Abstract
Trypanosomes that cause sleeping sickness endocytose Apolipoprotein L1 (APOL1)-containing Trypanolytic Factors from human serum, leading to trypanolytic death through generation of APOL1-associated lytic pores in trypanosomal membranes. The trypanosome T. brucei rhodesiense counteracts trypanolysis by expressing the surface protein SRA, which can bind APOL1 common variant G0 to block its trypanolytic activity. However, two missense variants in the C-terminal predicted coiled-coil (CC) domain of human APOL1, G1 (S342G/I384M) and G2 (ΔN388Y389), decrease or abrogate APOL1 binding to T. brucei rhodesiense SRA, preserving APOL1 trypanolytic activity. These evolutionarily selected APOL1 missense variants, found at high frequency in some populations of African descent, also confer elevated risk of kidney disease. Understanding the SRA-APOL1 interaction and the role of APOL1 G1 and G2 variants in kidney disease demands structural characterization of the APOL1 CC domain. Using CD, heteronuclear NMR, and MD simulation on structural homology models, we report here unique and dynamic solution conformations of nephropathy variants G1 and G2 as compared with common variant G0. Conformational plasticity in G1 and G2 CC domains led to inter-helical α1-α2 approximation coupled with secondary structural changes and delimited motional properties absent in the G0 CC domain. The G1 substitutions conferred local structural changes principally along helix α1, whereas the G2 deletion altered structure of both helix α2 and nearby helix α1. These dynamic features of APOL1 CC variants likely reflect their intrinsic structural properties, and should help interpret future APOL1 structural studies and define the contribution of APOL1 risk variants to kidney disease.
Keywords: Kidney disease, T. brucei rhodesiense, nuclear magnetic resonance, Circular dichroism, molecular dynamics simulation
Introduction
Apolipoprotein L1 (APOL1) is a minor serum component synthesized at highest levels in liver, pancreas, and kidney and circulating in HDL-independent complex with APOA1 and haptoglobin-related protein [2]. The APOL1 gene, one of six contiguous APOL genes (APOL1-6) believed to have arisen through gene duplication, is present only in humans and a few other higher primate species. APOL1 allelic variants commonly found in Africa and in Americans of African descent encode human innate immunity against Trypanosoma brucei rhodesiense, the causative agent of human sleeping sickness. APOL1-mediated trypanolysis requires trypanosomal endocytic internalization of human serum Trypanosome Lytic Factor TLF1, containing APOL1 in complex with haptoglobin-related protein and haptoglobin [3]. The internalized APOL1 is believed to kill trypanosomes through one or more conformational changes conferring the ability to form lytic pores in tryanosomal membranes due either to intrinsic channel-like activity of APOL1 [4] [5] and/or to activation of oxidative processes [6].
T. brucei rhodesiense expresses a protein (Serum Response-Associated, SRA) that neutralizes the trypanolytic activity of APOL1 common variant G0 through direct interaction with the APOL1 C-terminal putative coiled-coil (CC) domain [4, 7-9]. The APOL1 renal disease risk variants G1 (S342G/I384M) and G2 (Δ388-389) commonly found in populations of West African descent retain trypanocidal potency by virtue of their greatly reduced binding to SRA [10]. However, the protection against sleeping sickness is accompanied by increased risk of non-diabetic kidney disease later in life, accompanied by accelerated progression to end-stage renal disease (ESRD), with increased risk of ischemic cardiovascular disease and all-cause mortality [11]. The incompletely penetrant nephropathic effects of APOL1 variants G1 and G2 remain little understood. In particular, the structures of critical regions involved in the APOL1-SRA interaction remain unknown.
Both renal disease risk variant G1 and G2 polymorphisms lie within the C-terminal APOL1 putative CC domain. Insight into the structural consequences of the G1 and G2 sequence variations will be important in understanding both APOL1 trypanolytic function and the contribution of APOL1 variants to renal disease risk. As a first step towards complete structure determination of the APOL1 CC domain, we report here CD spectra, thermal denaturation curves, and molecular homology modeling and molecular dynamics simulation of the coiled-coil domains from APOL1 common variant G0 and renal disease risk variants G1 and G2. We also report solution NMR spectra of C-terminal APOL1 CC domains from variants G0 and G1. The results of these experiments highlight differences in structure and stability associated with disease risk variants of APOL1 CC domain, and will inform future studies defining SRA-binding surfaces of the APOL1 CC domain.
Results
Molecular cloning of C-terminal APOL1 CC variants
APOL1 protein is modeled as three structural sub-domains (Figure 1A): an N-terminal putative “pore-forming domain” (PFD) (aa 1-235 including the N-terminal signal sequence), a putative “membrane-addressing domain” (MAD) (aa 238-304), and a C-terminal CC domain consisting of a predicted α-helical hairpin (within C-terminal aa 304-398) [12], previously shown to bind to the N-terminal predicted CC region of SRA [7, 8]. However, the aa boundaries encompassing the APOL1 C-terminal CC domain are not precisely defined. APOL1 aa 336-398 are predicted to encompass the putative CC of APOL1 common variant G0, and are aligned with those of renal disease risk variants G1 (S342G/I384M), and G2 (ΔN388/Y389) in Fig. 1A.
Figure 1.
(A) Proposed subdomains of APOL1: (ss), signal sequence; PFD, pore-forming domain; MAD, membrane-addressing domain; CC, coiled-coil domain (SRA-binding domain). Below, amino acid (aa) sequence of aa 336-398 of the common variant (G0) CC domain, aligned with the corresponding CC domains of renal disease risk variants G1 (S342G/I384M) and G2 (ΔN388/Y389). Sequence differences are in red. (B-D) Mass spectrogram and (inset) Coomassie Blue-stained 16% (w/v) reducing SDS-PAGE gel of recombinant APOL1 CC domains of G0, G1 & G2. (B) Mass of 13C/15N-labeled G0 CC. Gel shows G0 CC band of expected monomeric mass (9.8 kDa): lane 1, protein standards; lane 2, sonicated bacterial lysate; lane 3, Ni-NTA agarose eluate; lane 4, size exclusion chromatography (SEC) G0 CC fraction eluted in PBS, pH 7.0. (C) Mass of 13C/15N-labeled G1 CC. Gel shows G1 CC band of expected monomeric mass: lane 1, protein standards; lane 2, sonicated bacterial lysate; lane 3, supernatant fraction after sonication; lane 4, dialyzed eluates from Ni-NTA agarose. (D) Mass of isotopically unlabeled G2 CC. Gel shows G2 CC band of expected monomeric mass: lanes as in panel C.
Recombinant APOL1 polypeptides including aa 304-398, aa 313-398, aa 323-398, and aa 339-398 were each expressed with N-terminal His6-tags. Tag-free CC variants exhibited higher likelihood of precipitation than tagged variants. CC domain constructs G0 and G1 yielded recombinant protein in amounts suitable for both CD and NMR investigation. Yields of purified G2 sufficed only for CD spectroscopy (see below). Studies are ongoing to increase G2 CC domain yields to levels sufficient for NMR study.
2D 1H-15N heteronuclear HSQC spectra collected on 15N labeled purified CC samples of APOL1 G0 and G1 were well resolved only for the aa 339-398 constructs of APOL1 variants G0 and G1. HSQC spectra of other G0 and G1 constructs showed poor spectral resolution due to significant linewidth broadening. We therefore focused subsequent structural characterization on CC G0 and CC G1 constructs encompassing aa 339-398. The purified APOL1 CC variant polypeptides were homogeneous by SDS-PAGE (Fig. 1) and remained so after anion exchange at 4° C. Yields of purified tag-free G0 and G1 polypeptides exceeded 3 mg/L of culture, but were less than 2 mg/L for G2. However, each purified variant polypeptide, exhibited time-dependent precipitation.
MALDI-TOF MS
MALDI-TOF-determined molecular mass of N-terminally His6-tagged proteins may be variable due to loss of the initiator Met [13, 14]. The experimental mass of each evaluated APOL1 CC polypeptide indeed reflected loss of one Met, as shown in Fig. 1B for 13C/15N-labeled His6-tagged APOL1 G0 CC domain (9741.12 Da), in agreement with its (minus Met) theoretical mass (9746.45 Da). MALDI-TOF masses also matched the theoretical masses for purified 13C/15N-labeled APOL1 G1 and isotopically unlabeled APOL1 G2 CC domains in His6-tagged (Figs. 1C and 1D, respectively) and untagged forms (data not shown).
CD spectroscopy
Far-UV CD spectra of G0, G1, and G2 CC domains are shown in Fig. 2A. CD spectra with characteristic double ellipticity minima at 209 (208 for G2) and 220 nm were consistent with well-folded structures. Molar ellipticity ratios between 222 and 208 nm (θ222: θ208) of 1.03 for G0, 0.95 for G1, and 0.95 for G2 suggested stabilization of these domains by coiled coil inter-helical contacts [15-17]. However, the lower ratios of the G1 and G2 CC domains might indicate fewer and/or weaker inter-helical contacts. The data suggest greater helical stability of the G0 CC domain than of the G1 and G2 CC domains. This suggestion is reinforced by thermal denaturation curves (Fig. 2B) showing decreases in MRE220 (mean residue ellipticity at 220 nm) of 18% for the G0 CC domain, 30% for the G1 CC domain, and 26% for the G2 CC domain. The results together suggest that helical stability of the APOL1 G0 CC domain is higher than that of the G1 and G2 CC domains.
