Abstract
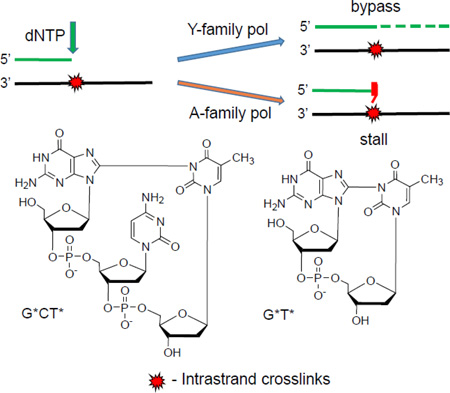
Oxidatively generated guanine radicals in DNA can undergo various nucleophilic reactions including the formation of C8-guanine cross-links with adjacent or nearby N3-thymines in DNA in the presence of O2. These G[8-3]T lesions have been identified in the DNA of human cells exposed to oxidative stress, and are most likely genotoxic if not removed by cellular defence mechanisms. The abilities of several representative polymerases to bypass the G[8-3]T lesions in two different sequence contexts, G*T* and G*CT* were assessed in vitro. The polymerase BF (bacillus fragment) from Bacillus stearothermophilus, the Y-family archaeal polymerases Dpo4 from Sulfolobus sulfataricus P2, and human DNA pol κ and pol η were selected for study. The A-family polymerase BF was strongly blocked, while relatively weak translesion synthesis was observed in the case of the Y-family polymerases Dpo4 and pol κ. Primer extension catalyzed by pol η was also partially stalled at various positions at or near the G[8-3]T cross-linked bases, but significant and distributive primer extension was observed beyond the sites of the lesions with the efficiency being consistently greater in the case of the G*CT* than in the case of the G*T* lesions. The results obtained with pol η are compared with translesion synthesis past other intrastrand cross-linked lesions with previously published results by others that include the isomeric G[8-5m]T lesions generated by ionizing radiation, the cis-syn cyclobutane pyrimidine dimer and the 6-4 photoproduct generated by UV radiation, and the Pt-G*G* lesions derived from the reactions of the chemotherapeutic agent cisplatin with DNA.
Graphical abstract
Guanine(C8)-thymine(N3) intrastrand cross-links are bypassed with varying efficiencies by Y-family polymerases, but the A-family polymerase BF is strongly blocked.
1 Introduction
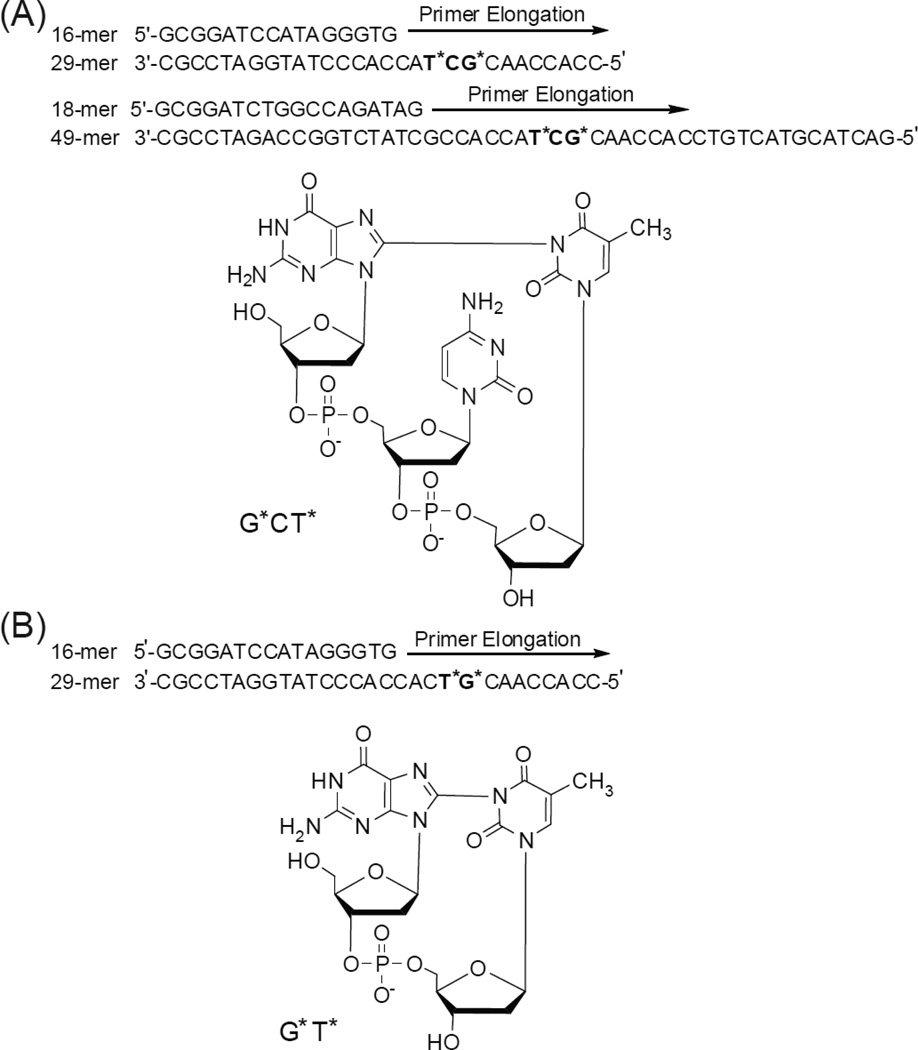
