Abstract
The ribosome has historically been considered to have no cell-specific function but rather serve in a “housekeeping” capacity. This view is being challenged by evidence showing that heterogeneity in the protein composition of the ribosome can lead to the functional specialization of the ribosome. Expression profiling of different tissues revealed that ribosomal protein large 3-like (Rpl3l) is exclusively expressed in striated muscle. In response to a hypertrophic stimulus, Rpl3l expression in skeletal muscle was significantly decreased by 82% whereas expression of the ubiquitous paralog Rpl3 was significantly increased by ~5-fold. Based on these findings, we developed the hypothesis that Rpl3l functions as a negative regulator of muscle growth. To test this hypothesis, we used the Tet-On system to express Rpl3l in myoblasts during myotube formation. In support of our hypothesis, RPL3L expression significantly impaired myotube growth as assessed by myotube diameter (−23%) and protein content (−14%). Further analysis showed that the basis of this impairment was caused by a significant decrease in myoblast fusion as the fusion index was significantly lower (−17%) with RPL3L expression. These findings are the first evidence to support the novel concept of ribosome specialization in skeletal muscle and its role in the regulation of skeletal muscle growth.
Keywords: ribosome, ribosome specialization, hypertrophy
INTRODUCTION
The ribosome is a supramolecular ribonucleoprotein complex that is comprised of two subunits: the small 40S subunit which is composed of 18S ribosomal RNA (rRNA) and 32 ribosomal proteins (RPs) and the large 60S subunit, which consists of 5S, 5.8S, 28S rRNAs and 47 RPs. As the sole molecular machine responsible for translating mRNA into protein, ribosome function, not surprisingly, is intimately linked to cell growth, proliferation and homeostasis (Grummt 2013, Campbell and White 2014).
The ribosome has historically been viewed as functioning in a constitutive manner, decoding ribonucleotide sequences into amino acid sequences without any real regulative properties. Contrary to this perspective, Mauro and Edelman proposed the ribosome filter hypothesis which states that through specialization, the ribosome can preferentially translates different subsets of mRNAs (Mauro and Edelman 2002). The specialization of ribosome function is thought to occur through differences in ribosomal protein composition, post-translational modification of ribosomal proteins and alternative forms of rRNA and their post-transcriptional modification (Xue and Barna 2012, Sauert, Temmel et al. 2014). Based on their investigation into the potential functional differences of ribosomal protein paralogs, Komili and colleagues put forth the concept of the “ribosome code”, drawing parallels to the histone code (Komili, Farny et al. 2007). The ribosome code posits that functionally distinct classes of ribosomes are generated through the combined effect of the different forms of specialization, providing a new level of gene regulation through the selective translation of target mRNAs (Komili, Farny et al. 2007). Importantly, a study by Kondrashov and coworkers extended the concept of ribosome specialization to mammals, demonstrating that the tissue-specific expression of Rpl38 during mouse embryonic development was required for proper patterning via translation of specific Hox transcripts (Kondrashov, Pusic et al. 2011).
One way in which specialization of the ribosome is thought to occur is through changes in the protein composition of the ribosome by paralog substitution (Xue and Barna 2012). During the course of characterizing the polycystic kidney disease type I gene region, Burn and colleagues identified a gene with 74% sequence identity to the Rpl3 (ribosomal protein large 3) gene which was designated Rpl3-like (Rpl3l) (Burn, Connors et al. 1996). In a follow-up study, they unexpectedly discovered that Rpl3l expression, in contrast to the ubiquitous expression of Rpl3, was restricted to skeletal muscle and the heart (Van Raay, Connors et al. 1996). Studies on genome evolution, and the development of gene families, revealed that, unlike almost every other ribosomal protein gene duplicate, Rpl3l arose as the result of a DNA-mediated duplication event (Jun, Ryvkin et al. 2009). The muscle-specific expression of Rpl3l is consistent with the finding that genes derived by such a duplication mechanism are often subject to more complex regulation because upstream regulatory sequences tend to be copied along with the protein coding region (Dharia, Obla et al. 2014). A recent analysis of human RNA-seq data by Gupta and Warner confirmed the muscle-specific expression of Rpl3l and further showed Rpl3l to be one of only a few ribosomal proteins with tissue-specific expression (Gupta and Warner 2014). Collectively, the findings from these studies highlight just how unique Rpl3l is among ribosomal proteins and raise the intriguing question as to why skeletal muscle has evolved its own version of the Rpl3 protein.
A hint as to what Rpl3l might be doing in skeletal muscle came from our transcriptome analysis of skeletal muscle hypertrophy (Chaillou, Lee et al. 2013). We found that Rpl3l expression was dramatically down-regulated in response to a hypertrophic stimulus while Rpl3 expression was concomitantly up-regulated, suggesting Rpl3l has a role in regulating skeletal muscle mass. The purpose of this study was to test the hypothesis that Rpl3l functions as a negative regulator of skeletal muscle growth. To test this hypothesis, we generated a Rpl3l stable clone in C2C12 myogenic cells employing a Tet-On system to allow for the inducible expression of RPL3L protein during myogenic differentiation. In agreement with our hypothesis, we found that expression of RPL3L caused a significant decrease in myotube diameter that was associated with reduced myoblast fusion. While our results do not exclude an extra-ribosomal function for Rpl3l, they are consistent with the idea that Rpl3l alters ribosome function in a yet-to-be determined way such that myoblast fusion was significantly decreased. Moreover, the findings of this study represent the first evidence to support the novel concept of ribosome specialization in skeletal muscle and its role in the regulation of skeletal muscle growth.
MATERIALS AND METHODS
Animal care and use
All procedures involving the use of animals were approved by the University of Kentucky Institutional Animal Care and Use Committee. Male C57BL/6J mice (The Jackson Laboratory, Bar Harbor, ME), 5 months of age were housed in a temperature-and humidity-controlled facility on a 14:10 h light:dark cycle with access to food and water ad libitum.
Materials
The following reagents were used in the experiments described below: C2C12 myoblasts (ATCC; Manassas, VA, USA); high-glucose DMEM, fetal bovine serum, horse serum, Lipofectamine 2000, G418, protein G Dynabeads, Taqman probe sets (Rpl3, Mm02342628_g1; Rpl3l, Mm01299911_g1; Gapdh, Mm99999915_g1; Rpl38, Mm03015864_g1; Ddit4, Mm00512504_g1) and Taqman master mix (Life Technologies; Grand Island, NY, USA); protease inhibitor cocktail, doxycycline hyclate and puromycin dihydrochloride (Sigma-Aldrich; St Louis, MO, USA); DC protein assay (Bio-Rad Laboratories; Hercules, CA, USA); ECL (GE Healthcare; Piscataway, NJ, USA); RNeasy Micro kit (Qiagen; Valencia, CA, USA).
