR plugin (Fig. 8) is to allow query-by-example in the PACS archive using as query input DICOM images provided by the user. This plugin is implemented as a PluginSet and makes use of the four types of plugin. It extends the user interface by providing a manager that allows a user to select query profiles and a context menu in the result window that allows a user to select an image and perform a query-by-example with it using the profile defined in the CBIR manager.
Fig. 8.
CBIR interface and validation tool
The CBIR index plugin uses OpenCV to extract imaging features from DICOM objects and indexes them using Lucene as the persistency backend. When a query is requested, the CBIR query plugin accesses the Lucene database and returns a list of candidate images sorted by relevancy to the query image. Finally, we expose that functionality as a RESTful webservice, that takes as arguments an image URI and a similarity profile.
In order to perform the clinical validation of the retrieval engine, we created a separate plugin that analyses the accuracy and performance of the CBIR component. This plugin extends only the graphical plugin. However, it makes extensive use of the RESTful web services provided by CBIR to obtain similarity results, validating them using as ground truth DICOM data mined by the textual indexing engines.
Dose Information System
With a default Dicoogle distribution, it is possible to extract and aggregate a plethora of information from the DICOM headers that can be statistically analyzed or exported in tabular format to be used by external tools, for instance, Excel or SPSS. Several studies have been made using Dicoogle, mainly in radiation dosage monitoring and population studies [29, 38].
In order to monitor radiation dose in radiology departments, it was necessary to develop a Dicoogle plugin. The developed dose information system is compliant with DICOM standard and supports the IHE radiation dose profile. It is integrated with Dicoogle repository and provides centralized access and dose analysis for the patient, study and population scopes. Two data sources were used to populate this system: the DICOM metadata associated with radiation dose and the dose reports stored in the pixel data. In order to extract them, a new indexing plugin was also developed that extracts the values using optical character recognition (OCR) and stores them in a dose report (a new DICOM object).
Discussion
Open source software has been building momentum and gaining acceptance in the medical arena. High-quality open source software, such as OsiriX, has shown that it is possible and practical to use these solutions effectively in certain contexts with reduced operational costs. Furthermore, the availability of source code makes those solutions very attractive for fast development of functionalities or even prototyping and validation of new concepts. In [39], for instance, it is stated that open source “is particularly promising concerning advanced image display and analysis applications where the rapid increase in demand cannot be matched by traditional expensive commercial solutions.” Those advantages are not exclusive. As referenced in the “Background” section, there are other examples where open source solutions can benefit large segments of the medical healthcare industry, for instance, when we need to test, evaluate, and deploy new inter-institutional protocols, adapt solutions to new data sources, and explore new venues on data storage and management.
In spite of the before-mentioned advantages, some shortcomings apply to the open source development and distribution model. Lack of support is commonly pointed out in this regard. This is an issue that is prevalent in the academia, where providing commercial grade support for an end-user is often not its focus. A related issue, documentation, is also an Achilles heel for many projects. Whether by being non-existent or irrelevant, such as class diagrams and source documentation for end-users of an application, failure to have proper documentation repels many potential users and contributors.
The availability of the source code, while encouraging the development custom tailored solutions, often leads to forks whose code does not find its way upstream to the main community. While the distribution model is not at fault here, it is still a non-optimal situation which may lead other interested parties into duplicating functionality, hence wasting time and resources.
These issues can be ameliorated by having a vibrant community centered on the project as seen on successful projects like the linux kernel or the Apache Software Foundation projects. Open source works best when distinct entities contribute time and skills on the topics where they are strongest.
Probably the most well-known example of an open source extensible platform operating in the medical arena is OsiriX [5, 40]. This project provides a fully fleshed DICOM visualization system to either mobile platforms or desktops as well. This tool is widely used and, due to its open source nature, it was extended and adapted beyond its initial scope [41]. If OsiriX is mostly focused on DICOM data visualization, Dicoogle is focused on being a multi-modal data repository and DICOM services platform. The “Results” section has presented two state-of-the-art extensions for Dicoogle: a CBIR engine fully integrated with the PACS repository and workflows, and a dose information system that extracts information from DICOM headers metadata and pixel data. Many other examples could be described to validate the Dicoogle concept and its plugin-based architecture presented in this article. For instance, we are finishing a plugin capable of synthetizing DICOM studies and workflows. This module will allow us to test other Dicoogle components (and associated technologies) in stress conditions or big data scenarios.
Proper design of the solution is crucial to the future developments in PACS. At the moment, there is replication of work and there is a lack of extensible platforms capable of providing high-functionality features to the developers, supplying transparency to the developer. With Dicoogle, we intended to connect the dots between clinical researchers and software engineers. Researchers can easily test and validate their solutions minimizing their concerns with DICOM environments, and clinical researchers can take advantage of the developed algorithms much faster and seamlessly, without drastic changes in their workflow.