The formation of oxidatively generated interstrand and intrastrand cross-linked DNA lesions have received growing attention because of their genotoxic properties.1, 2 Interstrand lesions generated by the reactions of endogenous or exogenous cross-linking agents with double-stranded DNA are particularly difficult to remove by cellular repair mechanisms.3 However, intrastrand cross-linked DNA lesions (IntraCL) such as the cisplatin cross-linked Pt-G*G* and Pt-G*TG* lesions (the asterisks denote the cross-linked bases) are also genotoxic if not removed by cellular DNA repair systems.4 Other well-known DNA IntraCL are the cis-syn cyclobutane thymine-thymine dimers (T*T*) formed by UV radiation,5 and the 8,5’-cyclopurine-2’-deoxynucleotides formed by free radical-mediated mechanisms.6, 7 The G[8-5m]T and G[8-5]C IntraCL are formed by the combination of C-centered pyrimidine 5-(2-deoxyuridylyl) methyl or 6-hydroxy-5,6-dihydro-2’-deoxycytid-5-yl radicals generated by exposing double-stranded DNA to ionizing radiation in dioxygen – free solutions.8–18 Another, more recently documented mechanism of oxidatively generated DNA intrastrand cross-linking reactions19–23 involves the one-electron oxidation of guanine in DNA by reactive oxygen and nitrogen species that are formed in vivo under persistent oxidative stress developed in response to inflammation in tissues.24 The guanine radical cations formed by this oxidation mechanism result in the formation of 8-oxo-7,8-dihydroguanine and a variety of deeper guanine oxidation products.25 We have shown earlier that the guanine radicals derived from deprotonation of guanine radical cations can react by the nucleophilic addition of its C8 position to the N3-atom of a nearby thymine base to form G[8-3]T cross-links.19–23 This oxygen-dependent cross-linking reaction occurs readily in aerated aqueous solutions,19–23 while G[8-5m]T and G[8-5]C lesions are formed under anaerobic conditions.7 Two different G[8-3]T intrastrand crosslinks have been found, one involves a cross-link between adjacent G and T bases (G*T*), and the other has an intervening C base between G* and T* (G*CT*), as shown in Fig. 1. The G[8-3]T lesions have been detected in human HeLa cells subjected to intense frequency-tripled 266 nm Nd:YAG laser pulses that generate guanine radicals that form chemical bonds with neighboring thymine bases in DNA in solution.26
Fig. 1.
Structures of the G*[8-3]T* intrastrand cross-links and the sequences of the DNA duplexes used in the experimental studies. The asterisks denote the cross-linked bases. Panel A: G*CT* lesion. Panel B: G*T* lesion.