Antibodies
Hemagglutinin (HA; H6908) and fast myosin heavy chain (MyHC; H6908), (Sigma-Aldrich; St. Louis, MO, USA); goat anti-rabbit Alexa Fluor 488 (A-11070) (Life Technologies; Grand Island, NY, USA); goat anti-mouse Alexa Fluor 494 (#610-109-121) (Rockland; Gilbertsville, PA, USA); Rps6 (2317) and HA (2367) (Cell Signaling Technology; Danvers, MA, USA); puromycin (MABE343) (Millipore, Temecula, CA, USA); α-tubulin (Ab52866) (Abcam; Cambridge, MA, USA); anti-rabbit or anti-mouse secondary antibodies conjugated to horseradish peroxidase (Vector Laboratories; Burlingame, CA, USA).
Plasmids
pENTR/D-TOPO entry vector (Life Technologies, Grand Island, NY, USA); pINDUCER20 vector kind gift of Dr. Hu (National Institute of Environmental Health and Sciences, NC, USA)(Meerbrey, Hu et al. 2011).
Synergist ablation
The cohort of mice used for the muscle hypertrophy study have been previously described (Chaillou, Lee et al. 2013). A bilateral synergist ablation surgical procedure was used to induce hypertrophy of the plantaris muscle as previously described by us (3). Briefly, following a small incision on the dorsal aspect of the lower hind limb of a fully anesthetized mouse (2% isoflurane at 0.5 L·min−1), the gastrocnemius and soleus muscles were excised without disturbing the blood supply or innervation to the plantaris muscle. Plantaris muscles were collected at one, three and five days after the surgery (n = 4–6 at each time point). Control plantaris muscle was collected from mice subjected to a sham synergist ablation surgery.
Ribosomal protein mRNA expression
To confirm the reported muscle-specific expression of Rpl3l, different tissues were collected from a second cohort of mice (n = 4); the tissues collected were brain, liver, diaphragm, plantaris, heart, intestine and lung. Upon collection, tissue was immediately frozen in liquid nitrogen and then stored at −80 °C.
Generation of stable cell lines
The Rpl3l coding sequence containing a hemagglutinin (HA) tag at the 3′-end (Rpl3l-HA) was generated by PCR from cDNA and cloned into the pENTR/D-TOPO entry vector. A LR recombination reaction was then performed to transfer Rpl3l-HA into the pINDUCER20 vector to generate pI20-Rpl3l-HA plasmid (Meerbrey, Hu et al. 2011). To generate the empty vector (EV) control plasmid the LR recombination reaction was carried out between the pENTR/D-TOPO entry vector and the pINDUCER20 vector to generate the pI20-EV plasmid. The pINDUCER is an inducible Tet-On system that employs a bicistronic vector containing the Ubc gene promoter driving constitutive expression of a third-generation reverse-tet transactivator (rtTA3). Upon doxycycline binding, the rtTA3 protein binds the TRE promoter to induce expression of a downstream gene of interest (Fig. 2A). The pI20-EV was used to assess the potential toxicity of expression and/or activation of rtTA3 and doxycycline.
Figure. 2.
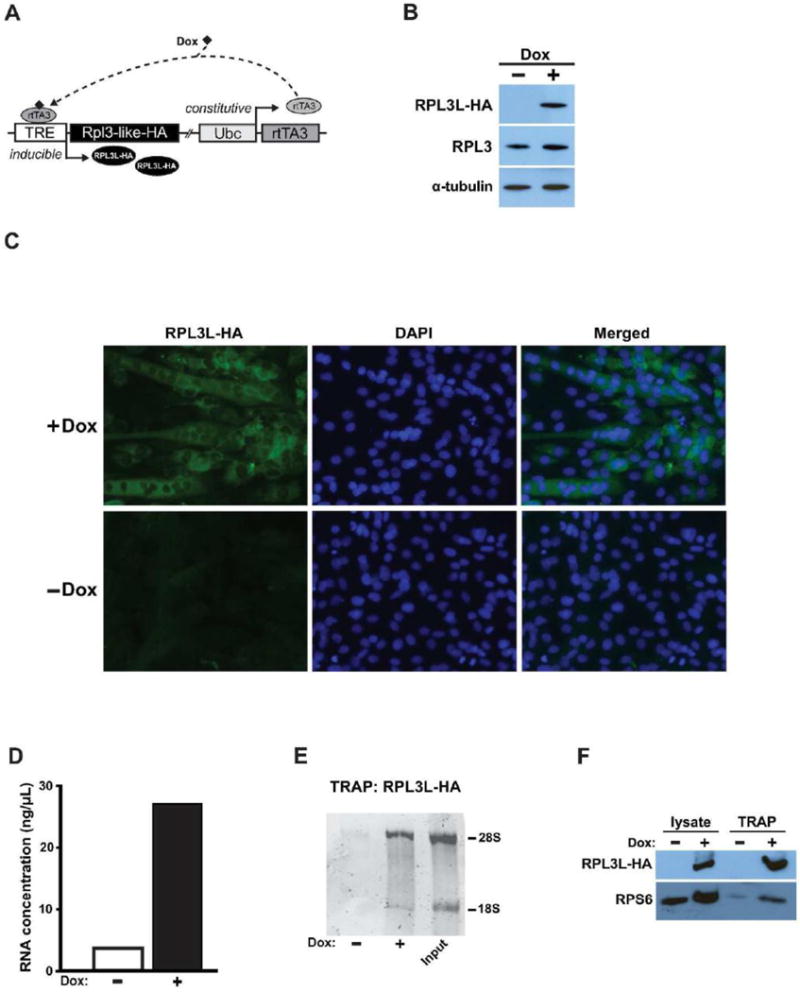
An in vitro model system to express RPL3L in C2C12 myogenic cells. A. Schematic of the pINDUCER Tet-On system used to express of RPL3L in C2C12 cells. Upon doxycycline (Dox) binding, constitutively expressed rtTA becomes activated with subsequent binding to the tetracycline response element (TRE), thereby inducing expression of HA-tagged RPL3L. B. Western blot showing exogenous RPL3L expression in myotubes in response to Dox treatment. RPL3L-C2C12 cells were differentiated four days in differentiation media containing 1 μg·mL−1 Dox with RPL3L expression detected using a HA antibody. Alpha-tubulin was used as a loading control for Western blot analysis. C. Immunostaining showing the presence of exogenous RPL3L in myotubes in response to Dox treatment. RPL3L-C2C12 cells were differentiated four days in differentiation media with or without Dox (1 μg·mL−1) and incubated with an antibody against HA. Nuclei were labelled by DAPI staining. D–E. RNA concentration and electrophoresis showing 18S and 28S rRNA following RPL3L-HA immunoprecipitation from 4-day differentiated myotubes treated with or without Dox (1 μg·mL−1). F. Immunoblotting for exogenous RPL3L and RPS6 proteins from protein lysates and immunoprecipitate samples. TRAP: translating ribosome affinity purification.