Figure 2.
(A) Far-UV 25 °C CD spectra of APOL1 CC common variant G0 (blue) and of disease risk variants G1 (red) and G2 (green; aa 339-398) reveal well-folded conformations in solution. Each construct includes an N-terminal His6-tag. (B) Change in Molar Residue Ellipticity at 220 nm (MRE) of G0 (blue), G1 (red) and G2 (blue) CC domains across the temperature range from 25 to 95 °C suggests that thermal stability of G1 and G2 CC domains is lower than that of G0. (C-E) APOL1 CC domain binding to SRA-GST fusion protein by Glutathione Sepharose pulldown, as evaluated by Coomassie Blue-stained SDS–PAGE (“Any kD”). (C) APOL1 CC variant polypeptides loaded into binding reactions. (D) Loaded GST-SRA at Mr ∼52 kDa (upper panel) and GST-SRA-bound APOL1 CC domains (G0, G1 and G2) at Mr ∼10.5 kDa (lower panel). (E) GST (upper panel and lack of GST-bound APOL1 CC domain binding (lower panel). One of three experiments with similar results. (F) Overlay of 2D 1H-15N HSQC spectra (298 K) of N-terminally (His)6-tagged APOL1 G0 (blue) and G1 (red) CC domain constructs (aa 339-398). The well dispersed crosspeaks indicate a folded protein conformation in solution. The crosspeak differences are encircled (in G1 variant only) or boxed (in G0 only).
SRA-APOL1 (CC) Binding Experiments
The pulldown experiments of Fig. 2C show that APOL1 CC polypeptides were bound by GST-SRA fusion protein, but not by GST alone. Binding of APOL1 G1 variant CC was 48 (± 9.7) % that of APOL1 G0, whereas binding of APOL1 G2 was only 9 (± 12.25) % that of G0 (n=3). This variant-dependent binding of APOL1 CC constructs is consistent with previous results using shorter APOL1 CC domain peptides encompassing only its α2 helix [4] (see below), and is compatible with the conformational rearrangements observed in MD simulations of the modeled structures of APOL1 variants G0, G1 and G2 (Fig. 3E-F).
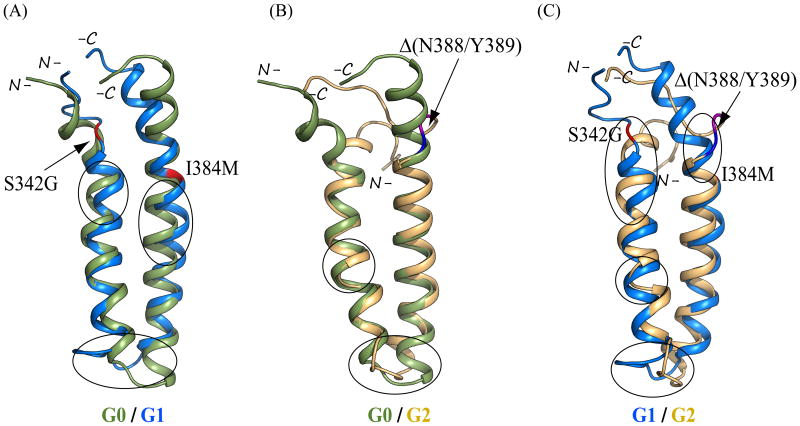
Figure 3.
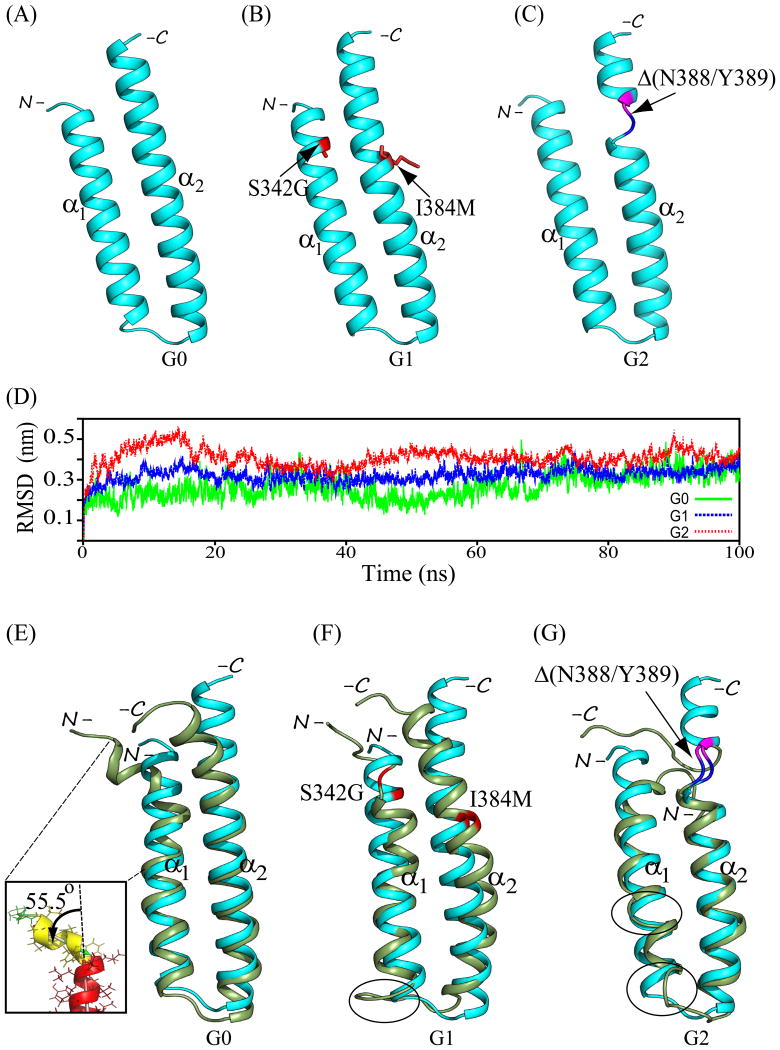
Backbone ribbon structural models of APOL1 CC domains (aa 336-398) of (A) common variant G0, (B) disease risk variant G1 harboring dual missense polymorphisms S342G and I384M (red stick format), and (C) disease risk variant G2 harboring the deletion polymorphism Δ(N388/Y389) which in all G2 models introduces loop conformations to residues N387 (blue) and K390 (purple) juxtaposed by the intervening deletion. (D) RMSD experienced by the protein backbone atoms relative to the starting structures during the simulation trajectory. Data were averaged from the 3 best chosen models of variant CC domains. The data suggest that G2 and G1 experience greater molecular motion than G0. (E-G) Overlays of the pre- (cyan) and post-simulation (smudge/(green)) representative backbone ribbon structures of APOL1 CC domains of common variant G0 (B), and disease risk variants G1 (C) and G2 (D). Structures were superposed by aligning backbone atoms (N, Cα, and C′) in PyMOL. Inset in (E) shows an enlarged view of the significant deviation of the N-terminal 7 aa from the remaining 17 aa constituting the principal axis of helix α1 in the G0 CC domain. Backbone sites of G1 missense polymorphisms are in red. G2 deletion site is bracketed in blue (residue 387) and purple (residue 390). Encircled regions emphasize additional inter-variant structural differences away from sites of aa sequence variation, see text for details.
Two-dimensional 1H-15N HSQC NMR spectroscopy
Following His6-tag cleavage, APOL1 CC domain samples prepared in NMR buffer (150 mM NaCl, 25 mM K phosphate, pH 7.0) exhibited poor stability with >70% loss over 24h-72h storage at 25 °C, 4 °C, -20 °C, or -80 °C. Serial 2D 1H-15N HSQC experiments (12 h acquisition) performed over a 3 day period revealed several crosspeaks of broadened linewidth with progressively decreased signal intensities. Sample stability was not improved by addition of 50 mM each of arginine and glutamic acid. APOL1 CC domains with uncleaved N-terminal His6-tags also showed progressive precipitation in NMR buffer, with 45% protein loss over 72h at 4 °C. However, these His6-tagged samples of APOL1 CC G0 and G1 exhibited patterns of well-dispersed crosspeaks as observed in 2D 1H-15N HSQC spectra (Fig. 2C). The numbers of crosspeaks resolved in these spectra match the total number of residues in each construct. An overlay of the 2D 1H-15N HSQC spectra of APOL1 CC domains G0 and G1 (Fig. 2C) reveals subtle differences in protein conformation. In agreement with the CD results, several broadened crosspeaks in the G1 central spectral region suggest reduced conformational stability compared to that of G0. Several newly emerged, conformation-specific 1H-15N correlation crosspeaks are observed only in spectra of G0 (boxed) or only in spectra of G1 (encircled), reflecting polymorphism-induced conformational differences.