The G[8-5m]T and G[8-5]C lesions are known to block translesion synthesis (TLS) in vitro catalyzed by high-fidelity DNA polymerases.13, 17, 18 On the other hand, the low-fidelity yeast and human DNA polymerases η (pol η) are able to bypass G[8-5]C and G[8-5m]T lesions in an error-prone manner, but with lower efficiencies than in the case of unmodified controls.12–15 The SOS induced polymerases IV (pol IV) and V (pol V) were also found to bypass G[8-5m]T cross-links in an error-prone manner.16 However, nothing is known about the impact of the G*T* and G*CT* IntraCL on translesion bypass.
In this communication, we explored the overall effects of the G[8-3]T lesions embedded in the related G*T* and G*CT* sequence contexts on the primer extension activities catalyzed by the thermostable bacterial high-fidelity replicative A-family polymerase BF,27, 28 the archaeal thermostable DinB-like Y-family TLS polymerase from S. solfataricus P2BF (Dpo4),29 and two human Y-family TLS DNA polymerases (pol η and pol κ).30 The objectives of this exploratory work were to determine if (1) the TLS characteristics of the same G[8-3]T lesion in the different G*T* and G*CT* sequence contexts that are known to distort DNA to different extents,31, 32 and (2) to compare the observed TLS phenomena with those observed in the case of the isomeric G[8-5m]T and G[8-5]T lesions,15, 27, 33, 34 and other well-known IntraCL lesions derived from UV irradiation and cisplatin reactions with DNA.
2 Materials and methods
2.1 Materials
The 17-mer 5’-CCACCAACG*CT*ACCACC and 5’-CCACCAACG*T*CACCACC-3’ oligonucleotides containing the G*CT* or G*T* lesions were synthesized by oxidation of the parent sequences with photochemically generated carbonate radical anions.19, 23, 32 The integrity of the G*CT* and G*T* modified oligonucleotides purified by anion-exchange HPLC was confirmed by MALDI-TOF/MS spectroscopy and enzymatic excision of dG*-dT* (from G*CT* sequence) and d(G*pT*) (from G*T* sequence) dinucleotides with nuclease P1 and alkaline phosphatase as described by Crean et al.19 The 17-mers were extended to the 26-mer or 49-mer oligonucleotides (Fig. 1) using standard ligation methods with complementary strands as templates and purified by denaturing gel electrophoresis as described earlier.32
2.2 Polymerases
The high-fidelity A-family27 and the archaeal polymerase IV from Sulfolobus sulfataricus (Dpo4)33 were kindly provided by Drs. Lorena Beese and Roger Woodgate, respectively. The human TLS polymerases and pol κ were purchased from Enzymax LLC (Lexington, KY, USA).
2.3 Running start primer extension assays
The 16-mer or 18-mer primers were 5’-32P-end-labeled using T4 polynucleotide kinase and [γ-32P]ATP, and then annealed to the unmodified 29-mer or 49-mer oligonucleotides without or with the G*T* or G*CT* lesions (Fig. 1). The annealing step was conducted with a 10% excess of the template strand by heating the sample solution to 90 °C, followed by slow cooling of the solution to room temperature. The primer extension reactions were conducted at temperatures of 37 and 55 °C in the case of the thermophilic polymerases BF and Dpo4, and at 30 °C in the case of pol η and pol κ. The template-primer DNA concentrations were ~10 nM in solutions of 25 mM potassium phosphate (pH 7.0), 5 mM MgCl2, 5 mM dithiothreitol, 100 µg/mL bovine serum albumin, 10% glycerol, 100 µM of each of the four dNTPs. The concentrations of DNA polymerases were 2 nM (BF), 20 nM (Dpo4), 60 nM (pol η), and 10 nM (pol κ), and these experiments were performed in triplicates.
2.4 Denaturing gel electrophoresis assays
The polymerase-catalyzed primer extension reactions were quenched by aliquots of denaturing loading buffer (95% formamide with 20 mM EDTA, 45 mM Tris borate, 0.1% bromphenol blue, and 0.1% xylene cyanol). The reaction products were resolved on a 20% polyacrylamide denaturing gel in the presence of 7 M urea. The dried gels were exposed to Storage Phosphor Screens contained in appropriate cassettes overnight and then analyzed using a Storm 840 Phosphorimager (Molecular Dynamics, Inc.) and quantitated using ImageQuant software.
