Abstract
Pathogen-associated molecular pattern (PAMP) recognition leads to TANK-binding kinase (TBK1) polyubiquitination and activation by trans-autophosphorylation, resulting in IFN-β production. Here we describe a mouse model of optineurin insufficiency (OptnΔ157) in which the TBK1-interacting N-terminus of optineurin was deleted. PAMP-stimulated cells from OptnΔ157 mice had reduced TBK1 activity, no phosphorylation of optineurin Ser187, and mounted low IFN-β responses. In contrast to pull-down assays where the presence of N-terminus was sufficient for TBK1 binding, both the N-terminal and the ubiquitin-binding regions of optineurin were needed for PAMP-induced binding. This report establishes optineurin as a positive regulator TBK1 via a bipartite interaction between these molecules.
Keywords: type I interferon, TBK1, optineurin
Introduction
Optineurin is a ubiquitin-binding protein whose mutations have been found in two neurodegenerative diseases: primary open angle/normal-tension glaucoma (POAG/NTG) and amyotrophic lateral sclerosis (ALS) [1, 2]. The exact role of optineurin in the pathogenesis of neurodegeneration is unclear. Moreover, the mutations found in ALS and POAG/NTG do not seem to overlap, suggesting that different regions of optineurin exert different and/or multiple functions, perhaps even in different cell types. Interestingly, in vitro studies have implicated optineurin in an unusually large number of cellular functions including regulation of inhibitor of κB kinase (IKK) and TANK binding kinase 1 (TBK1), autophagy, vesicle trafficking, cell division, regulation of transcription, and maintenance of the structure of Golgi apparatus [3–9]. During these processes optineurin is mostly thought to act as an adaptor, exerting its function by bridging various cellular proteins. The initial studies of optineurin interactions with other proteins revealed that its N-terminal region is indispensible for binding to TBK1, myosin VI, Rab8 and glutamate receptor GluR1a, whereas its C-terminal region is required for binding to RIP1, CYLD, myosin VI, and huntingtin [10, 11]. It was subsequently shown, though, that such binding is dynamic and contingent on various ubiquitination and phosphorylation events. For example, optineurin binding to LC3, a protein expressed on autophagosomal membranes, is enhanced upon TBK1-mediated phosphorylation of optineurin on serine 177 (S177) [5].
A prominent feature of optineurin is its ubiquitin binding. Optineurin contains two ubiquitin binding sites, highly homologous to those of NF-κB essential modulator (NEMO): the Ubiquitin-binding regions of ABIN proteins and NEMO (UBAN), and a zinc finger (ZF)[3, 12, 13]. This bipartite region maps to the C-terminal portion of optineurin and is necessary for its selective high-affinity binding to K63- and M1-polyubiquitinated proteins. Optineurin binding to such polyubiquitin chains was proposed to be important for both cell signaling and autophagy [3, 5]. During signaling, given the close homology between optineurin and NEMO ubiquitin-binding domains, it was proposed that optineurin binds to the same polyubiquitin-modified proximal signaling molecules to which NEMO binds during NF-κB and interferon regulatory factor 3 (IRF3) pathway activation [3, 14, 15]. Ubiquitination is indispensible in both signaling pathways as it allows the assembly of multimeric signaling complexes (signalosomes) necessary for kinase activation and signal propagation. However, whereas NEMO deficiency leads to complete shutdown of NF-κB and IRF3 activation in response to various pathogens or pathogen-mimicking ligands [16, 17], the role of optineurin is still controversial.
Serine-threonine kinase TBK1 is a central kinase regulating type I IFN secretion in response to pathogens [18, 19]. It does so by direct phosphorylation of IRF3, a transcriptional factor that then moves to the nucleus and binds promoter regions of type I IFN genes [20]. TBK1 is constitutively expressed in most cells as an inactive homodimer in which kinase domains (KD) face away from each other [21]. Upon PAMP recognition by Toll-like receptors (TLR) or intracellular DNA and RNA sensors, TBK1 is K63-ubiquitinated, allowing signalosome assembly and intradimer KD interaction, leading to activation by trans-autophosphorylation at Ser172 [22]. The role of optineurin as an adaptor for TBK1 signalosome assembly was addressed in several studies, but there have been disparate results. It was reported that overexpression of optineurin in HEK293-hTLR3 cells inhibited, whereas transient optineurin silencing promoted production of type I interferon IFN-β upon viral infection [15]. This suggested that optineurin was a negative regulator of TBK1 activation, perhaps acting as a competitive inhibitor of NEMO. However, two mouse models designed to abolish ubiquitin-binding activity of optineurin, i.e. one carrying a point mutation in the ubiquitin-binding domain (OptnD477N) and another lacking the entire C-terminal region encompassing the UBAN and ZF (Optn470T), argued the opposite, i.e. that the ubiquitin-binding function of optineurin was required for positive regulation of TBK1. Notably, in bone marrow derived macrophages (BMDM) and dendritic cells (BMDC) from both models optineurin was necessary for optimal TBK1 activation and IFN-β secretion upon TLR-3, -4, and -9 stimulation [4, 23]. Optn470T mice also had diminished IFN-β secretion during LPS-induced sepsis. Although this issue of was considered to be closed with the newer in vivo data, a recent report reiterated the role of optineurin as negative regulator of TBK1 in HeLa cells in a viral infection model [24]. Moreover, that study proposed a novel mechanism of optineurin-mediated TBK1 suppression, demonstrating that optineurin brings the CYLD deubiquitinase to polyubiquitinated TBK1, thus leading to signal shutdown. Given the discrepancy between in vivo and in vitro results, it is possible that OptnD477N and Optn470T, which like most mutations found in ALS patients lack the ubiquitin-binding function, act in a dominant-negative fashion. Importantly, the TBK1-binding site in optineurin maps to amino acids 1-127 [25], and is preserved in these models. To resolve this controversy and elucidate the physiologic role of optineurin in TBK1 activation, we have examined the contribution of optineurin to type I IFN activation by designing a mouse model in which the N-terminal region (residues 1-157; OptnΔ157 mice), encompassing the entire TBK1-binding site, was deleted.
Materials and methods
Mice
Mice with the N-terminal truncation of optineurin were generated by genetic recombineering as described [26]. Exons 2 and 3 of the mouse optineurin gene in 129 embryonic stem (ES) cells were floxed, and ES clones verified by Southern blot were injected into C57BL/6 blastocysts in the Mouse Cancer Genetics Program, NCI-Frederick. The chimeras were crossed to β-actin-Cre to generate whole body optineurin deletion (OptnΔ157). The OptnΔ157 mice used in these studies were backcrossed onto the B6 background for at least four and up to eleven generations; for all experiments, the controls from the same generation were used. The mice were kept in NCI animal facility and all studies were approved by the National Cancer Institute Animal Care and Use Committee.