Conclusion
Dicoogle has been employed successfully in very distinct use cases, both in production and research. For instance, it is being used by third entities to support regional PACS, as DICOM data-mining tool [28, 29, 38], and as an educational tool for students. Moreover, it is tackling other challenges of relevancy to healthcare institutions such as interfacing with XDS-i [24] or performing content-based image retrieval [30].
This range of distinct application with very heterogeneous requirements is only possible due to its software architecture. The extension mechanisms have allowed us to leverage existing DICOM functionality and explore new directions in a non-intrusive manner. A stable platform where most protocols are ready to use translates to faster development time due to a lower barrier to entry and places the focus on the task at hand whether it is an experimental feature or a data analysis task. Developers can leverage the existing DICOM functionality to quickly develop, adapt, or prototype features for their use cases.
Dicoogle presents advantages to both research institutes and small to medium medical institutions. Its Open Source nature, the low hardware requirements, and its facilitated deployment make this software readily available. To cope with the rapid pace of development in the PACS arena, the proposed extensible plugin-based software is an important requirement, as it allows for quick prototyping, experimentation, and validation while enabling a great deal of code and functionality reuse.
References
- 1.Huang HK: PACS and imaging informatics: basic principles and applications. Hoboken, NJ:Wiley, p 704, 2004
- 2.Oosterwijk H. Dicom basics, third edition, aubrey. TX: OTech, Inc; 2005. [Google Scholar]
- 3.Costa C, Silva A, Oliveira J: Current perspectives on PACS and a cardiology case study. Advanced Computational Intelligence Paradigms in Healthcare. Springer Berlin, Heidelberg, pp 79–108, 2007
- 4.Duncan LD, Gray K, Lewis JM, Bell JL, Bigge J, McKinney JM. Clinical integration of picture archiving and communication systems with pathology and hospital information system in oncology. Am Surg. 2010;76:982–986. [PubMed] [Google Scholar]
- 5.Ratib O, Rosset A. Open-source software in medical imaging: development of OsiriX. Int J Comput Assist Radiol Surg. 2006;1:187–196. doi: 10.1007/s11548-006-0056-2. [DOI] [Google Scholar]
- 6.Costa C, Ferreira C, Bastião L, Ribeiro L, Silva A, Oliveira JL: Dicoogle-an open source peer-to-peer PACS. Journal of Digit imaging 24(5):848–856, 2011 [DOI] [PMC free article] [PubMed]
- 7.McDonald CJ, et al. Open Source software in medical informatics—why, how and what. Int J Med Inform. 2003;69:175–184. doi: 10.1016/S1386-5056(02)00104-1. [DOI] [PubMed] [Google Scholar]
- 8.Mansoori B, Erhard KK, Sunshine JL. Picture Archiving and Communication System (PACS) implementation, integration benefits in an Integrated Health System. Acad Radiol. 2012;19:229–235. doi: 10.1016/j.acra.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 9.Myers B: U.S. Medical imaging informatics industry reconnects with growth in the enterprise image archiving market
- 10.Vossberg M, Tolxdorff T, Krefting D. DICOM image communication in Globus-based medical grids. IEEE Trans Inf Technol Biomed. 2008;12:145–153. doi: 10.1109/TITB.2007.905862. [DOI] [PubMed] [Google Scholar]
- 11.Yang C-T, Chen C-H, Yang M-F. Implementation of a medical image file accessing system in co-allocation data grids. Futur Gener Comput Syst. 2010;26:1127–1140. doi: 10.1016/j.future.2010.05.013. [DOI] [Google Scholar]
- 12.Teng CC, Mitchell J, Walker C, Swan A, Davila C, Howard D, Needham T: A medical image archive solution in the cloud. In Software Engineering and Service Sciences (ICSESS), 2010 I.E. International Conference on (pp. 431–434). IEEE, (2010, July)
- 13.Costa C, Freitas F, Pereira M, Silva A. Oliveira JeL: indexing and retrieving DICOM data in disperse and unstructured archives. Int J Comput Assist Radiol Surg. 2009;4:71–77. doi: 10.1007/s11548-008-0269-7. [DOI] [PubMed] [Google Scholar]
- 14.Pohjonen H, Ross P, Blickman JG, Kamman R. Pervasive access to images and data—the use of computing grids and mobile/wireless devices across healthcare enterprises. IEEE Trans Inf Technol Biomed. 2007;11:81–86. doi: 10.1109/TITB.2006.885548. [DOI] [PubMed] [Google Scholar]
- 15.Digital Imaging and Communication in Medicine (DICOM)-Part 1-20: National Electrical Manufacturers Association, 2015