3 Results
3.1 Replicative A-family polymerase BF
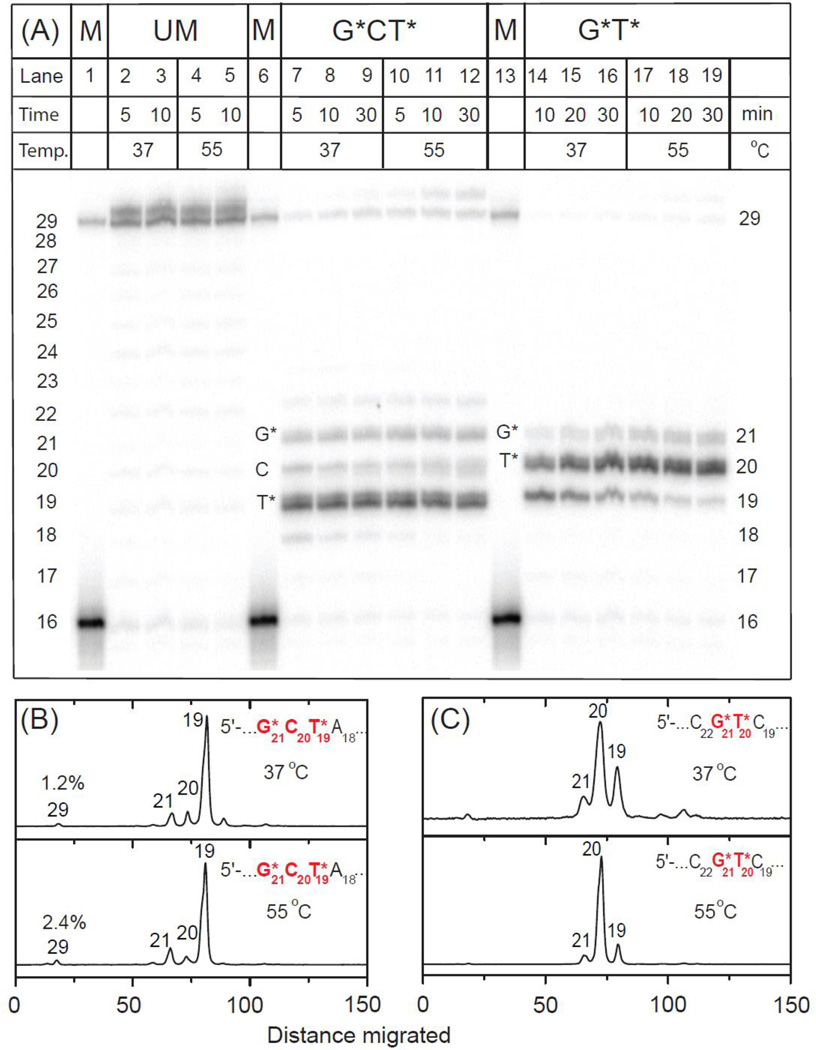
A typical extension experiment of the initial 16-mer primer strand catalyzed by the thermophilic A-family polymerase BF is shown in Fig. 2. It is evident that this replicative polymerase is mostly stalled after inserting a nucleotide opposite the first cross-linked T19* template base in the case of the G*CT19* lesion (Fig. 1B), thus resulting in a dominant 19-mer stalled extension product. A similar stalling phenomenon was also observed after insertion of a nucleotide opposite the cross-linked C* by the A-family polymerase T7− in the case of the G[8-5]C lesion.17 Much smaller extents of nucleotide incorporation are observed opposite the two subsequent template sites C20 and G21* (Fig. 2B). Increasing the temperature from 37 to 55 °C does not significantly change this product distribution, except that the fraction of fully extended 29-mer and somewhat longer extension products increase from ~ 1.2% at 37 °C to ~ 2.4% at 55 °C; the formation of smaller amounts of blunt-end 30-mer primer extension products is not unusual (Fig. 2A).16, 35, 36
Fig. 2.