Cell culture, plasmids, and reagents
Human embryonic kidney (HEK) 293 cells were obtained from the American Type Culture Collection (ATCC), and HEK293 cells that stably express human TLR3 (HEK293-hTLR3) from Invivogen. Cell lines were maintained in DMEM supplemented with 10% fetal calf serum, 5 mM glutamine, and antibiotics (complete medium). Bone marrow-derived macrophages (BMDMs) were generated from mouse bone marrow cells and cultured in complete RPMI supplemented with supernatant of L929 cell cultures (final concentration of 30%). The mouse optineurin gene was cloned from the cDNA generated from mouse embryonic fibroblast mRNA, and subcloned into pcDNA3.1+ expression vector. To find the alternative translation initiation site in mice with the N-terminal truncation, OptnΔ157 and OptnΔ192 DNA fragments were generated. To subclone WT, OptnΔ157, OptnΔ192, the following forward primers were used: ACCATGTCCCATCAACCTCTGAGC, ACCATGCGCCTTCGGGCTGAAAAGGC, ACCATGACCGAAGGAGAGACTGAAGGG, respectively. The reverse primer was: GGGCTCTAGATCAAATGATGCAGTCCATCACATGG. HEK293 cells were transfected by calcium phosphate method with pcDNA3.1+ harboring WT, OptnΔ157 or OptnΔ192, and lysates were prepared 20 hr later in RIPA buffer. The human optineurin gene, as well as E50K, Δ147 and D474N mutants were subcloned into bacterial pDest15 expression vector harboring a GST tag. The plasmid encoding myc-TBK1 plasmid was a kind gift from Rongtuan Lin (McGill University). Antibodies recognizing phospho-S172 TBK1, TBK1, IRF3, and phospho-S396 IRF3 were from Cell Signaling, anti-myc and anti-IκBα from Santa Cruz, and anti-optineurin (N-term and C-term) from Cayman. Anti-β-actin, N-ethylmaleimide (NEM) and LPS were from Sigma. Poly(I:C) was purchased from Invivogen. Phospho-S177 optineurin antibody was generated as reported [5]. Vesicular stomatitis virus (VSV) strain Indiana was kindly provided by Jonathan W. Yewdell (NIAID). Secondary antibodies conjugated with HRP were purchased from GE Healthcare. Nitrocellulose membranes were purchased from Biorad. Protease and phosphatase inhibitor cocktails and SuperScript® First-Strand Synthesis System were obtained from Roche, and enhanced chemiluminiscence reagent from Pierce. Power SYBR Green was obtained from Applied Bioscience. High-molecular weight poly(I:C) was from InvivoGen, NuPAGE®SDS-PAGE, and Lipofectamine 2000 from Invitrogen. Triton and RIPA buffer for mammalian cell lysis were obtained from Thermo Scientific.
Immunofluorescence Microscopy
BMDM were grown in 8 well Lab-Tec chamber slides (Thermo Fisher Scientific) for 24 hr and fixed with 4% paraformaldehyde for 15 min. After fixation, cells were washed three times with PBS, permeabilized with 0.1% Triton X-100 for 15 min, and blocked with 10% goat serum for 1 hr. The slides were then incubated overnight with anti-optineurin from Cayman (1:500) and anti-GM130 antibody from BD Transduction Laboratories (1:500) in PBS at 4°C, and after thorough washing incubated with secondary goat anti-rabbit IgG Alexa Fluor®594 and IgG-Alexa Fluor®488 (each at 1:2000) from Molecular Probes. The slides were mounted with DAPI-containing mounting medium (Vector laboratories) and images were acquired with a confocal microscope (LSM 510 META, Carl Zeiss) and processed using Zeiss LSM image browser.
In vitro protein pull-down assays
In vitro protein pull-down assays were performed as described [3]. Briefly, GST or GST-optineurin fusion proteins were expressed in BL21 E. Coli cells, purified on glutathione Sepharose 4B beads (Amersham Pharmacia), and equal amounts as verified by Coomassie blue staining were used in pull-down assays. HEK293 cells transfected with myc-TBK1 were lysed in Triton lysis buffer (20 mM Tris pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 30 mM NaF, 2 mM sodium pyrophosphate, 5 mM N-ethylmaleimide, and protease inhibitor cocktail). Purified GST proteins on beads were used to pull down HEK293 cell lysate with overexpressed myc-TBK1 in lysis buffer. The beads were washed with RIPA buffer and after protein elution resolved by SDS-PAGE and immunoblotted.
Immunoblotting and immunoprecipitation
Whole cell extracts of peripheral blood mononuclear cells (PBMC), BMDM, and HEK293 cells were prepared in RIPA lysis buffer for detection of phospho-IRF3 and IRF3, or Triton lysis buffer for other proteins. Proteins were resolved on NuPAGE®SDS-PAGE and transferred to nitrocellulose membranes and immunoblotted with the indicated primary and HRP-conjugated secondary antibodies. Signals were detected using enhanced chemiluminiscence. Immunoprecipitation was performed as described [3]. Briefly, BMDM were treated with 100 μg/ml poly(I:C) or 100 ng/ml LPS for the indicated times. The cells were washed once with PBS and lysed in Triton lysis buffer supplemented with protease and phosphatase inhibitors. For p-TBK1 blots after optineurin immunoprecipitation, HEK293-hTLR3 cells were lysed with the following lysis buffer: 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% Triton X-100, 2 mM EDTA, 2 mM sodium pyrophosphate, 25 mM β-glycerophosphate, 1 mM sodium orthovanadate and protease inhibitor cocktail (Thermo Scientific). The cell lysates were precleared with protein G-conjugated Sepharose-4B (Sigma) and incubated with anti-optineurin antibody for 16 hr at 4°C, followed by incubation with protein G-conjugated beads for another hour. The immunoprecipitates were resolved by SDS-PAGE and immunoblotted with anti-pTBK1, -TBK1, and -optineurin antibodies.
ELISA for IFN β
BMDM cultured in complete DMEM were stimulated with 100 ng/ml LPS or 30 μg/ml poly(I:C), and supernatants were collected at the indicated times. Mice were injected with 1010 plaque forming units (pfu) of VSV intraperitoneally (i.p.) and blood was drawn 16 hr later. Supernatants and sera were assayed for IFN-β with the IFN-β ELISA kit (PBL Interferon Source) following the manufacturer’s protocol.