Molecular modeling
The three best modeled structures of the CC domains of G0, G1, and G2 showed good stereochemical quality with 100% of residues occupying favored regions in the Ramachandran map (not shown). Figure 3(A-C) presents the best of the three modeled structures for APOL1 CC domain variants G0, G1, and G2 modeled on the LDLR-binding helical hairpin domain (aa 53-126) of apolipoprotein E3 (RCSB ID 1NFN; see Methods). The structural fold of the G0 CC domain models (aa 336-398) comprises two α-helices, α1 (aa 338-361) and α2 (aa 365-397), packed as an anti-parallel helical hairpin (Fig. 3A). Residues Asp337, Gly362, Ala363, and Lys364 (in addition to residues at N- and C-termini) adopt non-helical loop conformations. The APOL1 CC structural models of G1 and G2 exhibit secondary structured elements of similar length, as viewed in MOLMOL and PyMOL. All CC models of G1 and G2 reveal a helix α1 (aa 338-362) extended by one C-terminal residue as compared to the modeled G0 CC (Figs. 3B-C). The extent of helix α2 (aa 365-397) is uniform in G0, G1 and G2 (Figs. 3). In all G2 models the N388/Y389 deletion induces a helix-to-loop transition for the immediately adjacent residues Asn387 and Lys390 (conjoined only in G2) (Fig. 3C).
Superposition of the three best modeled G1 CC structures onto the modeled G0 backbone atoms yields RMSD values of 0.78-0.86 Å, consistent with preservation of the structural fold and secondary structure packing. However, superposition of the modeled G2 CC structure onto the modeled G0 backbone atoms resulted in higher values of RMSD of 1.77-2.14 Å. We next applied molecular dynamics (MD) simulation to analyze amino acids critical to eliciting local and global conformational changes driven by primary sequence variation of APOL1 CC domain variants G0, G1, and G2.
MD simulation of modeled APOL1 variant CC structures
Trajectory analysis
The narrow distribution of 3-model average values of radius of gyration (Rg) for CC G0, G1, and G2 suggests overall stability with persistence of conformational fold for the full duration of MD simulation. The modeled G0 polypeptide adopts a marginally more stable conformation than G1 which, in turn, appears more stable than G2, corroborating the RMSD, atomic fluctuation, and other simulation parameters governing conformational changes observed in localized CC regions as discussed below.
Figure 3D compares average RMSD evolution for the three best CC domain models of APOL1 G0, G1, and G2. The RMSDavg values vary across the simulation trajectory between 0.07-0.32 nm for G0 (mean 0.26 nm), 0.09-0.38 nm for G1 (mean 0.32 nm), and 0.09-0.43 nm for G2 (mean 0.41 nm). RMSDavg values progressively increase for each APOL1 CC variant during the initial 16 ns of simulation, likely reflecting conformational rearrangements adjusting structural restraints and non-covalent intramolecular interactions. RMSD variation during the following 84 ns is modest for G0 and G1, in contrast to the higher variability and magnitude of G2 RMSD throughout the simulation. These results are consistent with relatively lower conformational stability of G2 than of G0 and G1, an observation in agreement with Rg data (not shown).
The structures of G0, G1, and G2 following 100 ns of MD simulations (post-MD) are shown in Figure 4(B-D). The post-MD conformations exhibit substantial differences in anti-parallel inter-helical packing as compared to pre-MD conformations, with structural rearrangements evident near the N-terminus of helix α1 in both G1 and G2, and bracketing the deletion site in helix α2 in G2. Overlay of best model CC domain backbone atom ribbon structures pre- (0 ns) and post-simulation (100 ns) shows pre-post RMSD differences of 2.92 Å for G0, 3.75 Å for G1, and 4.90 Å for G2 (Fig. 3E-F), suggesting greater conformational changes in G1 and G2 than in G0 structures. G1 and G2 also exhibit larger deviations than G0 at and near the open-end termini of helices α1 and α2 (Figs. 3 & 4).
Figure 4.
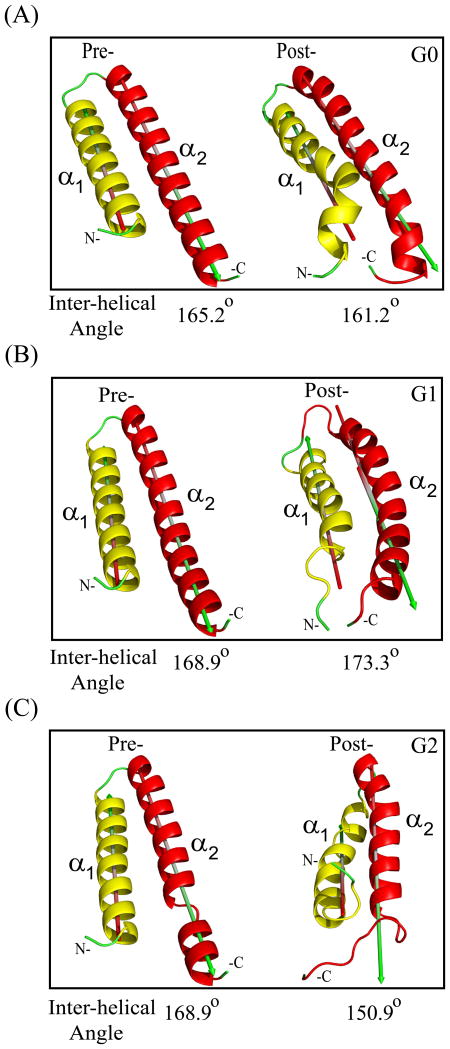
α1-α2 inter-helical angles of APOL1 C-terminal CC domain helical hairpin structures of variants G0 (A), G1 (B) and G2 (C) in pre- and post-simulation conformations (angles between major axes of helices α1 and α2 determined in PyMOL; 180° describes perfectly anti-parallel helices, and 0° describes perfectly parallel helices).
Loss of helicity in pathogenic variants
Post-MD structures revealed 12 CC domain residues in G1 that underwent helix-to-loop transition, including V338-G342, E361, and G362 in helix α1, and S365-T367, Q396, and E397 in helix α2. Similar helix-to-loop transitions occurred in 17 residues of the G2 CC domain, including V338-V341, E355, E361, and G362 in helix α1, and S365, N387, K390-E397 in helix α2, contributing to increased segmental flexibility at near-terminal regions of G2. These data suggest that G1 and G2 exhibit decreased conformational stability in solution, compared to G0.
Evolution of inter-helical angles
In the pre-MD simulation structure of G0, the first 7 residues (aa 338-344) of helix α1 deviated 5.9° from the principal axis of the remaining α1 residues (aa 345-361), oriented away from helix α2. In the post-MD structure, this deviation increased substantially to 55.5° from the principal axis of the remaining α1 residues (aa 345-361), away from helix α2 (Fig. 3E, Inset). This segment in the post-MD G1 structure deviated 31.5° from helix α2 (oriented as in G0). In contrast, in the post-MD G2 structure this initial α1 helical segment deviated 45.2° towards helix α2. Most of these 7 residues in G1 and G2, (but not G0) modeled as loop rather than α helix. Conformational deviation of the G2 C-terminal 9 aa of helix α2 (encompassing ΔN388/Y389) was larger (5.51 Å) than in G0 (2.29 Å) or G1 (2.53 Å). Similarly, pre-to-post MD conformational deviations of the 9 N-terminal α1 helix residues were 3.62 Å in G0, 4.80 Å in G1 and 5.68 Å in G2, again suggesting larger conformational changes in G1 and G2 than in G0.
The post-MD α1- α2 inter-helical angle of G0 increased by 4° (Fig. 4) likely reflecting increased deviation from helix α2 of the N-terminal 7 residue segment of helix α1 (Fig. 3E). The post-MD G1 α1- α2 inter-helical angle decreased by 4.4°, reflecting closer packing of terminal regions involving inclination of helix α2 towards helix α1 and inter-helical loop flexibility (Fig. 3F & 4B). The remarkable 18° post-simulation increase in the G2 α1-α2 inter-helical angle likely reflected summed localized helix-to-coil transitions (Fig. 4C). Thus, the inter-helical angle approximation in pathogenic variants G1 and G2 parallels the observed heterogeneity in their structural conformations.