Suspended C2C12 myoblasts were transfected by either pI20-Rpl3l-HA or pI20-EV to generate stable Rpl3l-HA and EV cell lines, respectively. After 24 h, the cells were cultured in growth media containing 0.6 mg·mL−1 G418. Seven to ten days later, 10–20 colonies were picked and expanded in growth medium containing 0.2 mg·mL−1 G418. Clones that differentiated into myotubes with a similar rate as wild-type C2C12 myoblasts were selected for further expansion. To assess induction of Rpl3l-HA expression, each stable cell line was treated with doxycycline (1 μg·mL−1) for four days in differentiation media; expression of RPL3L was determined by Western blot analysis using an antibody against the HA-tag. Preliminary experiments using different doxycycline concentrations (0.1, 0.25 and 1 μg·mL−1) showed that a concentration of 1 μg·mL−1 induced a high level of expression of RPL3L protein in myotubes (Fig. 2B) with no effects on the growth of EV-myotubes.
Cell culture
Cell culture experiments were performed in a humidified environment at 37 °C with 5% CO2. The stable cell lines generated from C2C12 myoblasts were maintained at low confluence in DMEM supplemented with 10% fetal bovine serum and 0.2 mg·mL−1 G418. Once cells were fully confluent, growth media was switched to differentiation media (DMEM containing 2% horse serum and 0.2 mg·mL−1 G418) and the myoblasts were induced to differentiate into myotubes for four days. The cells were only treated with doxycycline (1 μg·mL−1) during differentiation with the differentiation media changed every 24 h. For immunohistochemistry and translating ribosome affinity purification (TRAP), cells were grown in 24-well plastic plates and 10 cm dishes, respectively.
Protein synthesis
For the non-radioactive measurement of protein synthesis using the SUnSET method, four-day differentiated myotubes were incubated in serum free DMEM with or without doxycycline (1 μg·mL−1) for 17 h. Following this incubation period, 1 μM puromycin was added to the media and myotubes collected 30 min later (Goodman, Mabrey et al. 2011).
Protein quantification
Myotubes were washed in cold PBS and lysed with ice-cold RIPA buffer [1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM NaCl, 20 mM Tris-HCl (pH 7.6), 1 mM PMSF, 5 mM benzamidine, 1 mM EDTA, 5 mM N-ethylmaleimide, 50 mM NaF, 25 mM β-glycerophosphate, 1 mM sodium orthovanadate, and 10 μl·ml−1 protease inhibitor cocktail]. Homogenates were then centrifuged at 15,294 g for 10 min at 4 °C. Protein concentration was determined using the DC protein assay kit.
Immunohistochemistry
Myotubes were grown as described above and immunostained for HA (Sigma-Aldrich, 1/100 dilution) and fast MyHC (1/400 dilution). Cultured cells were fixed in PBS containing 4% paraformaldehyde, permeabilized in PBS containing 0.5% Triton X-100, blocked in PBS-1% BSA and then incubated overnight at 4 °C with primary antibodies against HA or fast MHC. Myotubes were then washed and incubated for 1 h at room temperature with goat anti-rabbit Alexa Fluor 488 (1/500 dilution) or goat anti-mouse Alexa Fluor 494 (1/200 dilution) secondary antibodies for HA and fast MyHC, respectively. Nuclei were stained using DAPI, with immunoreactivity visualized using a multichannel fluorescent microscope (Zeiss Axio Observer, Thornwood, NY) coupled with Axio Vision Rel software (version 4.8).
Analysis of myotube diameter and myogenic fusion
The myotube diameter was determined in cells visualized with an inverted phase contrast microscope (Telaval 31, Zeiss) coupled with a digital camera (Scopetek, Int., Hangzhou, China). For each experimental condition, five captured images (10× magnification) were analyzed in three biological replicates and the analysis was repeated in three independent experiments. The diameter of 6–9 myotubes was measured for each field of view using ImageJ software (NIH, Bethesda, MA) and the average diameter per myotube was calculated as the mean of three measurements taken along the length of the myotube. The fusion index (number of nuclei inside myosin heavy chain (MHC)-positive myotubes / total number of nuclei) was determined. For each experimental condition, three captured images (10× magnification) were analyzed in three biological replicates and the analysis was repeated in three independent experiments.
Western blot analysis
Protein homogenates obtained for the analysis of protein content (described above) and TRAP protein samples (described below) were subjected to SDS-PAGE and immunobloting. For the non-radioactive measurement of protein synthesis using the SUnSET method, the cells treated with puromycin were lysed with ice-cold RIPA buffer and the protein homogenates were centrifuged at 500 g for 5 min, as previously described (Goodman, Mabrey et al. 2011). Ten micrograms of protein homogenate was subjected to SDS-PAGE and transferred onto PVDF membranes. Membranes were incubated overnight at 4 °C with primary antibodies against HA (Sigma-Aldrich, 1/2,000), Rps6 (1/1,000), puromycin (1/20,000) or α-tubulin (1/15,000); for TRAP samples, HA antibody from Cell Signaling Technology (1/1000 dilution) was used. Following the washes with TBS-T (TBS, 01% TWEEN-20), membranes were incubated for 1 h at room temperature with the corresponding anti-rabbit or anti-mouse secondary HRP-conjugated antibody, washed again in TBS-T, then incubated for 5 min in ECL and exposed to X-ray film. Analysis of band intensity was performed using ImageJ software on scanned film. For the analysis of HA immunoblots, equal loading of protein in all lanes was verified using the reference protein α-tubulin. For the SUnSET analysis, the density of the whole lane (incorporating the entire molecular weight range of puromycin-labeled peptides) was assessed with membranes stained with Coomassie Blue to verify equal loading of protein in each lane.
Translating Ribosome Affinity Purification (TRAP)
To determine if exogenous RPL3L was associated with ribosomes, we used the translating ribosome affinity purification (TRAP) protocol as described by Heiman and colleagues (Heiman, Schaefer et al. 2008). Four-day differentiated Rpl3l-HA myotubes cultured in 10 cm dishes (four dishes per condition) were rinsed twice with ice-cold PBS. The myotubes were scraped in ice-cold PBS (2 mL per dish) and centrifuged for 5 min at 500 rpm at 4 °C. The pellet was then resuspended into 750 μl of polysome extraction buffer (10 mM HEPES [pH 7.4], 150 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol, 100 μg·mL−1 cycloheximide, 10 μL·mL−1 protease inhibitor, 25 uL RNase inhibitor) and homogenized on ice. After 10 min on ice, homogenates were centrifuged for 10 min at 2,000 g at 4 °C. The supernatant was then treated with 1% NP-40, mixed by vortexing, incubated on ice for 5 min, and centrifuged for 10 min at 13,000 g at 4 °C to pellet insolubilized material. Four micrograms of antibody against HA (Sigma-Aldrich) were added to the cleared supernatant and incubated with rotation for 4 h at 4 °C. One hundred microliters of slurry protein G Dynabeads was washed in the polysome extraction buffer (without protease and RNase inhibitors), resuspended with the antibody-antigen complex homogenate and rotated overnight at 4 °C. Captured immunoprecipitates were washed three times with high-salt polysome wash buffer (10 mM HEPES [pH 7.4], 350 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol, 100 μg·mL−1 cycloheximide, 1% NP-40). The bead-antibody-antigen complexes were lysed in 350 μL RLT lysis buffer containing 1% β-mercaptoethanol and RNA was extracted using an RNeasy Micro kit accordingly to the manufacturer’s instructions. One hundred and fifty nanograms of purified RNA was run out on a 3.5% polyacrylamide gel (acrylamide/bisacrylamide 29:1) to visualize ribosomal RNA. The flow-through obtained following centrifugation of immunoprecipitated samples was precipitated with acetone and the pellet resuspended in ice-cold RIPA buffer. Precipitated protein samples were then subjected to SDS-PAGE and immunoblotting.