Genotyping and quantitative RT-PCR (qRT-PCR)
OptnΔ157 mice were genotyped with the following primers: forward: CCTGCTTCCTCATGCAGTGATCCAGAG and reverse: AAGGAAAAAAGAGCTCGCGGCCGCGCTCCTGATAACAC; another forward primer was used as internal control: ACGCGTCGACGTCGGCCATAGCGGGGACACACACTTGT. Real-time PCR was performed with SYBR Green using a 7500 Real Time PCR System (Applied Bioscience). Housekeeping ribosomal 18S RNA was amplified to normalize RNA content of the lysate and obtain a ΔCT value. The primers used were, Optn-Forward: GCTCCGAAATCAAGATGGAG; Optn-Reverse: GCAGAGTGGCTAACCTGGAC; 18S-Forward: AAATCAGTTATGGTTCCTTTGGTC; 18S-Reverse: GCTCTAGAATTACCACAGTTATCCAA, Il6-Forward: GCTACCAAACTGGATATAATCAGGA and Il6-Reverse: CCAGGTAGCTATGGTACTCCAGAA-3′.
Statistical analysis and protein quantification
Statistical analysis was done using Student’s t test with GraphPad Prism software. Densitometry was done with Image J software (National Institutes of Health).
Results
Generation of mice lacking the optineurin TBK1 binding region
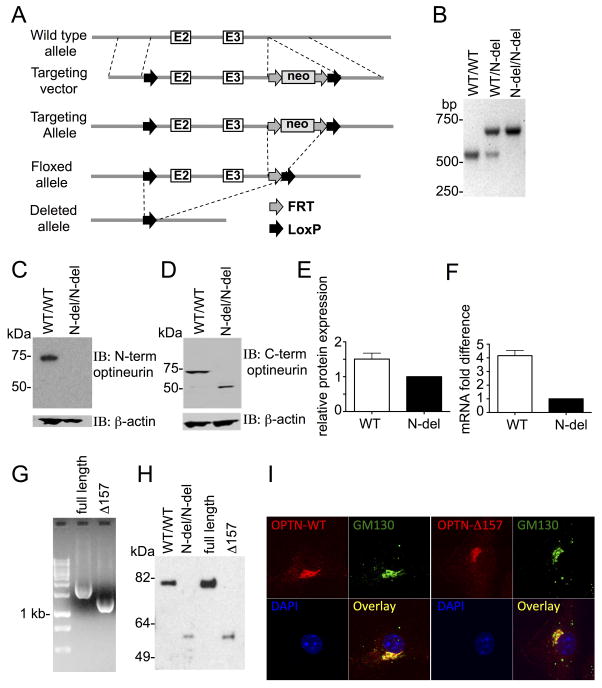
To generate mice lacking the optineurin TBK1-binding region, the first two coding exons of Optn were floxed (Fig 1A). Mice carrying the floxed allele were crossed to mice expressing Cre recombinase under the control of the β-actin promoter, achieving whole body deletion of this fragment. The excision of the floxed exons was verified by PCR of tail DNA (Fig 1B), and lysates of bone marrow derived macrophages (BMDM) from homozygous WT and the mice with N-terminal deletion that were immunoblotted for optineurin. An antibody against N-terminal optineurin detected a ~75 kDa WT protein, whereas no band was detected in mice where the N-terminus of optineurin was deleted (Fig 1C). In the latter, an antibody against the C-terminus of optineurin uncovered a ~50 kDa band (Fig 1D), demonstrating that the excision of the first coding exon did not result in complete optineurin deficiency, instead generating a truncated protein. The truncated protein was expressed at approximately 1.5 fold lower level than its WT counterpart, as measured by densitometry (Fig. 1E). The comparison of mRNA expression levels showed that N-terminal deletion also had several fold lesser mRNA levels (Fig. 1F). To examine which of the possible alternative open reading frames in the optineurin gene was being used to generate truncated protein, we created expression vectors encoding the possible truncated proteins transcribed from the two ATG codons that could likely generate a ~50 kDa protein. Constructs of the full-length optineurin and the two possible N-terminal truncations starting from the first and the second methionine in exon 4 (OptnΔ157 and OptnΔ192) were made, and confirmed by PCR analysis (Fig. 1G and data not shown). Western blot of lysates of HEK293 cells transfected with WT, OptnΔ157, or OptnΔ192 constructs demonstrated that the deletion of the first two coding exons resulted in the initiation of the optineurin translation at the first ATG of exon 4 (Fig. 1H and data not shown). The mice were hence designated OptnΔ157.
Figure 1. Characterization of mice lacking the optineurin TBK1-binding region.
(A) The generation of mice lacking the optineurin TBK1-binding region is shown. A targeting construct with LoxP-flanked exons 2 and 3 was inserted into the endogenous locus and deleted upon Cre-mediated recombination. Neo cassette, FRT, and LoxP recombination sites are indicated. (B) PCR distinguishing mice with WT optineurin and an N-terminal deletion. (C and D) Western blot of BMDM from the indicated mice with anti-optineurin antibodies raised against N-terminal and C-terminal epitopes, respectively. (E) Optineurin protein from BMDM from WT and N-terminal deletion mice was detected by an antibody against optineurin C-terminus. The level of optineurin expression was quantified from three replicate blots on Image J software and the WT protein level is expressed as % of the level of the deleted protein. (F) Optineurin mRNA was detected in BMDM by qRT-PCR. ΔΔCT of N-terminal deletion was designated as 1, and the difference between ΔΔCT of WT optineurin and N-terminal deletion is depicted as mean ± SEM for 2 independent experiments with duplicate samples. (G) PCR of the WT or the indicated optineurin mutants from pcDNA3.1+ expression plasmids. (H) Western blot with an antibody against the C-terminus of optineurin in PBMC from the indicated mice (first two lanes), and lysates of HEK293 cells transfected with the WT optineurin or the indicated mutants in pcDNA3.1+ vector (last two lanes). (I) BMDM from WT and OptnΔ157 mice were analysed by confocal microscopy after staining with the indicated antibodies and DAPI (for nuclei). Objective used for acquiring the images was Plan-Apochromat 63x/1.4 Oil DIC. One representative experiment out of three is shown.
The initial characterization did not show differences between WT and OptnΔ157 mice. OptnΔ157 mice were born at normal Mendelian frequencies and did not exhibit signs of disease. Moreover, OptnΔ157 had the same cellular localization as WT protein, both being found in the cytoplasm, and preferentially located at the Golgi apparatus, as demonstrated by their colocalization with Golgi matrix protein GM130 (Fig. 1I).