Panel A: Running-start primer extension on 29-mer unmodified or modified templates with G*CT* and G*T* lesions catalyzed by BF polymerase. In the presence of all four dNTPs 5’-32P-labeled 16-mer primer (Fig. 1) was extended by BF polymerase (see text for experimental conditions). GCT unmodified template (UM): lanes 2 and 3, incubation times 5 and 10 min at 37 °C; lanes 4 and 5, incubation time 5 and 10 min at 55 °C (the results for the unmodified GT are not shown because they are indistinguishable from those shown for the unmodified GCT sequence). G*CT* template: lanes 7 – 9, incubation time 5, 10 and 30 min at 37 °C; lanes 10 – 12, incubation time 5, 10 and 30 min at 55 °C. G*T* template: lanes 14 – 16, incubation time 10, 20 and 30 min at 37 °C; lanes 17 – 19, incubation time 10, 20 and 30 min at 55 °C. The 29-mer would constitute a fully extended primer and its position together with the unextended 16-mer are shown in the lanes 1, 6 and 13. Panels B and C: Histograms of the denaturing gel autoradiograph (Panel A) for extension of 16-mer primers on the G*CT* (B) and G*T* (C) templates catalyzed by BF polymerase. In each case, the profiles are derived from the respective lanes for 30 min incubation time at 37 and 55 °C. Repeat experiments showed similar inefficient primer extension yields to those indicated at the 30 min time points (1.5±0.3% at 37 °C and 2.6±0.4% at 55 °C in the case of G*CT*, while the yields were negligible in the case of G*T* under the same conditions).
In the case of the G*T20* lesion at 37 °C, primer extension generates mostly 19 and 20-mers, with the 20-mers being more abundant. These products arise from the insertion of nucleotides opposite C19 and T20* (Figs. 2A and 2C). Under the same conditions, but at 55 °C, the 19-mer product is almost fully converted to the 20-mer extension product indicating that the nucleotide insertion process opposite T20* is slower than opposite C19. However, nucleotide insertion opposite G21* to yield the 21-mer extension product is still inhibited at the higher temperature (Fig. 2C). Thus, the primer extension beyond the cross-linked template base T* is strongly inhibited in the G*T20* template, thus yielding negligible levels of full 29-mer primer extension products (<0.5%) (Figs 2A and 2C). In summary, in both the G*CT* and G*T* cases, the slowest insertion steps are the primer extension steps beyond the cross-linked thymines T19* and T20*, respectively.
3.2 Y-family polymerase Dpo4
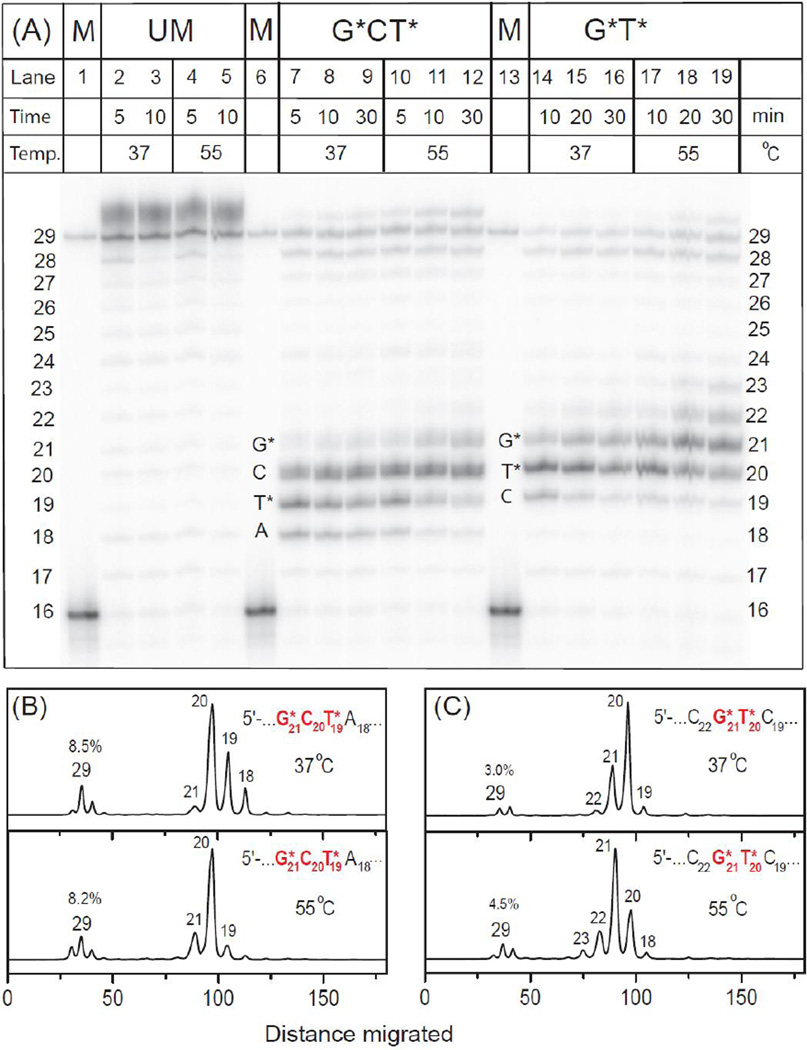