RNA isolation, cDNA synthesis, qPCR
Total RNA extraction and reverse transcription were performed as previously described (Chaillou, Lee et al. 2013). qPCR was performed using 5 μL of diluted cDNA (1/20 dilution from stock cDNA mixture) and 1 μL of primer mix in a 20 μL final volume. qPCR were performed using an ABI 7500 RT-PCR system. Quantification cycles (Cq) were determined by ABI 7500 software v2.0.1. Absolute quantification was achieved by exponential conversion of the Cq using the qPCR efficiency. qPCR efficiency was estimated from standard curves obtained by serial dilutions (1-log range) of a pooled sample for each RT set (Peinnequin, Mouret et al. 2004). The comparison of the mRNA abundance of Rpl3 and Rpl3l between several tissues was determined after the normalization with Rpl38. Relative quantification of Rpl3 and Rpl3l mRNAs in response to synergist ablation was obtained after the normalization with the geometric mean of exponential conversion of the Cq of three reference genes (Rpl38, Gapdh and Ddit4). The geometric mean based on these genes was not affected by synergist ablation over time.
Microarray
The microarray hybridation and processing were performed as previously described (Chaillou, Lee et al. 2013) with microarray data available at GEO, accession number GSE47098.
Statistical analysis
All data are expressed as mean ± SE. The changes in gene expression (qPCR) obtained from skeletal muscle were analyzed using a one-way ANOVA followed by a Dunnett’s post hoc test. The data obtained in vitro were analyzed using a two-tailed t-test for paired samples. For these statistical analyses, the level of significance was set at p < 0.05.
RESULTS
Striated muscle-specific expression of Rpl3l
To confirm the muscle-specific expression of Rpl3l, the relative expression level of Rpl3l and its paralog Rpl3 was measured by qPCR in several mouse tissues (Fig. 1A). The expression of Rpl3 mRNA was ubiquitous, with the lowest expression observed in the plantaris and diaphragm muscles. In contrast, Rpl3l mRNA was highly expressed, specifically in striated muscles, with the highest level observed in the plantaris muscle. Notably, expression of Rpl3l mRNA was extremely low in the intestine, a tissue containing smooth muscle cells. These results clearly show expression of Rpl3l is restricted to striated muscle, confirming earlier studies using microarray and Northern blot analyses (Van Raay, Connors et al. 1996, Thorrez, Van Deun et al. 2008).
Figure 1.
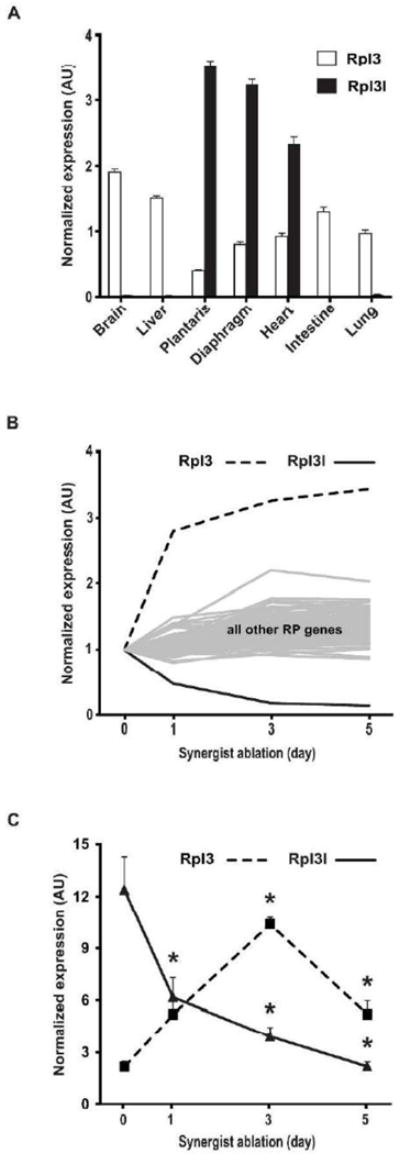
Rpl3l expression in adult mouse tissue and during skeletal muscle hypertrophy. A. Rpl3l and Rpl3 expression was determined by qPCR in several tissues of adult mice (n=4), normalized to Rpl38 expression. B. Microarray analysis of ribosomal protein gene expression during plantaris muscle hypertrophy induced by synergist ablation presented as fold-change relative to sham control; the microarray data accession number is GSE47098. C. The change in Rpl3l and Rpl3 expression observed by microarray during muscle hypertrophy was confirmed by qPCR; expression was normalized using the geometric mean based on the reference genes Rpl38, Gapdh and Ddit4. Data are expressed as mean +/− SE with significant (p < 0.05) difference from sham control (Sham) designated by asterisk.
Decreased expression of Rpl3l in response to a hypertrophic stimulus
Given our interest in skeletal muscle hypertrophy, we queried our published skeletal muscle hypertrophy microarray data set to determine if Rpl3l expression changed in response to a hypertrophic stimulus induced by synergist ablation (Chaillou, Lee et al. 2013). Expression of Rpl3l mRNA was decreased following a single day of synergist ablation and was further decreased after three and five days (Fig. 1B). In stark contrast to Rpl3l, expression of Rpl3 was increased in response to synergist ablation across the same time course (Fig. 1B). As shown in Fig. 1B, the dramatic change in expression of Rpl3 and Rpl3l with synergist ablation was unique compared to the other 77 ribosomal protein genes. qPCR analysis confirmed the microarray findings; Rpl3l mRNA expression progressively decreased across the time course, reaching 18% of sham control at day 5 whereas Rpl3 mRNA expression was significantly increased at each time point, peaking with a ~5-fold increase at day 3 (Fig. 1C).