Inducible binding of TBK1 by the optineurin N-terminal region
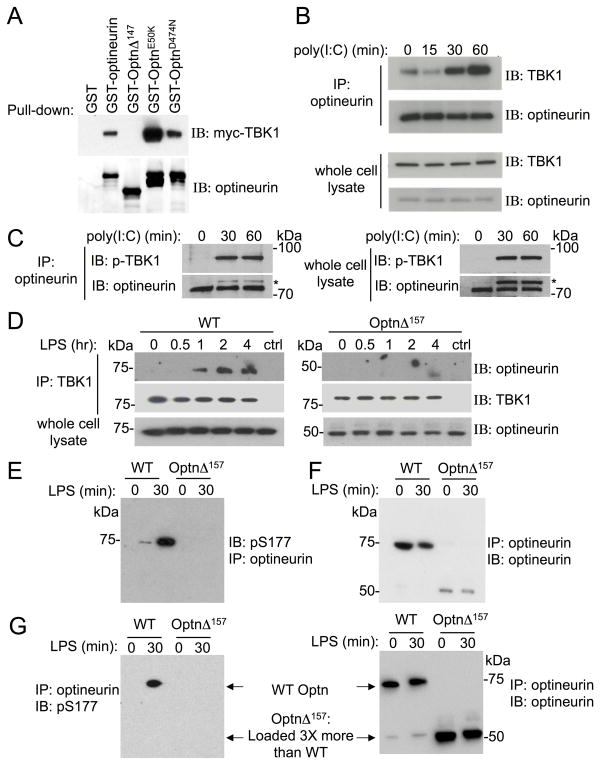
The region of optineurin responsible for TBK1-binding has been mapped to residues 1-127 [25], and thus should be missing in OptnΔ157 mice. To test this, myc-tagged TBK1 from HEK293 cell lysates was pulled down with either GST fusion proteins containing WT optineurin or OptnΔ147, the human truncation homologous to mouse OptnΔ157. As expected, GST-WT optineurin pulled down TBK1 (Fig 2A), as did the control optineurin E50K, a glaucoma-related mutation that was found to associate to TBK1 even more strongly than the WT protein [25]. In contrast, TBK1 was not pulled down with OptnΔ147, confirming that this region is crucial for this interaction. A ubiquitin-binding point mutant in the UBAN (optineurin D474N) bound to TBK1 comparably to the WT protein, strengthening the previous report that the ability of optineurin to bind ubiquitin is not a prerequisite for a TBK1 interaction [23]. The binding of TBK1 and optineurin was further tested in cell lysates from cells stimulated with a synthetic double-strand RNA mimicking agent, poly(I:C), which signals via TLR3 [27]. It has been reported that optineurin is constitutively bound to TBK1 [15, 23]. We found, however, that only a small fraction of optineurin co-immunoprecipitated with TBK1 in unstimulated HEK293-hTLR3 cells, and that binding greatly increased upon stimulation (Fig 2B). Moreover, TBK1 bound to optineurin was activated, i.e. phosphorylated on S172 (Fig 2C). Inducible binding was also compared in WT and OptnΔ157 BMDM after stimulation with LPS, a TLR4 ligand. Whereas WT optineurin did not co-immunoprecipitate with TBK1 in unstimulated cells, LPS induced an association that increased over time (Fig 2D). OptnΔ157 did not bind to TBK1 in any situation, confirming that optineurin binding to TBK1 is inducible and contingent on the presence its N-terminal TBK1-binding region.
Figure 2. The N-terminus of optineurin is important for TBK1 binding.
(A) Lysates of HEK293 cells overexpressing myc-TBK1 were incubated with purified GST fusion proteins containing WT optineurin or the indicated mutants. Bound proteins were subjected to immunoblotting using anti-myc and anti-optineurin antibodies. One of the three experiments is shown. (B) Lysates of HEK293-hTLR3 cells were stimulated with 100 μg/ml of poly(I:C) for indicated times and were immunoprecipitated with anti-optineurin. The immunoprecipitate and the whole cell lysate were resolved on SDS-PAGE and immunoblotted with anti-TBK1 and antioptineurin antibodies. (C) HEK293-hTLR3 cells were stimulated with 10 μg/mL of poly(I:C) for the indicated times and an immunoprecipitation was performed with an anti-optineurin antibody. Immunoprecipitates (left) and whole cell lysates (right) were blotted for phospho-TBK1 and optineurin; one representative experiment out of two is shown. *Designates phospho-TBK1 signal from the previous blot. (D) BMDM from WT and OptnΔ157 mice were stimulated with LPS for the indicated times and cells lysates were immunoprecipitated with anti-TBK1. Bound proteins were resolved on SDS-PAGE and immunoblotted using anti-optineurin and anti-TBK1 antibodies, respectively. (E) and (F) BMDMs were stimulated with LPS for 30 min and the lysates were immunoprecipitated using anti-optineurin antibody. Bound proteins were subjected to SDS-PAGE and immunoblotted either with anti-phosphoS177 that recognizes phosphorylation of mouse optineurin at residue S187 (left panel), or anti-optineurin antibody (right panel). (G) Optineurin immunoprecipitates shown in E and F, were loaded to SDS-PAGE gels in ratio of KI:WT lysate as 3:1. Membranes were immunoblotted as indicated above. All experiments were done 3 or more times, and a representative blot is shown.
TBK1 phosphorylates human optineurin on S177 [5]. Although this is thought to be important for autophagy, its relevance in TBK1-mediated signaling events is unclear. We tested if TBK1 association to optineurin via its N-terminus is necessary for this phosphorylation upon TLR-mediated stimulation. LPS stimulation of BMDM cells resulted in WT optineurin phosphorylation on S187, the murine homolog of S177 (Fig 2E). In contrast, no optineurin phosphorylation was observed in OptnΔ157 cells. Because OptnΔ157 is expressed at a lower level than the WT (Fig 2F), we also loaded 3-fold more OptnΔ157 immunoprecipitate, but still saw no phosphorylation of the truncated form (Fig. 2G). Together, these results demonstrate that optineurin binds to TBK1 in an inducible manner, and that the N-terminal region of optineurin is required for both TBK1 association and S187 phosphorylation.