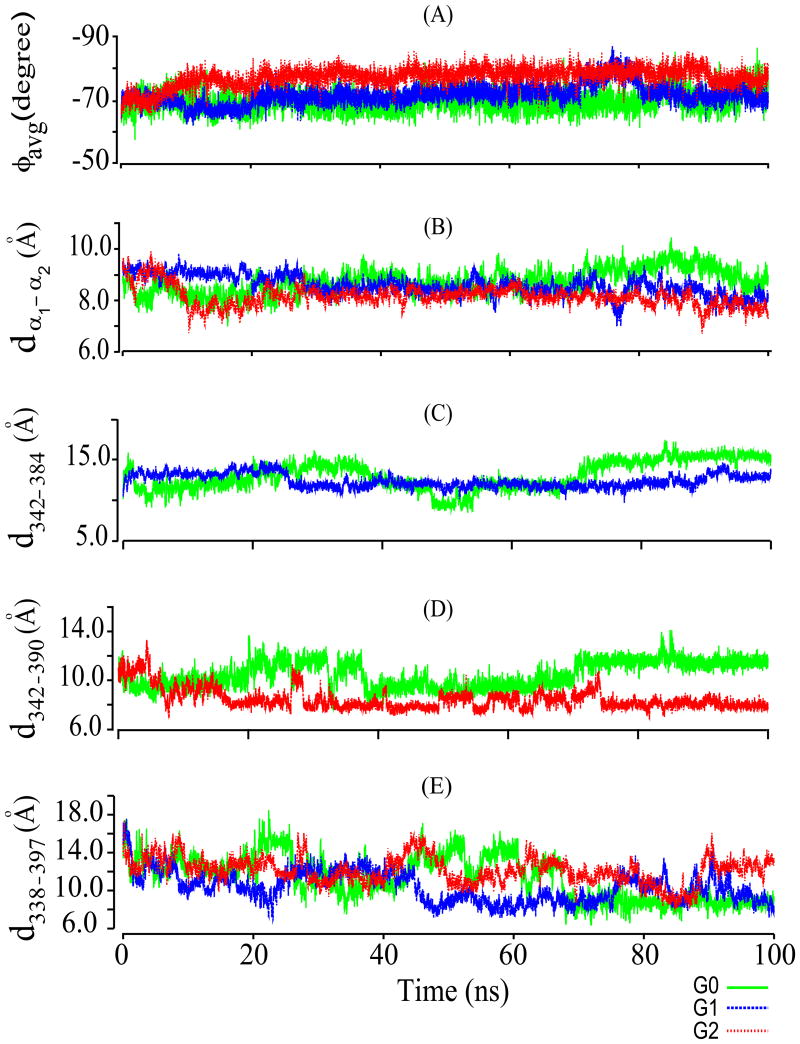
Time evolution of average ϕ angle for individual residues of CC domain helices α1 (Fig. 5A) and α2 (data not shown), varied within a narrow range for both helices of G0 (-69°for α1, -70° for α2) and G1 (-71° for α1 and α2), with larger ϕ angle variation in G2 (-77° for α1, -74° for α2). These increased average ϕ angles are consistent with helix-to-loop transitions in select residues that are more prominent in G2 than G1 (see above).
Figure 5.
(A) Time-dependent variation of average ϕ angle (ϕavg) of helix α1 (aa 338–362) of CC domains from APOL1 variants G0, G1, & G2 during the simulation. (B) Simulation time-dependent distance between Cα atoms of helix α1 and helix α2 (dα1-α2) in CC variants G0, G1, & G2. (C) Simulation time-dependent distance between Cα atoms of residue 342 (S in G0, G in G1) and residue 384 (I in G0, M in G1; d342-384) in CC variants G0 and G1. (D) Simulation time-dependent distance between Cα atoms of S342 and K390 (d342-390) in CC variants G0 and G2. Color scheme is below the figure. (E) Simulation time-dependent distance between Cα atoms V338 (first residue of helix α1) and E397 (last residue of helix α2; d338-397) in CC domain variants G0, G1, & G2. These data suggest that the CC domains of G1 and G2 have less compact structures with greater flexibility than that of G0. Color keys are at the bottom right.
RMSD time evolution of Cα atoms in APOL1 CC domain helices α1 and α2 revealed average RMSD values for G0 of 1.7 and 1.9 Å, for G1 of 2.5 and 2.0 Å, and for G2 of 2.3 and 3.1 Å (data not shown). Pre-post simulation RMSD differences for helix α1 Cα atoms were 2.2 Å for G0, 2.7 Å for G1, and 2.6 Å for G2. Those for helix α2 Cα atoms were 2.2 Å for G0, 2.9 Å for G2, and 2.8 Å for G2, suggesting greater conformational change in G1 and G2 CC domains during MD simulation than in G0. Helix α1 RMSD is highest in the G1 CC domain, whereas RMSD for helix α2 is highest in G2. Helix-specific RMSD variation across the simulation period follows similar trends in both helices of G0 and G1, with variation lowest in G0 and highest in G2, perhaps reflecting conformational readjustments accompanying α1-α2 inter-helical packing.
These inter-helical orientation changes, manifest as helical approximation and repacking, were reflected in altered inter-helical Cα-Cα distances (dCα-Cα) throughout the simulation trajectory (Fig. 5B).The dCα-Cα alterations between residues 342 and 384 in G0 and G1 (Fig. 5C) and between residue positions 342-390 in G0 and G2 (Fig. 5D) were consistent with the CC domain risk variant polymorphism sites. Inter-helical approximation also changed dCα-Cα between V338 (first α1 residue) and Q398 (last α2 residue) in G1 and G2 to a greater degree than in G0 (Fig. 5E).
Residue-wise MD properties
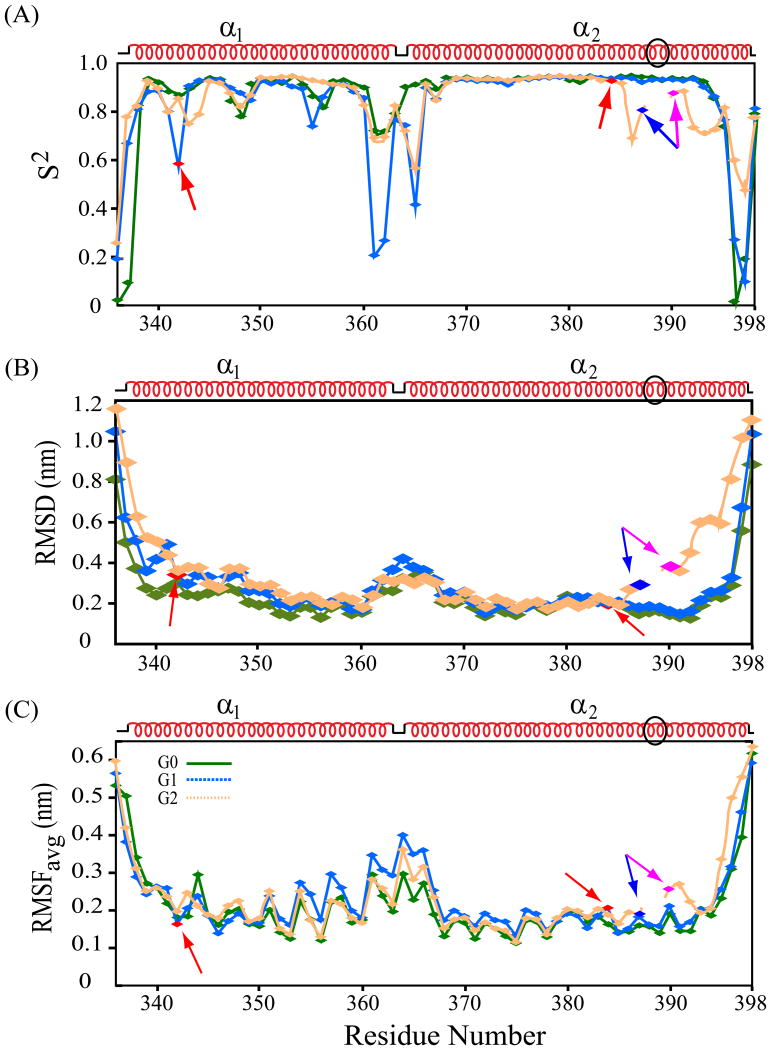
Residue-wise properties of dihedral order parameters (S2), average RMSD of backbone atoms (Cα, C′, and N), and RMSFavg were deduced from simulation trajectories of the 3 best models (Fig. 6). Similar S2 variation patterns across all residues for G0 (0.85), G1 (0.81) and G2 (0.82) (Fig. 6A) suggested modest decreases in overall conformational rigidity in G1 and G2 vs. G0. The average S2 value of 0.80 for G1 helix α1 was lower than for α1 in G0 or G2 (0.89 for each), suggesting locally decreased rigidity within helix α1 of G1 as compared to G0 or G2. Conversely, the average S2 value for G2 helix α2 of 0.83 was lower than for α2 in G0 (0.86) or G1 (0.85), such that decreased rigidity in G1 and G2 arises in each case from that helix harboring the pathogenic mutation (S342G in isolation suffices to phenocopy G1's predisposition to kidney disease [18] [4]. The low S2 value for G1 G342 (0.57), in particular, contrasts with those of S342 in G0 (0.87) and G2 (0.85), suggesting higher magnitude atomic motions of G342 in G1 compared to S342 in G0 and G2. Deletion of N388/389 in G2 induced an apparently more flexible environment for adjacent residues N386-A394 than in the same region of G0 and G1 (Fig. 6A).
Figure 6.
Post-simulation structural properties. (A) dihedral order parameter (S2), (B) average backbone RMSD (nm), and (C) RMSFavg (nm) of all protein atoms for each residue of APOL1 CC variants G0 (green), G1 (blue), and G2 (orange) superimposed in each panel. Each dataset was averaged from the 3 best selected models of each CC variant. G1 missense variant residues (S342G and I384M) are in red. G2 deletion site (ΔN388/Y389) flanked by blue (residue 387) and purple (residue 390). In (C) higher RMSF values indicate larger atomic motions. Schematic of pre-simulation secondary structure is above each panel. Black oval in secondary structure indicates residues deleted in G2. Lower S2, higher RMSD, & higher RMSF in select regions of risk variants G1 & G2 suggest their weaker conformational stability in solution than that of G0 variant.