Inducible expression of Rpl3l
The change in Rpl3l mRNA expression following synergist ablation suggested Rpl3l may have a role in regulating skeletal muscle growth. To study Rpl3l function required that we develop an in vitro system to express Rpl3l because it is not expressed in either C2C12 myoblasts or myotubes as assessed by RT-PCR (data not shown). Thus, we generated HA-tagged Rpl3l stable clone in C2C12 cells employing a Tet-On system to allow for inducible, gain-of-function studies. The Tet-On system we used was recently described by Meerbrey and colleagues and is referred to as pINDUCER; the functional elements of the pINDUCER system are depicted in Fig. 2A (Meerbrey, Hu et al. 2011). As shown in Fig. 2B, the expression of exogenous RPL3L (RPL3L with a HA-tag) after four days of differentiation was gradually increased with successively higher doxycycline concentration, reaching the highest level of expression using 1 μg·mL−1 doxycycline; importantly, myogenic differentiation or myotube size of the empty vector stable cell line was not affected at this doxycycline concentration and, therefore, was used for the remaining experiments. Immunohistochemistry using an antibody against the HA-tag confirmed induction of RPL3L protein in myotubes in response to doxycycline treatment (Fig. 2C).
To determine if the exogenous RPL3L protein was incorporated into the ribosome, the TRAP protocol, as described by Heiman and colleagues, was performed using the HA antibody (Heiman, Schaefer et al. 2008). The amount of RNA recovered by ribosome immunoprecipitation was comparable to that reported by Knight and coworkers and demonstrated the specificity of the precipitation reaction (Fig. 2D) (Knight, Tan et al. 2012). The successful incorporation of exogenous RPL3L into the ribosome was confirmed by enrichment of 18S and 28S rRNAs only in immunoprecipitate from doxycycline-treated myotube lysate (Fig. 2E), as well as the presence of RSP6, a protein of the 40S ribosome subunit, in addition to RPL3L (Fig. 2F).
Induction of RPL3L expression impairs myotube growth and myoblast fusion
As a first effort to understand the role of RPL3L in skeletal muscle hypertrophy, we determined the effect of RPL3L expression on myotube size. As shown in Fig. 3A–3B, myotube diameter was significantly smaller by 23% in RPL3L-myotubes treated with doxycycline compared to untreated myotubes. Consistent with the smaller myotube size, the protein content was significantly reduced by 14% in RPL3L-myotubes treated with doxycycline (Fig. 3C). Empty vector (EV) control myotubes showed no change in myotube size or protein content with doxycycline treatment (Fig. 3A–C). The reason for the lower protein content in the EV samples is not known but likely reflects the fact that the EV stable cell line represents a completely unique cell line; importantly, myotube size and protein content were not changed by doxycycline treatment in the EV myotubes indicating the significant change in these parameters was the result of RPL3L expression and not caused by constitutive expression of rtTA, rtTA activation or doxycycline toxicity.
Figure 3.
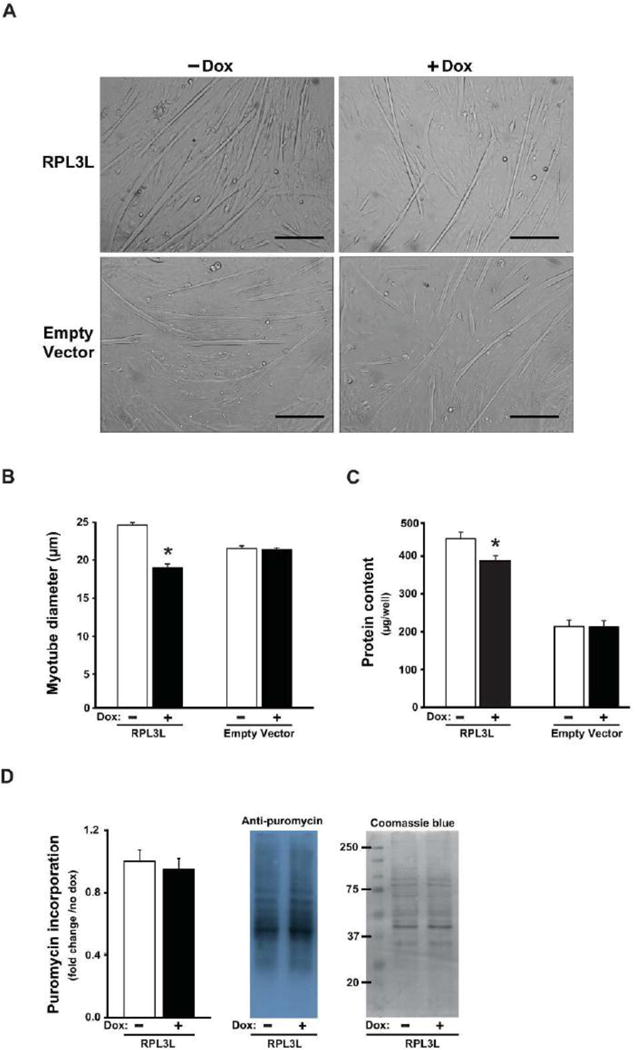
RPL3L expression impairs myotube growth after four days of differentiation. A. Representative photographs from RPL3L-myotubes and empty vector (EV)-myotubes. B. Myotube diameter measured in RPL3L and EV myotubes. C. Protein content determined from protein lysates of RPL3L and EV myotubes. D. Quantification of the puromycin-labeled peptides in RPL3L-myotubes and representative image of immunoblot analysis followed by Coomassie Blue staining (loading control). Histograms represent mean ± SE from three biological replicates obtained in three independent experiments with significant difference (p < 0.05) from untreated RPL3L-myotubes designated by asterisk.
Next, we used the SUnSET method, as described by Goodman and coworkers, to assess whether or not the decrease in myotube size by RPL3L expression was caused by an inhibition of protein synthesis (Goodman, Mabrey et al. 2011). The amount of puromycin incorporation, as determined by Western blot analysis, was the same with or without doxycycline treatment indicating that a change in the rate of protein synthesis was not responsible for the smaller myotube size associated with RPL3L expression (Fig. 3D).
In considering what other factors could impact myotube size, we thought RPL3L might be affecting the ability of myoblasts to fuse with developing myotubes during the differentiation process. To investigate this possibility, we determined the fusion index (number of myonuclei relative to total number of nuclei) using fast myosin heavy chain immunostaining to identify myonuclei. As shown in Fig. 4, the fusion index was significantly decreased by 17% upon RPL3L expression but remained unchanged in empty vector control with doxycycline treatment. This finding suggests that expression of RPL3L was inhibiting myoblast fusion through a yet-to-be determined mechanism involving the ribosome.
Fig. 4.