The N-terminal region of optineurin is required for TBK1 phosphorylation and IFN-β production
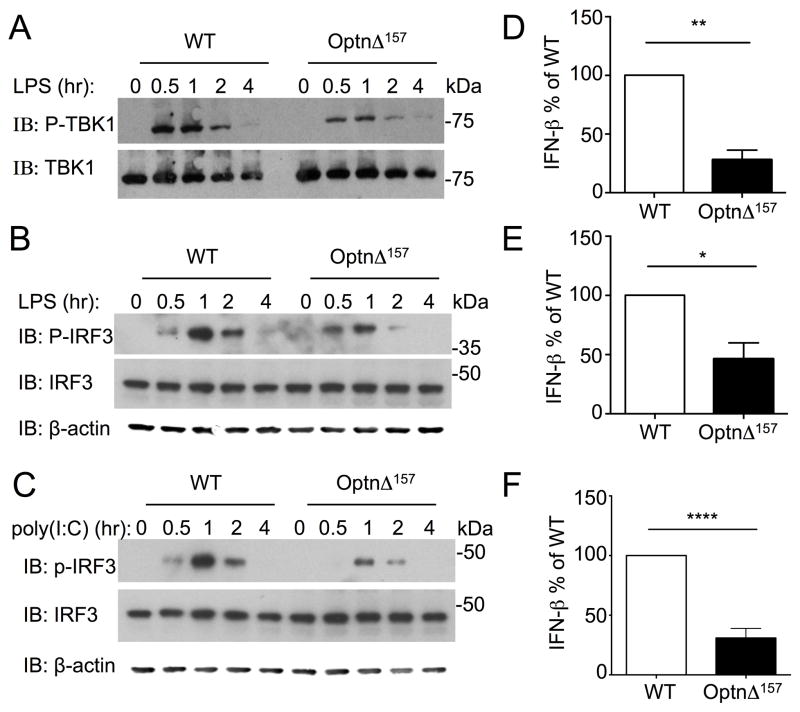
Upon TLR stimulation, TBK1 is activated by phosphorylation at S172, and subsequently phosphorylates the transcription factor IRF3, which is essential for IFN-β production [22]. To test the importance of N-terminal region of optineurin for TBK1 activation, phosphorylation of TBK1 at S172 was determined. Stimulation with LPS induced phosphorylation of TBK1 in WT BMDM that peaked at 30 min to 1 hr and waned thereafter (Fig 3A). TBK1 was phosphorylated to a lesser extent in OptnΔ157 cells. We next examined substrates of TBK1 in the TLR3/4 signaling cascades. IRF3 phosphorylation was detectable 30 min after LPS stimulation in WT BMDM, peaked at 1hr, and decreased thereafter (Fig 3B). Phosphorylation of IRF3 in OptnΔ157 BMDM was markedly reduced compared to WT cells. A similar reduction of IRF3 phosphorylation in OptnΔ157 BMDM was observed after poly(I:C) treatment (Fig 3C). Thus, TLR-induced TBK1 and IRF3 activation were compromised in BMDM in which an interaction between optineurin TBK1 is precluded.
Figure 3. The N-terminus of optineurin is important for TBK1 phosphorylation and IFN-β secretion.
(A) BMDMs from WT and OptnΔ157 mice were stimulated with LPS for the indicated times and cell extracts were subjected to immunoblotting using anti-phospho-TBK1 (upper panel), stripped, and reprobed with an antibody recognizing total TBK1 (lower panel). (B) Lysates from (A) were immunoblotted with anti-phospho-IRF3 antibody (upper panel), stripped, and reprobed with an antibody recognizing total IRF3 (middle panel). Immunoblot with β-actin antibody as loading control is shown for A and B. (C) BMDMs from WT and OptnΔ157 mice were stimulated with poly(I:C) and cell lysates immunoblotted with anti-phospho-IRF3 antibody (upper panel), stripped, and reprobed with an antibody recognizing total IRF3 (middle panel) or β-actin (lower panel). For A–C the experiments were done 3 or more times and the representative blot is shown. WT and OptnΔ157 mice were treated with 100 ng/ml LPS (D), 30 μg/ml poly(I:C) (E), and IFN-β was measured in supernatants after 7 hr of stimulation. The results from 3 separate experiments were normalized to WT IFN-β levels. (F) WT and OptnΔ157 mice were infected with VSV 1 × 1010 pfu for 16 hr and IFN-β was measured in the sera. Sera were taken from at least 7 mice per genotype from 2 separate experiments, and the results were normalized as in E. The average IFN-β from two experiments was 173 pg/mL and 61,4 in WT, and 41,6 and 12,5 pg/mL in OptnΔ157 mice. The data represent mean ± SE measured. * p < 0.05; ** p <0.001: **** p<0,0001.
TBK1-mediated IRF3 phosphorylation is crucial for IFN-β production in response to pathogens. BMDM were LPS and poly(I:C) treated and IFN-β secretion was monitored. In both cases, IFN-β production was diminished in OptnΔ157 cells (Fig 3D and 3E). To test the importance of optineurin in a viral model in vivo, WT and OptnΔ157 mice were infected with Vesicular Stomatitis Virus (VSV). Again, the IFN-β measured in the sera was decreased in OptnΔ157 mice (Fig 3F). These data argue that TBK1:optineurin association is required for optimal IFN-β responses, and confirm that optineurin is a positive regulator of TBK1 both in vitro and in vivo.
Several reports found reduced activation NF-κB upon optineurin overexpression, suggesting that it acts as a negative regulator [2, 3]. However, enhanced NF-κB activation was not found in the genetic models of optineurin insufficiency or deficiency [4, 23, 28]. To test if OptnΔ157 mitigated the activation of NF-κB, we stimulated BMDM with LPS or poly(I:C) and monitored proximal and distal signaling events. A similar pattern of IκB degradation was observed in both WT and OptnΔ157 cells for both stimuli (Suppl. Fig. 1A and B), and the NF-κB-regulated cytokine IL-6 was equally produced (Suppl. Fig. 1C and D). These results argue that OptnΔ157 does not interfere with LPS and poly(I:C) mediated NF-κB activation. Taken together, the optineurin N-terminus is dispensable for NF-κB pathway but necessary for optimal TBK1 activity and IFN-β production.
Discussion
Optineurin has been proposed to bind to TBK1 and regulate its kinase activity. However, the outcome of this interaction remains controversial. Here we found that in primary cells N-terminal truncated optineurin OptnΔ157 was unable to bind to TBK1, resulting in diminished TBK1 and IRF3 activity and IFN-β secretion. Although because OptnΔ157 was present at a 1.5 lower level, we cannot formally exclude the possibility that lower level of the truncated product contributed to the impaired IFN-β response, we believe that this relatively mild optineurin deficiency could not by itself explain the complete lack of PAMP-induced inducible TBK1 binding to OptnΔ157 and subsequent lack of optineurin phosphorylation. Our data contrast several reports in HEK293 and HeLa cell lines, one of which proposed that optineurin recruits the CYLD deubiquitinase to TBK1 in a cell-cycle dependent manner to shut down TBK1 activity [15, 24, 29]. We speculate that the reason for this discrepancy lies in the cell lines used and/or the fact that these studies were limited to overexpression or silencing. In addition, given that optineurin has been shown to interact to a large number of cellular proteins and its levels increase upon cell stimulation, it is also possible that it participates in additional regulatory pathways, and its activity is different at different times after stimulation. Our results, however, strongly support the findings of two other genetic models of optineurin insufficiency (OptnD477N and Optn470T) [4, 23]. Moreover, while this article was in preparation, another study found diminished TBK1 activity and IFN-β production in an optineurin knockout model [28]. Together these results strongly argue that optineurin is positive regulator of TBK1-mediated IFN-β responses. Although it is formally possible that OptnΔ157 acts as a dominant negative protein, we find it unlikely because pull-down assays showed that this truncation does not bind to TBK1, and more importantly, OptnΔ157 cells exhibited a similar loss of TBK1 activity upon PAMP stimulation as did other optineurin insufficiency models.