Overall average per-residue RMSD values were 2.4 Å (G0), 2.8 Å (G1), and 3.4 Å (G2) (Fig. 6B). Backbone atom RMSDs for helix α1 residues only were 2.2 Å (G0), 2.7 Å (G1), and 3.0 Å (G2), and for helix α2 only were 2.1 Å, (G0) 2.2 Å (G1), and 3.1 Å (G2). The larger deviations of G1 and G2 CC domains from pre-MD structures suggest larger conformational rearrangements in G1 and G2 than in G0. Individual residue RMSD values for G0 helix α1 were near average value (except for 2 terminal residues), whereas all helix α2 residues exhibited below-average RMSDs. In contrast, the 4 initial residues of helix α1 in G1 and G2 exhibited high RMSDs of 4.3 Å and 5.1 Å (Fig. 6B). G1 residues G362-T376 had higher-than-average RMSDs, in contrast to average values in G0 and G2, and consistent with low S2 values of the G1 inter-helical loop (Fig. 6A). The 7 C-terminal residues K390-Q396 beyond the G2 deletion site exhibited above-average RMSD (5.3 Å), in contrast to below-average values in the same regions of G0 and G1 (Q396 excepted). G1 residues 341-343 (including S342G) showed markedly increased RMSD, in contrast to the same regions in G0 and G2. G1 and G2 residues adjacent to I384 exhibited below-average RMSD, in contrast to G0. These results together demonstrate specific conformational changes in the APOL1 CC domain associated with disease risk missense variants G2 and (primarily induced through mutation S342G) G1.
RMSFavg ensemble values for all protein atoms are shown as averages of the 3 best models (Fig. 6C). Excluding loop residues T336 and D337 and Leu398 at the termini (with high values), overall RMSFavg was 1.9 Å for G0, 2.0 Å for G1, and 2.1 Å for G2. Several structured segments showed above-overall RMSFavg values. The G2 residues distal to the ΔN388/Y389 deletion site, K390-L398, exhibited (with the exception of Q393) higher-than-overall RMSFavg (Fig. 6C), in contrast to below-average values for corresponding residues in G0 and G1. G1 residues adjacent to pathogenic variant G342 showed elevated RMSFavg values absent in corresponding positions of G0 and G2, whereas residues near I384M exhibited below-average RMSFavg in all three CC domain variants. Inter-helical loop residues E361-T367 exhibited RMSFavg higher in G1 than in G0 and G2. G1 helix α1 residues Y351, Y354, E355 and K357 showed RMSFavg values markedly higher-than-overall RMSFavg for helix α1. RMSFavg for residues bracketing ΔN388/Y389 were larger than in G0 and G1. These RMSD data together suggest greater helix α1 atomic motion in the CC domain of G1 than in G0 or G2, and greater helix α2 atomic motions in CC G2 (see above). Most residues of higher-than-average motional amplitude localized to the distal half of helix α1 and near the G2 deletion or the helix termini.
H-bonding networks in pathogenic variants
APOL1 CC domain variants also differed in H-bonding patterns. The backbone and hydroxyl group atoms (Hγ and Oγ) of S342 in helix α1 of the G0 and G2 CC domains made several intra-helical H-bonds, as well as H-bonds to nearby residues of helix α2. These include H-bonds between backbone HN and sidechain Hγ of S342 with backbone COs of V338 and of A339 in both pre- and post-MD structures, H-bonds between S342 Oγ and sidechain amide proton (Hε2) of Q396 in G0 and between S342 Oγ and sidechain Hε2 of Q393 and backbone HN of A394 in G2. All these H-bonds were lost in G1 due to the S342G substitution. Moreover, the G1 CC H-bonds that were present pre-simulation between HN of G342 and CO of V338 and of A339 and between CO of G342 to HN of V346 were absent post-simulation. A single G1 CC domain H-bond between G342 backbone CO and HN of L345 was present throughout the simulation trajectory. G342's loss of H-bonds to preceding residues V338 and A339 likely contributed to loss of G1 helical structure in the region immediately preceding G342. In contrast, G1 preserved the 4 H-bonds between residue 384 backbone atoms and atoms of neighboring residues, despite sidechain protrusion and increased solvent exposure of G2 variant M384.
Backbone (HN and CO) atoms of N388 and Y389 in helix α2 of CC G0 and G1 formed H-bonds with neighbor residues, and Y389 sidechain hydroxyl atoms formed multiple H-bonds with nearby α1 helix backbone CO atoms of residues A339 and V341. In addition, 5 intra-helical H-bonds connected sidechain OH atoms and sidechain HN of Q393 with sidechain HN and backbone CO of Q396. Loss of these G2 CC domain H-bonds likely contributed to the destabilization of downstream residues reported by the G2 dynamic simulation structures.
Overlay of the best models of APOL1 CC domain post-simulation backbone atom ribbon structures revealed RMSD of 3.47 Å between G0 and G1, 5.97 Å between G0 and G2, and 5.99 Å between G1 and G2 (Figure 7). The higher RMSDs of G1 and G2 structures attributable to the above conformational changes recapitulate the structural heterogeneity among these CC variants. Taken together, the trajectory analyses demonstrate that major conformational changes in APOL1 CC domains of both G1 and G2 occurred early in simulation and were generally maintained through the rest of the simulation period (with subsequent minor fluctuations as noted). These results suggest that our post-simulation structures of the APOL1 CC domain variants are valid approximations of their most heavily populated solution conformations.
Figure 7.
Overlays of representative post-simulation backbone ribbon structures of APOL1 CC domains of (A) G0 (smudge/(green)) on G1 (marine), (B) G0 (smudge) on G2 (light orange), and (C) G1 (marine) on G2 (light orange). Structures were superposed by aligning backbone atoms (N, Cα, and C′) in PyMOL. In A, G0 and G1 residues 336-398 were used for superimposition. In B, G0 residues 338-398 were superposed on G2 residues 336-398 (with deletion of residues 388 and 389). In C, G1 residues 338-398 were superposed on G2 residues 336-398 (Δ388/389). G1 missense variants are in red. Site of G2 deletion is bracketed in blue (residue 387) and purple (residue 390). Encircled regions emphasize inter-variant structural differences d342-390outside the terminal regions. See text for details.
Discussion
We have reported biophysical data and MD simulation results for the “common variant” APOL1 G0 CC domain and for the APOL1 CC domains from renal disease risk-variants G1 and G2. The APOL1 missense polymorphisms that define renal disease risk variants G1 and G2 [1] are located in the APOL1 CC domain The same missense variants confer protection against African sleeping sickness [1] through reduction or loss of binding to T. brucei rhodesiense SRA [4]. Thus, knowledge of the APOL1 CC domain structure and of structural changes introduced by disease risk variants G1 and G2, is important for our understanding of innate resistance to some forms of African trypanosomiasis, as well as for our understanding of APOL1-associated renal disease.
No APOL1 structure, including that of its CC domain, has been reported to date, as the recombinant holoprotein and its subdomains have proven unstable in concentrated solution. We found conditions allowing demonstration of conformational and thermal sensitivity differences in CD spectra of APOL1 CC domains G0, G1, and G2 at neutral pH (Fig 2A,B). We also demonstrated that CC domains G0 and G1 exhibit multiple chemical shift differences in their 1H-15N HSQC NMR spectra recorded at neutral pH (Fig. 2D). We demonstrated, in addition, that APOL1 CC G0 bound to GST-SRA better than G1, and much better than G2 (Fig. 2C) in the same neutral pH conditions as used for acquisition of NMR spectra. These SRA-binding properties at neutral pH 7.0 were qualitatively consistent with earlier measurements of surface plasmon resonance binding measurements at pH values ranging from 6.1 to 4.5, using mobile phase SRA and immobilized short biotinylated APOL1 CC variant peptides. These short APOL1 peptides encompassed only aa 365-398 [4] (helix α2 without helix α1), such that G1 peptide included variant I384M but lacked the pathologically more important missense variant S342G. Interestingly, Thomson et al found that the inhibitory effect of SRA aa 24-267 on lipid bilayer cation channel activity attributed to recombinant APOL1 G0 holoprotein was absent at cis pH 5.3 but maximal at cis pH 7.4, in which conditions it was inactive against G2 holoprotein [4]. The pH-dependence of fluid-phase GST-SRA binding to the complete anti-parallel coiled-coil domains of APOL1 variants G0, G1, and G2 will be interesting to compare and interpret in light of the solved NMR structures of these α1-α2 helical hairpin domains as they become available.
In the absence of defined CC domain structures, we evaluated and evolved previously reported APOL1 structural models. An early model of the SRA-binding APOL1 CC domain included C-terminal aa 340-392 [7, 8]. A more recent model included APOL1 aa 326-398, encompassing a helical hairpin comprising two antiparallel helices, each ∼30 residues in length [4, 12, 19]. Our current model of the APOL1 CC domain also predicts a helical hairpin with a 25 residue α1 helix and a 33 residue α2 helix in anti-parallel arrangement. Our MD simulations suggest, within the models' constraints, that the dynamic conformational perturbations exhibited in solution by CC domains of APOL1 risk variants G1 and G2 differ substantially from those of G0.