- 16.Digital Imaging and Communication in Medicine (DICOM)-Part 4: National Electrical Manufacturers Association, 2015
- 17.Digital Imaging and Communication in Medicine (DICOM)-Part 18: National Electrical Manufacturers Association, 2015
- 18.Koutelakis GV, Lymberopoulos DK. WADA Service: an extension of DICOM WADO service. IEEE Trans Inf Technol Biomed. 2009;13:121–130. doi: 10.1109/TITB.2008.2007197. [DOI] [PubMed] [Google Scholar]
- 19.Valente F, Viana-Ferreira C, Costa C, Oliveira JL: A RESTful image gateway for multiple medical image repositories. Information Technology in Biomedicine, IEEE Transactions on, 16(3):356–364, 2012 [DOI] [PubMed]
- 20.Pattynama PMT. Legal aspects of cross-border teleradiology. Eur J Radiol. 2010;73:26–30. doi: 10.1016/j.ejrad.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 21.Witting K: Health Information Exchange: Integrating the Healthcare Enterprise (IHE). In Introduction to Nursing Informatics (pp. 79–96). Springer London, 2015
- 22.IHE Radiology (RAD) Technical Framework - Revision 14.0 Volume 4 (RAD TF-4): National Extensions, IHE International, Inc. 2015. url:http://www.ihe.net/uploadedFiles/Documents/Radiology/IHE_RAD_TF_Vol4.pdf
- 23.International I. Cross-enterprise document sharing for imaging (XDS-i) 2015. [Google Scholar]
- 24.Ribeiro LS, Rodrigues RP, Costa C, Oliveira JL: Enabling outsourcing XDS for imaging on the public cloud. Stud Health Technol Inform 192:33–37, 2013 [PubMed]
- 25.Ebbert TL: The state of teleradiology in 2003 and changes since 1999. Yale University 2006 [DOI] [PubMed]
- 26.Binkhuysen FHB, Ranschaert ER. Teleradiology: evolution and concepts. Eur J Radiol. 2011;78:205–209. doi: 10.1016/j.ejrad.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 27.Thrall JH. Teleradiology Part II. Limitations, risks, and opportunities. Radiology. 2007;244:325–328. doi: 10.1148/radiol.2442070676. [DOI] [PubMed] [Google Scholar]
- 28.Bastião L, Santos M, Costa C, Silva A, Rocha N. Dicoogle statistics: analyzing efficiency and service quality of digital imaging laboratories. Heildelberg: Springer; 2013. [Google Scholar]
- 29.Santos M, Basti??o L, Costa C, Silva A, Rocha N: DICOM and clinical data mining in a small hospital PACS: a pilot study: Springer Berlin Heidelberg, 2011
- 30.Valente F, Costa C, Silva A. Dicoogle, a Pacs featuring profiled content based image retrieval. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viana-Ferreira C, Costa C, Oliveira JL: Dicoogle relay-a cloud communications bridge for medical imaging, 2012
- 32.Viana-Ferreira C, Ferreira D, Valente F, Monteiro E, Costa C, Oliveira JL. Dicoogle mobile: a medical imaging platform for Android. Stud Health Technol Inform. 2012;180:502. [PubMed] [Google Scholar]
- 33.Silva LAB, Ribeiro LS, Santos M, Neves N, Francisco D, Costa C, Oliveira JL: Normalizing heterogeneous medical imaging data to measure the impact of radiation dose. J Digit Imaging 1–13, 2015 [DOI] [PMC free article] [PubMed]
- 34.Bastiao Silva L, Beroud L, Costa C, Oliveira JL: Medical imaging archiving: a comparison between several NoSQL solutions. In Biomedical and Health Informatics (BHI), 2014 IEEEEMBS International Conference on (pp. 65–68). IEEE. (2014, June)
- 35.Monteiro EJ, Costa C, Oliveira JL: A DICOM viewer based on web technology
- 36.Wang S, et al. An automated DICOM database capable of arbitrary data mining (including radiation dose indicators) for quality monitoring. J Digit Imaging. 2011;24:223–233. doi: 10.1007/s10278-010-9329-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santos M, Bastião L, Costa C, Silva A, Rocha N: DICOM and clinical data mining in a small hospital PACS: a pilot study: Springer Berlin Heidelberg, 2011
- 38.Santos M, de Francesco S, Silva LAB, Silva A, Costa C, Rocha N: Multi vendor DICOM metadata access—a multi site hospital approach using Dicoogle, 2013
- 39.Ratib O, Rosset A, Heuberger J. Open Source software and social networks: disruptive alternatives for medical imaging. Eur J Radiol. 2011;78:259–265. doi: 10.1016/j.ejrad.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 40.Choudhri A, Radvany M. Initial experience with a handheld device digital imaging and communications in medicine viewer: OsiriX mobile on the iPhone. J Digit Imaging. 2011;24:184–189. doi: 10.1007/s10278-010-9312-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Camarlinghi N, et al. Combination of computer-aided detection algorithms for automatic lung nodule identification. Int J Comput Assist Radiol Surg. 2012;7:455–464. doi: 10.1007/s11548-011-0637-6. [DOI] [PubMed] [Google Scholar]