Most mutations described in ALS patients either lack the C-terminal domain or harbor mutations in the ubiquitin binding region, but the mechanism of their noxious effect is still debated [2, 30]. The finding that both N-terminal and C-terminal optineurin insufficiency together with the knockout model had diminished TBK1 activity argues that optineurin mutations found in human ALS patients result in loss-of-function rather than gain-of-function due to dominant-negative activity. Further elucidation of the role of optineurin in ALS pathogenesis, currently ongoing in our laboratory, will show if this is directly linked to suboptimal TBK1 activity and/or IFN-β production, and perhaps leads to neuroinflammation, a major driving force in ALS. Of note, neither young OptnΔ157 nor OptnΔ470T mice develop signs of motor neuron degeneration (up to one year of life; IM and JDA unpublished observations), which is perhaps expected given the fact that ALS neurodegeneration is multifactorial, usually occurs late in life, and optineurin likely only has a protective role, as discussed here.
The N-terminus of optineurin was shown to bind TBK1 in vitro using recombinant GST fusion products [25]. Here we have extended this finding to show the in vivo relevance of this interaction in TBK1 activation. The proposed binding site can likely be narrowed to amino acids 78-121 in human optineurin, which are highly homologous to the TBK1-binding sites of three other TBK1 adaptor proteins, namely TANK, NAP1, SINTBAD, all of which have also been implicated in IRF3 and NF-κB activation. However, it was suggested that optineurin binding to TBK1 is constitutive and independent of ubiquitination [15, 23]. This is at odds with the proposed dynamic regulation of TBK1, in which PAMP stimulation results in the recruitment of inactive TBK1 to signalosomes via adaptor proteins, leading to its high local concentration and activation by trans-autophosphorylation [21]. Moreover, K63-linked polyubiquitination of TBK1 on lysines 30 and 401 is a prerequisite for its activation and optineurin, unlike the adaptors SINTBAD and NAP1, has an ubiquitin-binding region [22, 31]. Consistent with this, we found that in HEK293-hTLR3 cells and primary bone marrow macrophages, WT optineurin bound inducibly to TBK1 upon TLR stimulation and was phosphorylated on S187. Inducible binding suggests that the optineurin scaffolding function precedes TBK1 recruitment to signalosomes. We also demonstrate that the TBK1 associated with optineurin is phosphorylated at S172. In contrast, OptnΔ157 was unable to bind TBK1, was not phosphorylated on S187, and led to reduced TBK1 activation. Together with a published report on the role of the ubiquitin-binding region in the optineurin C-terminus [23], current findings suggest that neither the N-terminus nor the ubiquitin-binding regions alone are sufficient for stimulus-triggered binding, thus arguing for a bipartite binding model between TBK1 and optineurin: the interaction with N-terminus of optineurin is further strengthened upon TBK1 polyubiquitination. It is possible that this bipartite interaction allows a positive feedback mechanism between those two proteins, as recently proposed during mitophagy, whereby the phosphorylation of optineurin by TBK1 on S473 and S513 increases its binding to ubiquitinated mitochondria, which in turn further amplifies TBK1 activation [32]. The optineurin scaffolding function is further characterized in our submitted manuscript, where we show that virus-induced TBK1 polyubiquitination on lysines 30 and 401 targets TBK1 to optineurin at the Golgi apparatus (Pourcelot et al.).
Both TBK1 and optineurin are multifunctional proteins. Interestingly, they form a functional unit not just in TLR signaling, but also during autophagy. Optineurin becomes an efficient autophagy adaptor when phosphorylated by TBK1 on S177 (or murine S187); it subsequently bridges LC3 coated autophagosomal membranes to autophagy targets, such as ubiquitinated cytosolic Salmonella, protein aggregates or damaged mitochondria [5, 32–35]. Notably, TBK1 mutants that cannot bind optineurin were also recently found in ALS patients, and the preliminary data suggest that they cause functional impairment of TBK1 in autophagy, rather than in signaling [36, 37]. In line with this, it was recently described that PINK1/parkin-mediated ubiquitination of damaged mitochondria leads to optineurin recruitment and TBK1-mediated optineurin S177 phosphorylation, consequently leading to LC3 recruitment and mitophagy [32, 34, 35]. It is unclear if there is a direct crosstalk between autophagy or mitophagy and the pathways leading to IFN-β secretion and/or whether some of the resources, such as optineurin itself, are limited. It is also possible that the two events are spatially and/or temporally dissociated, and that inducible optineurin binding serves to distinguish between these different cellular events. Notably, S177 phosphorylation of optineurin by another kinase, Polo-like kinase 1 (Plk1), causes dissociation of optineurin from Golgi-associated Rab8, allowing its nuclear translocation and association with myosin phosphatase targeting subunit 1 (MYPT1), which then antagonizes Plk1 activity and permits cell division [9]. It is thus likely that other regulatory mechanisms are in place to efficiently transduce specific signals from the shared pool of optineurin, which can apparently even harbor similar posttranslational modifications when exerting different functions. Other reported posttranslational modifications of optineurin might contribute. A tumor suppressor ubiquitin ligase HACE-1 was shown to add K48-polyubiquitin to optineurin, prompting its association with another autophagy adaptor p62/SQSTM1, accelerating autophagy flux [38]. Notably, the bipartite interaction of optineurin to TBK1, via both N-terminus and ubiquitin-binding, is likely one of such specificity-ensuring mechanisms. Overall, the finding that TBK1 interaction to optineurin is inducible, and dependent on N-terminus, perhaps makes the inhibition of this interaction an interesting target in overactive TBK1-mediated responses such as in autoimmune disease or cancer.