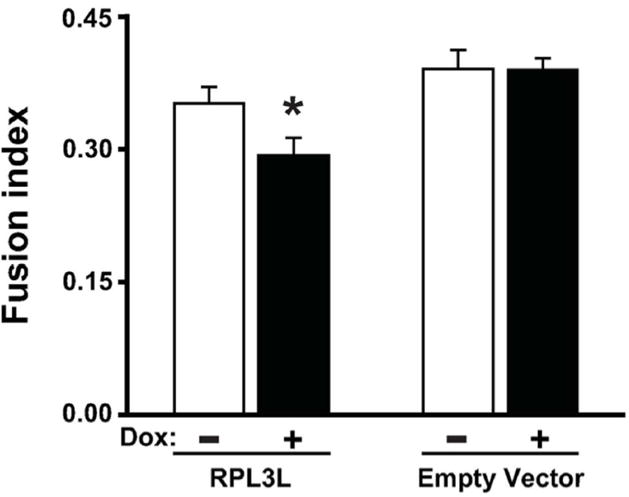
RPL3L expression impairs myotube fusion after four days of differentiation. The fusion index was determined by counting the number of nuclei inside myosin heavy chain (MHC)-positive myotubes, divided by the total number of nuclei. Histograms represents mean ± SE from three biological replicates obtained in three independent experiments with significant difference (p < 0.05) from untreated RPL3L-myotubes designated by asterisk.
DISCUSSION
The ribosome has generally been considered to have no cell-specific function but rather serves in a “housekeeping” capacity. This view has been challenged by evidence showing that heterogeneity in the protein composition of the ribosome can result in the functional specialization of the ribosome (Xue and Barna 2012, Sauert, Temmel et al. 2014). Ribosome specialization can alter the intrinsic translational activity of the ribosome in a number of different ways such that there is the preferential translation of specific mRNAs, a change in translational fidelity or the level of IRES-mediated translation. For example, during mouse embryonic development, specific Hox mRNAs were shown to be selectively translated in those tissues that are enriched in ribosomal protein L38 (Kondrashov, Pusic et al. 2011). The specialization of ribosome function represents a fundamental shift in how the ribosome is viewed – mRNA translation by the ribosome is not just a strictly constitutive process but can be regulative in nature. Moreover, the cellular specialization of ribosome function is an exciting discovery because it represents a completely new level of gene regulation, reminiscent of where the nascent field of microRNAs was in the early 2000s (Couzin 2002, Moss and Poethig 2002, Gilbert 2011, Xue and Barna 2012).
The Rpl3l gene was originally identified by Burn and colleagues and found to share high sequence homology with Rpl3 gene (Burn, Connors et al. 1996). In a subsequent study, the same group provided the first evidence that, in stark contrast to the ubiquitously expressed gene Rpl3, expression of Rpl3l was restricted to skeletal muscle and heart (Van Raay, Connors et al. 1996). Here, we confirmed the striated muscle-specific expression of Rpl3l, showing that it was highly expressed in diaphragm, plantaris and heart but almost undetectable by qPCR in non-muscle tissue. Moreover, the finding that Rpl3l expression was extremely low in the intestine, a tissue containing a high proportion of smooth muscle, further emphasizes the striated muscle-specific expression of Rpl3l.
We became aware of Rpl3l while analyzing microarray data from a study examining changes in gene expression of skeletal muscle in response to a robust hypertrophic stimulus induced by synergist ablation (Chaillou, Lee et al. 2013). This fortuitous finding prompted us to look at the expression profile of the complete set of 79 ribosomal protein genes following synergist ablation. Somewhat to our surprise, Rpl3l, and its paralog Rpl3, were the only two ribosomal protein genes to show such a large (> 2.5-fold) change in expression (see Fig. 1B). Even more curious was the apparent reciprocal change in Rpl3l and Rpl3 expression, being down- and up-regulated, respectively. This finding, coupled with the fact that Rpl3l is muscle-specific, raised the intriguing question, why has skeletal muscle evolved to have its own version of the ubiquitously expressed Rpl3 gene?
Beyond reports on its muscle-specific expression, there is nothing known about the function of Rpl3l (Van Raay, Connors et al. 1996, Thorrez, Van Deun et al. 2008). Based on the high-degree of amino acid conservation (~80%) between RPL3 and RPL3L, it is reasonable to suggest that RPL3L performs a similar, but distinct, function as RPL3 (Van Raay, Connors et al. 1996). As one of the largest ribosomal proteins, RPL3 has been shown to be required for peptidyltransferase activity of the ribosome as well as being one of only two ribosomal proteins able to initiate the assembly of the large ribosomal subunit (Nowotny and Nierhaus 1982, Schulze and Nierhaus 1982). More recently, genetic and biochemical studies by the Dinman laboratory have shown that RPL3 functions as the “gatekeeper” of the A site allowing for the synchronization of aa-tRNA binding and translocation (Petrov, Meskauskas et al. 2004, Meskauskas, Petrov et al. 2005, Meskauskas and Dinman 2007). Although the results of the current study do not offer any insight into just how Rpl3l might be altering ribosome function, our findings do provide the first evidence of a role for Rpl3l in the regulation of skeletal muscle hypertrophy. In particular, the results from our in vitro study support the idea that Rpl3l acts to inhibit myotube growth by limiting myoblast fusion through an unknown mechanism but one that presumably involves altering ribosome function. The magnitude of myotube atrophy caused by RPL3L expression is comparable to that reported following oxidative stress, cytokine exposure or glucocorticoid treatment (Sultan, Henkel et al. 2006, Menconi, Gonnella et al. 2008, Li, Moylan et al. 2009, McClung, Judge et al. 2009, Yamaki, Wu et al. 2012). Similarly, our data suggest the down-regulation of Rpl3l expression in response to synergist ablation may be necessary for effective satellite cell fusion that is known to occur during compensatory hypertrophy (McCarthy, Mula et al. 2011). The notion that Rpl3l may be acting as a break on hypertrophic growth through the regulation of satellite cell fusion is supported by microarray data showing that Rpl3l expression is progressively up-regulated in skeletal muscle during post-natal development, a period during which the rate of satellite cell fusion, and subsequently muscle growth, gradually levels off (Cheng, Merriam et al. 2004, White, Bierinx et al. 2010). The details of how a change in ribosome function by Rpl3l affects myoblast or satellite cell fusion remains unknown but is the focus of ongoing studies.
Several lines of evidence provide further support for the idea that Rpl3l regulates myoblast or satellite cell fusion. In an effort to gather additional clues about Rpl3l function, we searched online microarray databases with the hope of finding other conditions under which Rpl3l expression changed. First, we found that Rpl3l expression was barely detectable in C2C12 myoblasts and myotubes by qPCR analysis (data not shown), a finding consistent with those reported in both mouse and human myoblasts and myotubes (GSE10428 and GSE10435), in quiescent and activated mouse satellite cells (GSE15155) as well as in myogenic progenitors during embryogenesis (GSE42389) (Harel, Maezawa et al. 2012, Pallafacchina, Blaauw et al. 2013). Next, we found data showing that Rpl3l expression was decreased during the first days of skeletal muscle regeneration following cardiotoxin injection, as well as in skeletal muscle of patients with Duchenne muscular dystrophy, a myopathic disorder characterized by continuous cycles of degeneration-regeneration (Haslett, Sanoudou et al. 2002, Lukjanenko, Brachat et al. 2013). Finally, as mentioned previously, Rpl3l expression is very low during the early phase of post-natal development (Cheng, Merriam et al. 2004), a period characterized by a robust increase in muscle fiber size and myonuclear number as a result of significant satellite cell fusion (White, Bierinx et al. 2010). Collectively, these studies along with our data demonstrate a clear relationship between the down-regulation of Rpl3l expression and myoblast fusion. As a correlative, the high level of Rpl3l expression observed in adult skeletal muscle under basal conditions may function to make the muscle recalcitrant to myoblast fusion. Given the unique fusogenic property of skeletal muscle, it seems likely that it would be important to regulate this property to prevent the untimely fusion of myoblast or other fusogenic cells that come in contact with the muscle fiber.