Helical segments in G1 and G2 experienced greater atomic motions and deviations than in G0, and underwent greater conformational perturbations throughout the simulation. Inter-helical plasticity in these models was reflected in variation of the α1-α2 inter-helical angle. The CC domain helices of G0 were well preserved during the simulation, whereas helical structure was lost in several residues of helix α1 adjacent to the G1 polymorphism S342G. Loss of helicity was also evident in G2 helix α2 near the two residue deletion site, and in select, nearby N-terminal residues of helix α1. These observations suggest that the APOL1 G0 CC domain exhibits a more compact structural fold in solution than do the CC domains of APOL1 risk variants G1 and G2. The increased conformational stability of G0 vs. G1 or G2 was supported by thermal denaturation CD data (Fig. 2B), by SRA-binding data (Fig. 2C), and by reduced intensity 1H-15N correlation crosspeaks and select crosspeak line-width broadening in the 2D 1H-15N HSQC spectrum of G1, compared to that of G0 (Fig. 2D). Structural differences in the simulation models of G0 and G1 could reflect differences in HSQC crosspeaks (Fig. 2D). A low expression yield of APOL1 G2 variant CC domain precluded collection of NMR data. However, MD simulation with the G2 CC domain predicted conformational changes greater than those of CC domains G0 and G1, as exhibited in overall and regional RMSDs, inter-helical angular approximation, and selected inter-residue distances. Several G2 residues underwent helix-to-coil transition, reflecting a lower conformational stability and structural compactness than in G0 and G1 CC domains.
Residue-wise analysis of α1 helix dihedral order parameters, RMSD, and RMSFavg demonstrated higher amplitude variations in select residues of G1 than in homologous residues of G2 or G0. These were attributable to stronger influence of the S342G substitution on nearby residues as well as in more distant regions in helix α1 (Fig. 5C-D). The S342 side chain in G0 and G2 faces the core structural region, and its hydroxyl atoms make H-bonds with adjacent intra- and inter-helical residues. The S342G substitution in G1 resulted in loss of these H-bonds, loss of helicity, and compromised conformational stability at and around the substitution site. This loss of inter-helical hydrogen bonds was compensated by reorganization of helix α2 through bending towards helix α1.
The G1 I384M substitution, in contrast, introduced no major conformational changes into the G1 structure. The longer hydrophobic Ile sidechain at residue 384 in G0 and G2 is oriented almost perpendicular to the bi-helical plane, with only partial exposure to the core structural region, and without important inter-helical spatial contacts. The I384M substitution in G1 maintained helix α2 hydrophobicity in both G0 and G2, with nearly identical hydrophobic side chain packing for Ile and Met, without major conformational rearrangement.
The G1 and G2 risk variants evolved as mutually exclusive haplotypes. It was initially unclear whether or not both G1 missense mutations (S342G and I384M in cis) were essential to confer risk of renal disease [1]. However, the APOL1 S342G substitution in isolation sufficed to reduce SRA interaction and to sustain trypanolytic activity in mice [4], whereas the I384M substitution in isolation had only modestly reduced SRA binding and completely lacked trypanolytic activity in mice [1, 4]. This difference may reflect the extensive disruption of the intra- and inter-helical H-bond network accompanying the S342G mutation pre- and post-MD simulation, in contrast to the relatively greater H-bond stability associated with the I384M substitution. Baboons and other apes with robust APOL1 trypanolytic activity express Gly at the position of residue 342.
The post-simulation structure of APOL1 G2 CC domain structure differed considerably from the modeled pre-simulation structure. The two residue deletion in G2 leads to remarkable structural changes not only near the α2 helix deletion site, but also in the nearby region of helix α1 (Fig. 5). The Y389 side chain in G0 and G1 is exposed to the protein core region, and its hydroxyl sidechain atoms make several intra- and inter-helical H-bonds (see Results). Loss of Tyr389 in G2 completely altered the H-bonding network at this position compared to those in G0 and G1, producing local conformational rearrangements in helix α2 and the adjacent region of helix α1, and weakening overall conformational stability compared to G0. This region of helix α2 is part of a conserved heptad repeat region of APOL1. Deletion of N388/Y389 in G2 helix α2 shifts K390 from heptad position “b” to “g”, bringing it into register with K388 of baboon APOL1, a critical residue for prevention of SRA binding to insure preservation of trypanolytic activity [4].
Our data from CD and NMR spectroscopy and MD simulation together suggest that the APOL1 CC domain conformations of trypanolytic variants G1 and G2 exhibit substantial and important differences from that of G0. The consequences of G1 and G2 sequence variation are propagated along the structural fold. The structural models presented here could provide insight into the mechanism of APOL1 function. CC domain helix α2 has been shown to suffice for SRA binding [4]. The results suggest that the interaction interface could involve, in addition to C-terminal aa 385-398 of helix α2, cooperative interaction with N-terminal aa 336-349 of helix α1 and aa 360-367 at the junction of helix α1 and the inter-helical loop region. Helical unwinding in the G1 CC domain near S342G, with changes adjacent inter-helical packing, likely contribute to decreased SRA binding by G1. The conformational plasticity predicted in the CC domains of APOL1 variants G1 and G2 likely contributes both to dominant resistance to trypanosomal SRA and to recessive susceptibility to later onset glomerular disease [1].
Materials and Methods
Molecular cloning of C-terminal APOL1 CC variants
APOL1 aa 304-398 has been predicted to fold as a coiled-coil. Based on APOL1 secondary structural coiled-coil predictions of PSIPRED [20], COILS [21], Paircoil2 [22], and MARCOIL [23], E. coli codon-optimized synthetic APOL1 cDNAs (Genscript, Piscataway, NJ) encoding APOL1 C-terminal polypeptides aa 339-398, 323-398, 313-398, and 304-398 were subcloned into pET-28a-c(+) (EMD Millipore, Billerica, MA) in-frame with an N-terminal His6-tag (thrombin cleavable, Amersham) using NheI and EcoRI, and with a C-terminal His6-tag using NcoI and XhoI (New England BioLabs, Beverly, MA). N-terminal His6-tags exhibited higher protein expression yields, and so were used for biophysical experiments. A GST fusion protein cDNA encoding SRA residues 24-267 (Uniprot Q8T309) was constructed in pGEX-6P1. Ligation products were transformed into E. coli BL21(DE3) cells (EMD), and DNA sequence integrity was confirmed. DNA quantitation was performed using a Nanodrop spectrophotometer (Thermo Scientific, Waltham, MA).
Protein expression and purification
E. coli cells harboring recombinant APOL1 CC variants were grown for protein expression in LB-kanamycin medium. At OD 0.6–0.8, G0 and G1 cells and GST-SRA cells were induced with 0.7 mM IPTG for 12 hrs at 23 °C. G2 cells were induced with 1.0 mM IPTG for 12 hrs at 23°C. Cells were centrifuged 6000 rpm for 10 min at 4 °C (GSA rotor, Sorvall RC 5B) and stored at -80°C until further use. Freeze–thawed cells were suspended on ice in buffer A (150 mM NaCl, 10 mM imidazole, 25 mM Tris-HCl, pH 8.0) containing Protease Inhibitor Cocktail tablets (Roche), 0.01% PMSF, and 10 μl / L culture of lysonase (Novagen), agitated 30 min at 4 °C, then subjected at 4°C to 7 sonication cycles of 7 s duration at 35% amplitude (Branson S-450D) with 1 min inter-cycle time. Sonicated lysates were centrifuged 17000 rpm for 45 min at 4 °C to separate supernatant and particulate inclusion body fractions. Recombinant G0, G1, and G2 CC polypeptides were purified from soluble fractions, quantitated, and tested for homogeneity by SDS–PAGE. Upon scale-up, yield of recombinant G2 polypeptide was insufficient for NMR analysis. Isotopic labeling was therefore limited to G0 and G1 constructs.
Uniformly 15N- or 13C/15N-enriched APOL1 G0 and G1 CC polypeptides were expressed in M9 minimal medium containing 15NH4Cl (1 g/l) and 13C glucose (2 g/l) as sole nitrogen and carbon sources, and 10% of uniformally 15N or 13C/15N labeled Bioexpress cell growth media (Cambridge Isotopes Laboratories, Tewksbury, MA), in a manner identical to that used for their unlabeled counterparts. Clarified supernatants were equilibrated with buffer A and loaded at 4° C onto Ni-NTA-Sepharose column (Amersham) pre-equilibrated with buffer A containing 1% Triton X-100, using a peristaltic Econo-pump (Bio-Rad). Protein-bound matrix was washed with > 25 column volumes (cv) of buffer B (500 mM NaCl, 40 mM imidazole, 25 mM Tris-HCl, pH 8.0). His6-Protein was eluted with > 6 cv of buffer C (150 mM NaCl, 500 mM imidazole, 25 mM Tris-HCl, pH 8.0).
Eluate fractions were evaluated by SDS-PAGE. APOL1 CC domain-containing fractions were pooled, dialyzed in buffer D (50 mM NaCl, 25 mM Tris-HCl, pH 8.0), and further purified by anion-exchange chromatography (Q-Sepharose fast flow matrix, GE Healthcare, Marlborough, MA). Purified polypeptides concentrated by Millipore stirred cell membranes or Amicon 3 kDa NMWL membrane centrifugal devices were quantitated by Bradford assay [24].