Supplementary Material
Suppl. Fig 1. (A and B) BMDMs from WT and OptnΔ157 mice were stimulated with 100 ng/mL of LPS or 10 μg/mL of poly(I:C) for the indicated times. Lysates were subjected to SDS-PAGE and blotted with an anti-IκBα antibody. (C and D) IL-6 mRNA was analysed by qRT-PCR in BMDM stimulated for 4hr with the indicated concentrations of LPS and poly(I:C). ΔΔCT of WT was designated as 1, and the difference between ΔΔCT of IL-6 mRNA from WT and OptnΔ157 cells is depicted as mean ± SEM for three experiments for LPS stimulation and mean ± SD for two experiments for poly(I:C), both with duplicate samples.
Acknowledgments
This work was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health, the Croatian Science Foundation grant #7459 and the support of the University of Rijeka to IM, grants from Fondation ARC (Association pour La Recherche contre le Cancer) and the ANRS (Agence Nationale pour la Recherche sur le SIDA) to DA, and fellowship to MP from Université Paris-Sud.
We are grateful to Lino Tessarollo and Eileen Southon (Mouse Cancer Genetics Program, NCI-Frederick, Frederick, MD) for assistance during generation of OptnΔ157 mice, Dr. Ivan Dikic for anti-phospho-S177 optineurin antibody, and Ehydel Castro and Bei Dong for expert technical assistance.
References
- 1.Rezaie T, Child A, Hitchings R, Brice G, Miller L, Coca-Prados M, Heon E, Krupin T, Ritch R, Kreutzer D, Crick RP, Sarfarazi M. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295:1077–1079. doi: 10.1126/science.1066901. [DOI] [PubMed] [Google Scholar]
- 2.Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, Komure O, Matsuura S, Kobatake K, Morimoto N, Abe K, Suzuki N, Aoki M, Kawata A, Hirai T, Kato T, Ogasawara K, Hirano A, Takumi T, Kusaka H, Hagiwara K, Kaji R, Kawakami H. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- 3.Zhu G, Wu CJ, Zhao Y, Ashwell JD. Optineurin negatively regulates TNFalpha-induced NF-kappaB activation by competing with NEMO for ubiquitinated RIP. Curr Biol. 2007;17:1438–1443. doi: 10.1016/j.cub.2007.07.041. [DOI] [PubMed] [Google Scholar]
- 4.Munitic I, Giardino Torchia ML, Meena NP, Zhu G, Li CC, Ashwell JD. Optineurin insufficiency impairs IRF3 but not NF-kappaB activation in immune cells. J Immunol. 2013;191:6231–6240. doi: 10.4049/jimmunol.1301696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dötsch V, Bumann D, Dikic I. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tumbarello DA, Waxse BJ, Arden SD, Bright NA, Kendrick-Jones J, Buss F. Autophagy receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome. Nat Cell Biol. 2012;14:1024–1035. doi: 10.1038/ncb2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sahlender DA, Roberts RC, Arden SD, Spudich G, Taylor MJ, Luzio JP, Kendrick-Jones J, Buss F. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J Cell Biol. 2005;169:285–295. doi: 10.1083/jcb.200501162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreland RJ, Dresser ME, Rodgers JS, Roe BA, Conaway JW, Conaway RC, Hanas JS. Identification of a transcription factor IIIA-interacting protein. Nucleic Acids Res. 2000;28:1986–1993. doi: 10.1093/nar/28.9.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kachaner D, Filipe J, Laplantine E, Bauch A, Bennett KL, Superti-Furga G, Israel A, Weil R. Plk1-dependent phosphorylation of optineurin provides a negative feedback mechanism for mitotic progression. Mol Cell. 2012;45:553–566. doi: 10.1016/j.molcel.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 10.Chalasani ML, Swarup G, Balasubramanian D. Optineurin and Its Mutants: Molecules Associated with Some Forms of Glaucoma. Ophthalmic Res. 2009;42:176–184. doi: 10.1159/000232400. [DOI] [PubMed] [Google Scholar]
- 11.Swarup G, Nagabhushana A. Optineurin, a multifunctional protein involved in glaucoma, amyotrophic lateral sclerosis and antiviral signalling. J Biosci. 2010;35:501–505. doi: 10.1007/s12038-010-0056-9. [DOI] [PubMed] [Google Scholar]
- 12.Wagner S, Carpentier I, Rogov V, Kreike M, Ikeda F, Lohr F, Wu CJ, Ashwell JD, Dotsch V, Dikic I, Beyaert R. Ubiquitin binding mediates the NF-kappaB inhibitory potential of ABIN proteins. Oncogene. 2008;27:3739–3745. doi: 10.1038/sj.onc.1211042. [DOI] [PubMed] [Google Scholar]
- 13.Laplantine E, Fontan E, Chiaravalli J, Lopez T, Lakisic G, Véron M, Agou F, Israël A. NEMO specifically recognizes K63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. EMBO J. 2009;28:2885–2895. doi: 10.1038/emboj.2009.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwamborn K, Weil R, Courtois G, Whiteside ST, Israel A. Phorbol esters and cytokines regulate the expression of the NEMO-related protein, a molecule involved in a NF-kappa B-independent pathway. J Biol Chem. 2000;275:22780–22789. doi: 10.1074/jbc.M001500200. [DOI] [PubMed] [Google Scholar]
- 15.Mankouri J, Fragkoudis R, Richards KH, Wetherill LF, Harris M, Kohl A, Elliott RM, Macdonald A. Optineurin negatively regulates the induction of IFNbeta in response to RNA virus infection. PLoS Pathog. 2010;6:e1000778. doi: 10.1371/journal.ppat.1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt-Supprian M, Bloch W, Courtois G, Addicks K, Israel A, Rajewsky K, Pasparakis M. NEMO/IKK gamma-deficient mice model incontinentia pigmenti. Mol Cell. 2000;5:981–992. doi: 10.1016/s1097-2765(00)80263-4. [DOI] [PubMed] [Google Scholar]
- 17.Zhao T, Yang L, Sun Q, Arguello M, Ballard DW, Hiscott J, Lin R. The NEMO adaptor bridges the nuclear factor-kappaB and interferon regulatory factor signaling pathways. Nat Immunol. 2007;8:592–600. doi: 10.1038/ni1465. [DOI] [PubMed] [Google Scholar]