The inverse pattern of expression of Rpl3 and Rpl3l reported in this study during skeletal muscle hypertrophy as well as during post-natal development (Cheng, Merriam et al. 2004) and skeletal muscle regeneration (Lukjanenko, Brachat et al. 2013) indicates there is coordinated regulation of these paralogs. In plants, and more recently in the mouse, there is evidence that the expression of ribosomal protein paralogs are in fact coordinately regulated, suggesting the cell tries to maintain a reciprocal level of paralog expression (Popescu and Tumer 2004, O’Leary, Schreiber et al. 2013). Unlike what we observed during skeletal muscle hypertrophy, we did not observe any change in RPL3 protein expression in myotubes upon induction of RPL3L expression (data not shown), suggesting that RPL3L does not regulate RPL3 expression in myotubes. Future studies are necessary to determine the mechanism responsible for the regulation of Rpl3 and Rpl3l during skeletal muscle hypertrophy.
How might ribosome specialization by RPL3L regulate myoblast fusion? Although there are a number of different scenarios for how RPL3L might be altering ribosome function, we favor the notion that the association of RPL3L with the ribosome increases the efficiency of −1 programmed ribosomal frameshifting (−1 PRF). This idea is based on the work of the Dinman laboratory which has shown that mutation in RPL3 can increase the efficiency of −1 PRF (Meskauskas, Petrov et al. 2005). Further, particular transcripts are more suspectible to −1 PRF because of the presence of a “slippery” site that, in turn, can lead to non-sense mediated decay (NMD) as the result of a premature stop codon in the −1 reading frame. Thus, according to our working model, RPL3L reduces the abundance of transcripts encoding proteins required for myoblast fusion via NMD as the result of increased −1 PRF. Ongoing experiments are assessing the ability of RPL3L to alter the efficiency of −1 PRF and to identify relevant transcripts harboring a “slippery” site and a downstream premature stop codon.
In conclusion, we have shown in this study that the striated muscle-specific ribosomal protein gene Rpl3l was down-regulated during skeletal muscle hypertrophy. In an effort to investigate the role of this gene during skeletal muscle growth, we expressed RPL3L protein in C2C12 myogenic cells during myotube formation. Our results demonstrated that RPL3L expression decreased myotube size as a result of decreased myoblast fusion, suggesting the down-regulation of Rpl3l during skeletal muscle hypertrophy promotes satellite cell fusion. The results of this study provide the first evidence of ribosome specialization in adult tissue, skeletal muscle in particular. Furthermore, the findings of this study support the hypothesis that ribosome specialization has a role in the regulation of skeletal muscle hypertrophy. Future studies will investigate the necessity of ribosome specialization in skeletal muscle hypertrophy in the adult and during post-natal development and the mechanism through which this specialization regulates skeletal muscle growth.
Supplementary Material
Acknowledgments
This work was supported by grants from NIH to JJM (AR061939 and AR064896).
Footnotes
This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: [10.1002/jcp.25294]
References
- Burn TC, Connors TD, Van Raay TJ, Dackowski WR, Millholland JM, Klinger KW, Landes GM. Generation of a transcriptional map for a 700-kb region surrounding the polycystic kidney disease type 1 (PKD1) and tuberous sclerosis type 2 (TSC2) disease genes on human chromosome 16p3.3. Genome Res. 1996;6(6):525–537. doi: 10.1101/gr.6.6.525. [DOI] [PubMed] [Google Scholar]
- Campbell KJ, White RJ. MYC regulation of cell growth through control of transcription by RNA polymerases I and III. Cold Spring Harb Perspect Med. 2014;4(5) doi: 10.1101/cshperspect.a018408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaillou T, Lee JD, England JH, McCarthy KA, Esser JJ. Time course of gene expression during mouse skeletal muscle hypertrophy. J Appl Physiol (1985) 2013;115(7):1065–1074. doi: 10.1152/japplphysiol.00611.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G, Merriam AP, Gong B, Leahy P, Khanna S, Porter JD. Conserved and muscle-group-specific gene expression patterns shape postnatal development of the novel extraocular muscle phenotype. Physiol Genomics. 2004;18(2):184–195. doi: 10.1152/physiolgenomics.00222.2003. [DOI] [PubMed] [Google Scholar]
- Couzin J. Breakthrough of the year. Small RNAs make big splash. Science. 2002;298(5602):2296–2297. doi: 10.1126/science.298.5602.2296. [DOI] [PubMed] [Google Scholar]
- Dharia AP, Obla A, Gajdosik MD, Simon A, Nelson CE. Tempo and mode of gene duplication in mammalian ribosomal protein evolution. PLoS One. 2014;9(11):e111721. doi: 10.1371/journal.pone.0111721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert WV. Functional specialization of ribosomes? Trends Biochem Sci. 2011;36(3):127–132. doi: 10.1016/j.tibs.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CA, Mabrey DM, Frey JW, Miu MH, Schmidt EK, Pierre P, Hornberger TA. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J. 2011;25(3):1028–1039. doi: 10.1096/fj.10-168799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I. The nucleolus-guardian of cellular homeostasis and genome integrity. Chromosoma. 2013;122(6):487–497. doi: 10.1007/s00412-013-0430-0. [DOI] [PubMed] [Google Scholar]
- Gupta V, Warner JR. Ribosome-omics of the human ribosome. RNA. 2014;20(7):1004–1013. doi: 10.1261/rna.043653.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel I, Maezawa Y, Avraham R, Rinon A, Ma HY, Cross JW, Leviatan N, Hegesh J, Roy A, Jacob-Hirsch J, Rechavi G, Carvajal J, Tole S, Kioussi C, Quaggin S, Tzahor E. Pharyngeal mesoderm regulatory network controls cardiac and head muscle morphogenesis. Proc Natl Acad Sci U S A. 2012;109(46):18839–18844. doi: 10.1073/pnas.1208690109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslett JN, Sanoudou D, Kho AT, Bennett RR, Greenberg SA, Kohane IS, Beggs AH, Kunkel LM. Gene expression comparison of biopsies from Duchenne muscular dystrophy (DMD) and normal skeletal muscle. Proc Natl Acad Sci U S A. 2002;99(23):15000–15005. doi: 10.1073/pnas.192571199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, Greengard P, Heintz N. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135(4):738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J, Ryvkin P, Hemphill E, Mandoiu I, Nelson C. The birth of new genes by RNA- and DNA-mediated duplication during mammalian evolution. J Comput Biol. 2009;16(10):1429–1444. doi: 10.1089/cmb.2009.0073. [DOI] [PubMed] [Google Scholar]