CD spectroscopy
Protein folding and intrinsic secondary structural characteristics of recombinant APOL1 CC polypeptides (purified by anion exchange or SEC in 150 mM NaCl, 25 mM K phosphate, pH 7.4, then clarified by centrifugation) were assessed by Far-UV (260–190 nm) CD measurements at 25°C on a Jasco-810 spectropolarimeter in a 1 mm path length quartz cuvette (Starna Cells, Atascadero, CA) with 1 nm bandwidth, 2 s response time, 20 nm/min scan speed, and 4 scans in continuous mode. Buffer-corrected spectra were recorded for 18 μM APOL1 CC domain. Data were converted into Molar Residue Ellipticity (MRE, θ×10-3) (deg cm2 dmol-1) as described [14]. Thermal stability of CC variants was measured by CD thermal denaturation experiments conducted at 220 nm (ellipticity minima) by heating the samples from 25 °C to 95 °C at a rate of 50 °C/hour, with 3 min thermal equilibration periods. Triplicate data were averaged for analysis.
MALDI-TOF MS
Polypeptide identity and homogeneity were confirmed by MALDI-TOF MS by determination of the m/z ratio on an AB/MDS Sciex 4800 Plus MALDI TOF/TOF Analyzer (Applied Biosystems, Carlsbad, CA) as previously described [14].
SRA-APOL1 (CC) Binding Experiments
GST pulldown experiments were conducted to test SRA binding potential of APOL1 CC variants G0, G1, and G2 in the conditions in which CD and NMR spectra were recorded. GST-SRA (aa 24-267) fusion protein was purified using Glutathione-Sepharose 4B beads (GE HealthCare) from the soluble fraction prepared from lysates of 500 mL cultures, as previously described [25]. Glutathione beads loaded with GST-SRA or with GST were extensively washed with phosphate-buffered saline, pH 7.4 then eluted with glutathione. Purity and homogeneity of GST-SRA as assessed by 16% (w/v) SDS-PAGE showed no protein bands below 25 kDa. Aliquots of Lysate supernatant aliquots containing equivalent amounts of APOL1 CC domain variants G0, G1, and G2 (Fig. 2C) were separately incubated at 4 °C with equal volumes of GST-SRA beads or GST beads for 12 hrs with rocking. The beads were then transferred into mini-columns and washed with 50 cv of 150 mM NaCl, 25 mM K phosphate, pH 7.0. Binding of APOL1 CC variants to GST-SRA (Fig. 2D) and to GST (Fig. 2E) was assessed by SDS-PAGE and quantitated using ImageJ (http://imagej.nih.gov/ij/).
Solution NMR spectroscopy
Two-dimensional 1H–15N HSQC NMR experiments [26] were performed at 25°C or 10°C on a Bruker Avance 600 MHz spectrometer equipped with a triple resonance (z-axis) pulsed-field gradient probe. NMR samples of 15N-enriched APOL1 CC domains (aa 339-398l; 0.3 mM G0 or 0.25 mM G1) were prepared in NMR buffer of 150 mM NaCl, 25 mM K phosphate, pH 7.0. All samples contained 10% (v/v) D2O, 1 mM DTT-d10, 0.25 mM DSS as internal standard, 0.05% (w/v) NaN3, and 1X protease inhibitor. 2D 1H–15N HSQC spectra were acquired using a 2048×256 data matrix with 128 scans, and a 12 h total acquisition time. Data were processed on an Intel PC workstation running OpenSuse11.1 using NMRPipe/NMRDraw [27]. Chemical shifts were referenced to the internal standard [28].
Molecular Modeling
Our initial secondary structure prediction for APOL1 G0 aa 304-398 by PsiPred [20] was subjected to subsequent three-dimensional structural prediction by Phyre2 [29], RaptorX [30, 31], I-Tasser [32], and Robetta [33]. Robetta identified as a potential template the LDLR-binding domain of apolipoprotein E3 (APOE3; PDB 1NFN), [34] (aa residue span 23-132), then built and aligned a comparative structural model for APOL1 G0 coiled-coil domain using HHSearch [35], RaptorX [30], and Sparks-X [36]. The resulting structural models identified a consensus region of aa 336-398 that exhibited preserved secondary structure in agreement with PsiPred. In contrast, predicted secondary structures within aa 304-335 were highly variable. Back-and-forth evaluation of structural models predicted by Robetta with those obtained from the other programs set criteria for selection of the best models of APOL1 G0 aa 304-398. The best Robetta model of high stereochemical quality was chosen as a final template for rebuilding homology models for APOL1 CC G0 encompassing aa 336-398 in Modeller v9.14 [37], while maintaining a modeling environment consistent with available structural information (PDB 1NFN). DOPE-based modeling was optimized to determine 50 structural models by satisfying spatial restraints of bond lengths, bond angles, and improper dihedral angles. A subset of models was subsequently refined through selection based on global low DOPE potentials, energy minimization to avoid steric clashes, and 500 ps MD simulation in Gromacs v5.0. The three models that best preserved secondary and tertiary structures and exhibited high structure stability, narrow oscillation window of RMS deviation, and good stereochemical quality were selected for further study.
These APOL1 G0 CC domain models served as templates for building structural models of the APOL1 G1 and G2 disease risk variant CC domains in Modeller [37]. The three best CC domain models of variants G1 and G2 were selected from 50 structural models generated for each using the same selection criteria as described above for G0. All structural models were energy-minimized in GROMACS v5.0 [38] using the steepest-descent method with 1,000 steps of step size 0.01 nm. Stereochemical quality of these models was judged based on low z-score, best (ϕ, Ψ) score in the Ramachandran map, and fewest atomic clashes as assessed by PROCHECK [39], PROSA [40], VERIFY3D [41], and PSVS server (http://psvs-1_5-dev.nesg.org/). Refined structures were subjected to molecular dynamics (MD) simulation study.
MD simulations of modeled APOL1 variant CC structures
100 ns MD simulations were performed for each of three best structural models selected for APOL1 CC domains of G0 and disease risk variants G1 and G2 with GROMACS v5.0 [38] using OPLS-AA/L all-atom force field [42] on either an 8-processor Linux PC running Intel Xeon or on a 12-processor linux PC on Orchestra clusters. Results of these single simulation runs were validated by comparing with those interpreted from separate trajectories of varied simulation times. Protein structures were passed pre-simulation to the “WHAT If” server (http://swift.cmbi.ru.nl/servers/html/index.html) for completion of missing side-chain atom(s). Protein molecules solvated explicitly with SPC water molecules were inserted in a periodic, cubical box of minimum distance 1.0 nm from center.
Standard ionization states of Lys, Arg, Glu, and Asp residues and the non-protonated state of Gln residues were used. Individual His residue protonation states were chosen based on hydrogen bonding patterns (on Nδ1, Nε1, or both/none) identified in the models. Structural inspection was performed by Coot v0.6.2 [43], H++ v3.1 [44], MOE[45], and SCWRL [46]. Protein termini were kept uncharged. Appropriate numbers of water molecules were replaced with Na+ and Cl- ions to achieve system electroneutrality at 150 mM NaCl. All structures were energy-minimized using a steepest-descent algorithm.
A 300 K reference temperature was set throughout the simulation using a Berendsen thermostat with a 0.1 ps time constant of 0.1 ps. Bond-lengths were constrained by the LINCS algorithm [47]. Before the production run, the system was equilibrated under position restraints with 250 ps NVT simulations followed by 250 ps NPT simulations in the presence of Perrinello-Rahman pressure-coupling. Time step was 2 fs, and electrostatic and van der Waals cutoffs were set to 1.0 nm. The PME algorithm [48] was used for long range electrostatic interactions.
Post-equilibration, full molecular dynamics simulations were executed for 100 ns, with coordinates written at 2 ps intervals. Simulation trajectories were analyzed by GROMACS routine utility and in-house scripts. Averaged data from 3 best models of each CC domain was visualized by xmgrace (http://plasma-gate.weizmann.ac.il/Grace/) and gnuplot v4.6 (http://www.gnuplot.info/). Backbone RMSD values (in Å) were calculated in MolMol [49]. Structure models were aligned and visualized in VMD [50] and PyMOL (Molecular Graphics System, v1.5.0.4 Schrodinger, LLC). Figures were prepared in PyMOL and gnuplot.
Acknowledgments
This work was supported by NIH grants MD007092 (to MP), MD007898 (to MP and SLA), and by the NephCure Foundation (to MP). We thank Drs. Marianne A Grant, Gabriel Birrane, and Simon Dillon (Beth Israel Deaconess Medical Center and Harvard Medical School) for helpful discussion. Access to Linux PCs on the Orchestra clusters was provided by the Harvard Medical School Computing Facility. The authors acknowledge Dr. Alan C. Rigby (Synta Pharmaceuticals Inc.) for additional computational support and Dr. James D. Baleja (Tufts University School of Medicine, Boston, MA) for providing access to CD instrument.
Abbreviations
- APOL1
apolipoprotein L1
- CC
coiled coil
- HSQC
heteronuclear single quantum correlation
- MD
molecular dynamics
- SRA
Serum Response-Associated
- RMSD
root mean square deviation
- RMSF
root mean square fluctuation
Footnotes
Conflicts of Interest: MRP and DJF have ownership interest in ApoloBio LLC.
Author Contributions: AKS and SLA conceived and designed the research with input from MRP and DJF. AKS performed experiments, spectroscopy, and simulations. AKS and SLA analyzed and interpreted results. AKS drafted the manuscript. AKS, SLA, DJF, and MRP revised and edited the manuscript.
Author contributions: All authors conceived and designed the experiments., DG, AP, JPB, LB, EC, BDH, AR performed the, experiments. All authors analyzed the data and contributed reagents/materials/analysis tools. DG, AP, DE, MN, GS, MB, CV wrote the paper.