- 18.McWhirter SM, Fitzgerald KA, Rosains J, Rowe DC, Golenbock DT, Maniatis T. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc Natl Acad Sci U S A. 2004;101:233–238. doi: 10.1073/pnas.2237236100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hemmi H, Takeuchi O, Sato S, Yamamoto M, Kaisho T, Sanjo H, Kawai T, Hoshino K, Takeda K, Akira S. The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J Exp Med. 2004;199:1641–1650. doi: 10.1084/jem.20040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 21.Helgason E, Phung QT, Dueber EC. Recent insights into the complexity of Tank-binding kinase 1 signaling networks: the emerging role of cellular localization in the activation and substrate specificity of TBK1. FEBS Lett. 2013;587:1230–1237. doi: 10.1016/j.febslet.2013.01.059. [DOI] [PubMed] [Google Scholar]
- 22.Tu D, Zhu Z, Zhou AY, Yun CH, Lee KE, Toms AV, Li Y, Dunn GP, Chan E, Thai T, Yang S, Ficarro SB, Marto JA, Jeon H, Hahn WC, Barbie DA, Eck MJ. Structure and ubiquitination-dependent activation of TANK-binding kinase 1. Cell Rep. 2013;3:747–758. doi: 10.1016/j.celrep.2013.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gleason CE, Ordureau A, Gourlay R, Arthur JS, Cohen P. Polyubiquitin Binding to Optineurin Is Required for Optimal Activation of TANK-binding Kinase 1 and Production of Interferon {beta} J Biol Chem. 2011;286:35663–35674. doi: 10.1074/jbc.M111.267567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Genin P, Cuvelier F, Lambin S, Corte-Real Filipe J, Autrusseau E, Laurent C, Laplantine E, Weil R. Optineurin regulates the interferon response in a cell cycle-dependent manner. PLoS Pathog. 2015;11:e1004877. doi: 10.1371/journal.ppat.1004877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morton S, Hesson L, Peggie M, Cohen P. Enhanced binding of TBK1 by an optineurin mutant that causes a familial form of primary open angle glaucoma. FEBS Lett. 2008;582:997–1002. doi: 10.1016/j.febslet.2008.02.047. [DOI] [PubMed] [Google Scholar]
- 26.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall-Clarke S, Downes JE, Haga IR, Bowie AG, Borrow P, Pennock JL, Grencis RK, Rothwell P. Polyinosinic acid is a ligand for toll-like receptor 3. J Biol Chem. 2007;282:24759–24766. doi: 10.1074/jbc.M700188200. [DOI] [PubMed] [Google Scholar]
- 28.Slowicka K, Vereecke L, McGuire C, Sze M, Maelfait J, Kolpe A, Saelens X, Beyaert R, van Loo G. Optineurin deficiency in mice is associated with increased sensitivity to Salmonella but does not affect proinflammatory NF-κB signaling. Eur J Immunol. 2015 doi: 10.1002/eji.201545863. [DOI] [PubMed] [Google Scholar]
- 29.Sakaguchi T, Irie T, Kawabata R, Yoshida A, Maruyama H, Kawakami H. Optineurin with amyotrophic lateral sclerosis-related mutations abrogates inhibition of interferon regulatory factor-3 activation. Neurosci Lett. 2011;505:279–281. doi: 10.1016/j.neulet.2011.10.040. [DOI] [PubMed] [Google Scholar]
- 30.Millecamps S, Boillee S, Chabrol E, Camu W, Cazeneuve C, Salachas F, Pradat PF, Danel-Brunaud V, Vandenberghe N, Corcia P, Le Forestier N, Lacomblez L, Bruneteau G, Seilhean D, Brice A, Feingold J, Meininger V, Leguern E. Screening of OPTN in French familial amyotrophic lateral sclerosis. Neurobiol Aging. 2011;32:557, e11–3. doi: 10.1016/j.neurobiolaging.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 32.Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Mol Cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korac J, Schaeffer V, Kovacevic I, Clement AM, Jungblut B, Behl C, Terzic J, Dikic I. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J Cell Sci. 2013;126:580–592. doi: 10.1242/jcs.114926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A. 2014;111:E4439–48. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freischmidt A, Wieland T, Richter B, Ruf W, Schaeffer V, Muller K, Marroquin N, Nordin F, Hubers A, Weydt P, Pinto S, Press R, Millecamps S, Molko N, Bernard E, Desnuelle C, Soriani MH, Dorst J, Graf E, Nordstrom U, Feiler MS, Putz S, Boeckers TM, Meyer T, Winkler AS, Winkelman J, de Carvalho M, Thal DR, Otto M, Brannstrom T, Volk AE, Kursula P, Danzer KM, Lichtner P, Dikic I, Meitinger T, Ludolph AC, Strom TM, Andersen PM, Weishaupt JH. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat Neurosci. 2015;18:631–636. doi: 10.1038/nn.4000. [DOI] [PubMed] [Google Scholar]
- 37.Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS, Couthouis J, Lu YF, Wang Q, Krueger BJ, Ren Z, Keebler J, Han Y, Levy SE, Boone BE, Wimbish JR, Waite LL, Jones AL, Carulli JP, Day-Williams AG, Staropoli JF, Xin WW, Chesi A, Raphael AR, McKenna-Yasek D, Cady J, Vianney de Jong JM, Kenna KP, Smith BN, Topp S, Miller J, Gkazi A, Al-Chalabi A, van den Berg LH, Veldink J, Silani V, Ticozzi N, Shaw CE, Baloh RH, Appel S, Simpson E, Lagier-Tourenne C, Pulst SM, Gibson S, Trojanowski JQ, Elman L, McCluskey L, Grossman M, Shneider NA, Chung WK, Ravits JM, Glass JD, Sims KB, Van Deerlin VM, Maniatis T, Hayes SD, Ordureau A, Swarup S, Landers J, Baas F, Allen AS, Bedlack RS, Harper JW, Gitler AD, Rouleau GA, Brown R, Harms MB, Cooper GM, Harris T, Myers RM, Goldstein DB. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–1441. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Z, Chen P, Gao H, Gu Y, Yang J, Peng H, Xu X, Wang H, Yang M, Liu X, Fan L, Chen S, Zhou J, Sun Y, Ruan K, Cheng S, Komatsu M, White E, Li L, Ji H, Finley D, Hu R. Ubiquitylation of Autophagy Receptor Optineurin by HACE1 Activates Selective Autophagy for Tumor Suppression. Cancer Cell. 2014;26:106–120. doi: 10.1016/j.ccr.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Fig 1. (A and B) BMDMs from WT and OptnΔ157 mice were stimulated with 100 ng/mL of LPS or 10 μg/mL of poly(I:C) for the indicated times. Lysates were subjected to SDS-PAGE and blotted with an anti-IκBα antibody. (C and D) IL-6 mRNA was analysed by qRT-PCR in BMDM stimulated for 4hr with the indicated concentrations of LPS and poly(I:C). ΔΔCT of WT was designated as 1, and the difference between ΔΔCT of IL-6 mRNA from WT and OptnΔ157 cells is depicted as mean ± SEM for three experiments for LPS stimulation and mean ± SD for two experiments for poly(I:C), both with duplicate samples.