- Knight ZA, Tan K, Birsoy K, Schmidt S, Garrison JL, Wysocki RW, Emiliano A, Ekstrand MI, Friedman JM. Molecular profiling of activated neurons by phosphorylated ribosome capture. Cell. 2012;151(5):1126–1137. doi: 10.1016/j.cell.2012.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131(3):557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145(3):383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Moylan JS, Chambers MA, Smith J, Reid MB. Interleukin-1 stimulates catabolism in C2C12 myotubes. Am J Physiol Cell Physiol. 2009;297(3):C706–714. doi: 10.1152/ajpcell.00626.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukjanenko L, Brachat S, Pierrel E, Lach-Trifilieff E, Feige JN. Genomic profiling reveals that transient adipogenic activation is a hallmark of mouse models of skeletal muscle regeneration. PLoS One. 2013;8(8):e71084. doi: 10.1371/journal.pone.0071084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro VP, Edelman GM. The ribosome filter hypothesis. Proc Natl Acad Sci U S A. 2002;99(19):12031–12036. doi: 10.1073/pnas.192442499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JJ, Mula J, Miyazaki M, Erfani R, Garrison K, Farooqui AB, Srikuea R, Lawson BA, Grimes B, Keller C, Van Zant G, Campbell KS, Esser KA, Dupont-Versteegden EE, Peterson CA. Effective fiber hypertrophy in satellite cell-depleted skeletal muscle. Development. 2011;138(17):3657–3666. doi: 10.1242/dev.068858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung JM, Judge AR, Talbert EE, Powers SK. Calpain-1 is required for hydrogen peroxide-induced myotube atrophy. Am J Physiol Cell Physiol. 2009;296(2):C363–371. doi: 10.1152/ajpcell.00497.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, Herschkowitz JI, Burrows AE, Ciccia A, Sun T, Schmitt EM, Bernardi RJ, Fu X, Bland CS, Cooper TA, Schiff R, Rosen JM, Westbrook TF, Elledge SJ. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci U S A. 2011;108(9):3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menconi M, Gonnella P, Petkova V, Lecker S, Hasselgren PO. Dexamethasone and corticosterone induce similar, but not identical, muscle wasting responses in cultured L6 and C2C12 myotubes. J Cell Biochem. 2008;105(2):353–364. doi: 10.1002/jcb.21833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskas A, Dinman JD. Ribosomal protein L3: gatekeeper to the A site. Mol Cell. 2007;25(6):877–888. doi: 10.1016/j.molcel.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskas A, Petrov AN, Dinman JD. Identification of functionally important amino acids of ribosomal protein L3 by saturation mutagenesis. Mol Cell Biol. 2005;25(24):10863–10874. doi: 10.1128/MCB.25.24.10863-10874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Poethig RS. MicroRNAs: something new under the sun. Curr Biol. 2002;12(20):R688–690. doi: 10.1016/s0960-9822(02)01206-x. [DOI] [PubMed] [Google Scholar]
- Nowotny V, Nierhaus KH. Initiator proteins for the assembly of the 50S subunit from Escherichia coli ribosomes. Proc Natl Acad Sci U S A. 1982;79(23):7238–7242. doi: 10.1073/pnas.79.23.7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary MN, Schreiber KH, Zhang Y, Duc AC, Rao S, Hale JS, Academia EC, Shah SR, Morton JF, Holstein CA, Martin DB, Kaeberlein M, Ladiges WC, Fink PJ, Mackay VL, Wiest DL, Kennedy BK. The ribosomal protein Rpl22 controls ribosome composition by directly repressing expression of its own paralog, Rpl22l1. PLoS Genet. 2013;9(8):e1003708. doi: 10.1371/journal.pgen.1003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallafacchina G, Blaauw B, Schiaffino S. Role of satellite cells in muscle growth and maintenance of muscle mass. Nutr Metab Cardiovasc Dis. 2013;23(Suppl 1):S12–18. doi: 10.1016/j.numecd.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Peinnequin A, Mouret C, Birot O, Alonso A, Mathieu J, Clarencon D, Agay D, Chancerelle Y, Multon E. Rat pro-inflammatory cytokine and cytokine related mRNA quantification by real-time polymerase chain reaction using SYBR green. BMC Immunol. 2004;5:3. doi: 10.1186/1471-2172-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov A, Meskauskas A, Dinman JD. Ribosomal protein L3: influence on ribosome structure and function. RNA Biol. 2004;1(1):59–65. doi: 10.4161/rna.1.1.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu SC, Tumer NE. Silencing of ribosomal protein L3 genes in N. tabacum reveals coordinate expression and significant alterations in plant growth, development and ribosome biogenesis. Plant J. 2004;39(1):29–44. doi: 10.1111/j.1365-313X.2004.02109.x. [DOI] [PubMed] [Google Scholar]
- Sauert M, Temmel H, Moll I. Heterogeneity of the translational machinery: Variations on a common theme. Biochimie. 2014 doi: 10.1016/j.biochi.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze H, Nierhaus KH. Minimal set of ribosomal components for reconstitution of the peptidyltransferase activity. EMBO J. 1982;1(5):609–613. doi: 10.1002/j.1460-2075.1982.tb01216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan KR, Henkel B, Terlou M, Haagsman HP. Quantification of hormone-induced atrophy of large myotubes from C2C12 and L6 cells: atrophy-inducible and atrophy-resistant C2C12 myotubes. Am J Physiol Cell Physiol. 2006;290(2):C650–659. doi: 10.1152/ajpcell.00163.2005. [DOI] [PubMed] [Google Scholar]
- Thorrez L, Van Deun K, Tranchevent LC, Van Lommel L, Engelen K, Marchal K, Moreau Y, Van Mechelen I, Schuit F. Using ribosomal protein genes as reference: a tale of caution. PLoS One. 2008;3(3):e1854. doi: 10.1371/journal.pone.0001854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raay TJ, Connors TD, Klinger KW, Landes GM, Burn TC. A novel ribosomal protein L3-like gene (RPL3L) maps to the autosomal dominant polycystic kidney disease gene region. Genomics. 1996;37(2):172–176. doi: 10.1006/geno.1996.0538. [DOI] [PubMed] [Google Scholar]
- White RB, Bierinx AS, Gnocchi VF, Zammit PS. Dynamics of muscle fibre growth during postnatal mouse development. BMC Dev Biol. 2010;10:21. doi: 10.1186/1471-213X-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13(6):355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaki T, Wu CL, Gustin M, Lim J, Jackman RW, Kandarian SC. Rel A/p65 is required for cytokine-induced myotube atrophy. Am J Physiol Cell Physiol. 2012;303(2):C135–142. doi: 10.1152/ajpcell.00111.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