References
- 1.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science (New York, NY. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weckerle A, Snipes JA, Cheng D, Gebre AK, Reisz JA, Murea M, Shelness GS, Hawkins GA, Furdui CM, Freedman BI, Parks JS, Ma L. Characterization of circulating APOL1 protein complexes in African Americans. Journal of lipid research. 2016;57:120–130. doi: 10.1194/jlr.M063453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomson R, Molina-Portela P, Mott H, Carrington M, Raper J. Hydrodynamic gene delivery of baboon trypanosome lytic factor eliminates both animal and human-infective African trypanosomes. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19509–19514. doi: 10.1073/pnas.0905669106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomson R, Genovese G, Canon C, Kovacsics D, Higgins MK, Carrington M, Winkler CA, Kopp J, Rotimi C, Adeyemo A, Doumatey A, Ayodo G, Alper SL, Pollak MR, Friedman DJ, Raper J. Evolution of the primate trypanolytic factor APOL1. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E2130–2139. doi: 10.1073/pnas.1400699111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heneghan JF, Vandorpe DH, Shmukler BE, Giovinnazo JA, Raper J, Friedman DJ, Pollak MR, Alper SL. BH3 domain-independent apolipoprotein L1 toxicity rescued by BCL2 prosurvival proteins. American journal of physiology. 2015;309:C332–347. doi: 10.1152/ajpcell.00142.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Styer AL, Hajduk SL. Trypanosome Lytic Factor-1 Initiates Oxidation-Stimulated Osmotic Lysis of Trypanosoma brucei brucei. The Journal of biological chemistry. 2015 doi: 10.1074/jbc.M115.680371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pays EVB, Vanhamme L, Paturiaux-Hanocq F, Nolan DP, Perez-Morga D. The trypanolytic factor of human serum. Nature reviews. 2006;4:477–486. doi: 10.1038/nrmicro1428. [DOI] [PubMed] [Google Scholar]
- 8.Vanhamme LP-HF, Poelvoorde P, Nolan DP, Lins L, Van Den Abbeele J, Pays A, Tebabi P, Van Xong H, Jacquet A, Moguilevsky N, Dieu M, Kane JP, De Baetselier P, Brasseur R, Pays E. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature. 2003;422:83–87. doi: 10.1038/nature01461. [DOI] [PubMed] [Google Scholar]
- 9.Lecordier L, Vanhollebeke B, Poelvoorde P, Tebabi P, Paturiaux-Hanocq F, Andris F, Lins L, Pays E. C-terminal mutants of apolipoprotein L-I efficiently kill both Trypanosoma brucei brucei and Trypanosoma brucei rhodesiense. PLoS pathogens. 2009;5:e1000685. doi: 10.1371/journal.ppat.1000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollak MR, Genovese G, Friedman DJ. APOL1 and kidney disease. Current opinion in nephrology and hypertension. 2012;21:179–182. doi: 10.1097/MNH.0b013e32835012ab. [DOI] [PubMed] [Google Scholar]
- 11.Mukamal KJ, Tremaglio J, Friedman DJ, Ix JH, Kuller LH, Tracy RP, Pollak MR. APOL1 Genotype, Kidney and Cardiovascular Disease, and Death in Older Adults. Arteriosclerosis, thrombosis, and vascular biology. 2015 doi: 10.1161/ATVBAHA.115.305970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pays E, Vanhollebeke B, Uzureau P, Lecordier L, Perez-Morga D. The molecular arms race between African trypanosomes and humans. Nat Rev Microbiol. 2014;12:575–584. doi: 10.1038/nrmicro3298. [DOI] [PubMed] [Google Scholar]
- 13.Rajamohan F, Nelms L, Joslin DL, Lu B, Reagan WJ, Lawton M. cDNA cloning, expression, purification, distribution, and characterization of biologically active canine alanine aminotransferase-1. Protein expression and purification. 2006;48:81–89. doi: 10.1016/j.pep.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Sharma AK, Sawhney P, Memisoglu G, Rigby AC. Expression, purification, and characterization of coiled coil and leucine zipper domains of C-terminal myosin binding subunit of myosin phosphatase for solution NMR studies. Protein expression and purification. 2012;81:126–135. doi: 10.1016/j.pep.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Ishigaki T, Ohki I, Utsunomiya-Tate N, Tate SI. Chimeric structural stabilities in the coiled-coil structure of the NECK domain in human lectin-like oxidized low-density lipoprotein receptor 1 (LOX-1) Journal of biochemistry. 2007;141:855–866. doi: 10.1093/jb/mvm093. [DOI] [PubMed] [Google Scholar]
- 16.Zhou NE, Kay CM, Hodges RS. Synthetic model proteins. Positional effects of interchain hydrophobic interactions on stability of two-stranded alpha-helical coiled-coils. The Journal of biological chemistry. 1992;267:2664–2670. [PubMed] [Google Scholar]
- 17.Cooper TM, Woody RW. The effect of conformation on the CD of interacting helices: a theoretical study of tropomyosin. Biopolymers. 1990;30:657–676. doi: 10.1002/bip.360300703. [DOI] [PubMed] [Google Scholar]
- 18.Kopp JB, Nelson GW, Sampath K, Johnson RC, Genovese G, An P, Friedman D, Briggs W, Dart R, Korbet S, Mokrzycki MH, Kimmel PL, Limou S, Ahuja TS, Berns JS, Fryc J, Simon EE, Smith MC, Trachtman H, Michel DM, Schelling JR, Vlahov D, Pollak M, Winkler CA. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol. 2011;22:2129–2137. doi: 10.1681/ASN.2011040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosset STS, Behar DM, Wasser WG, Skorecki K. The population genetics of chronic kidney disease: insights from the MYH9–APOL1 locus. Nat Rev Nephrol. 2011;7:313–326. doi: 10.1038/nrneph.2011.52. [DOI] [PubMed] [Google Scholar]
- 20.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic acids research. 2013;41:W349–357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science (New York, NY. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 22.McDonnell AV, Jiang T, Keating AE, Berger B. Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics. 2006;22:356–358. doi: 10.1093/bioinformatics/bti797. [DOI] [PubMed] [Google Scholar]
- 23.Delorenzi M, Speed T. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics. 2002;18:617–625. doi: 10.1093/bioinformatics/18.4.617. [DOI] [PubMed] [Google Scholar]
- 24.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 25.Sharma AK, Zhou GP, Kupferman J, Surks HK, Christensen EN, Chou JJ, Mendelsohn ME, Rigby AC. Probing the interaction between the coiled coil leucine zipper of cGMP-dependent protein kinase Ialpha and the C terminus of the myosin binding subunit of the myosin light chain phosphatase. The Journal of biological chemistry. 2008;283:32860–32869. doi: 10.1074/jbc.M804916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mori S, A C, Johnson MO, van Zijl PC. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J Magn Reson B. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 27.Delaglio F, G S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 28.Wishart DS, B CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, JL Markley, S BD. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 29.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nature protocols. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morten Källberg HW, Sheng Wang, Jian Peng, Zhiyong Wang, Hui Lu, Jinbo Xu. Template-based protein structure modeling using the RaptorX web server. Nature protocols. 2012;7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J, Pa J. RaptorX: exploiting structure information for protein alignment by statistical inference. PROTEINS 79 Suppl. 2011;10:161–171. doi: 10.1002/prot.23175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Y R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: Protein structure and function prediction. Nature Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song YDF, Wang RY, Kim D, Miles C, Brunette T, Thompson J, Baker D. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong LM, Parkin S, Trakhanov SD, Rupp B, Simmons T, Arnold KS, Newhouse YM, Innerarity TL, Weisgraber KH. Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia. Nature structural biology. 1996;3:718–722. doi: 10.1038/nsb0896-718. [DOI] [PubMed] [Google Scholar]
- 35.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 36.Yuedong Yang EF, Zhao Huiying, Zhou Yaoqi. Improving protein fold recognition and template-based modeling by employing probabilistic-based matching between predicted one-dimensional structural properties of the query and corresponding native properties of templates. Bioinformatics. 2011;27:2076–2082. doi: 10.1093/bioinformatics/btr350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 38.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski RA, M MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 40.Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic acids research. 2007;35:W407–410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowie JU, Luthy R, Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science (New York, NY. 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 42.Kaminski GA, F RA, Tirado-Rives J, Jorgensen WL. Evaluation and Reparametrization of the OPLS-AA Force Field for Proteins via Comparison with Accurate Quantum Chemical Calculations on Peptides. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]
- 43.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anandakrishnan R, Aguilar B, Onufriev AV. H++ 3.0: automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic acids research. 2012;40:W537–541. doi: 10.1093/nar/gks375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Molecular Operating Environment (MOE): Chemical Computing Group Inc Sherbooke St. West,Suite #910, Montreal, QC, Canada, H3A 2R7. 2013 [Google Scholar]
- 46.Krivov GG, Shapovalov MV, Dunbrack RL., Jr Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 2009;77:778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hess Berk, B H, Berendsen Herman JC, Fraaije Johannes GEM. LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry. 1997;18:1463–1472. [Google Scholar]
- 48.Essmann Ulrich, P L, Berkowitz Max L, Darden Tom, Lee Hsing, Pedersen Lee G. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 49.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 50.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]