Typical gel autoradiograph of primer extension phenomena are shown in Fig. 3A. The histograms derived from the denaturing gel autoradiograph show that in the case of the G*CT* template the yields of the fully extended 29-mer primer are higher than in the experiments with the high fidelity polymerase BF, and attain ~ 8.0% at 37 – 55 °C (Fig. 3B). The appearance of intermediate primer extension products catalyzed by Dpo4 is distributive as shown, for example, by Vyas et al.37, 38 Thus, the fainter bands of products shown in Fig. 3A can be identified by counting the bands generated by the successive nucleotide insertion steps and primer extension product within the range of 18 – 29 nucleotides in lengths. The primer extension product accumulation is greatest when the primer 3’-terminal nucleotide is positioned opposite the C20 template bases in the G21*C20T19* strand at 37 °C and at 55 °C, thus suggesting that nucleotide incorporation opposite G21* is the rate-determining step (Fig. 3B).
Fig. 3.
Panel A: Running-start primer extension on 29-mer unmodified or modified templates with G*CT* and G*T* lesions catalyzed by Dpo4 polymerase. In the presence of all four dNTPs 5’-32P-labeled 16-mer primer (Fig. 1) was extended by Dpo4 polymerase (see text for experimental conditions). GCT unmodified template (UM): lanes 2 and 3, incubation times 5 and 10 min at 37 °C; lanes 4 and 5, incubation time 5 and 10 min at 55 °C. G*CT* template: lanes 7 – 9, incubation time 5, 10 and 30 min at 37 °C; lanes 10 – 12, incubation time 5, 10 and 30 min at 55 °C. G*T* template: lanes 14 – 16, incubation time 10, 20 and 30 min at 37 °C; lanes 17 – 19, incubation time 10, 20 and 30 min at 55 °C. The 29-mer would constitute a fully extended primer and its position together with the unextended 16-mer are shown in the lanes 1, 6 and 13. Panels B and C: Histograms of the denaturing gel autoradiograph (Panel A) for extension of 16-mer primers on the G*CT* (B) and G*T* (C) templates catalyzed by Dpo4 polymerase. In each case, the profiles are derived from the respective lanes for 30 min incubation time at 37 and 55 °C. The averages of three trials were similar at both temperature (7.0±2.0% and 3.0±0.6% in the case of G*CT* and G*T*, respectively).
In the case of the G*T* template at 37 °C, Dpo4 is partially stalled opposite T20 at 37 °C and opposite G21* at 55 °C (Fig. 3A). Thus, there is an accumulation of primer extension products after insertion of a nucleotide opposite T20* or G21*, respectively. The yields of the fully extended 29-mer primers are 3.0% at 37 °C and 4.5% at 55 °C (Fig. 3C), which is lower than in the case of the G*CT* template strand under the same conditions.
3.3 The human polymerases pol η and pol κ
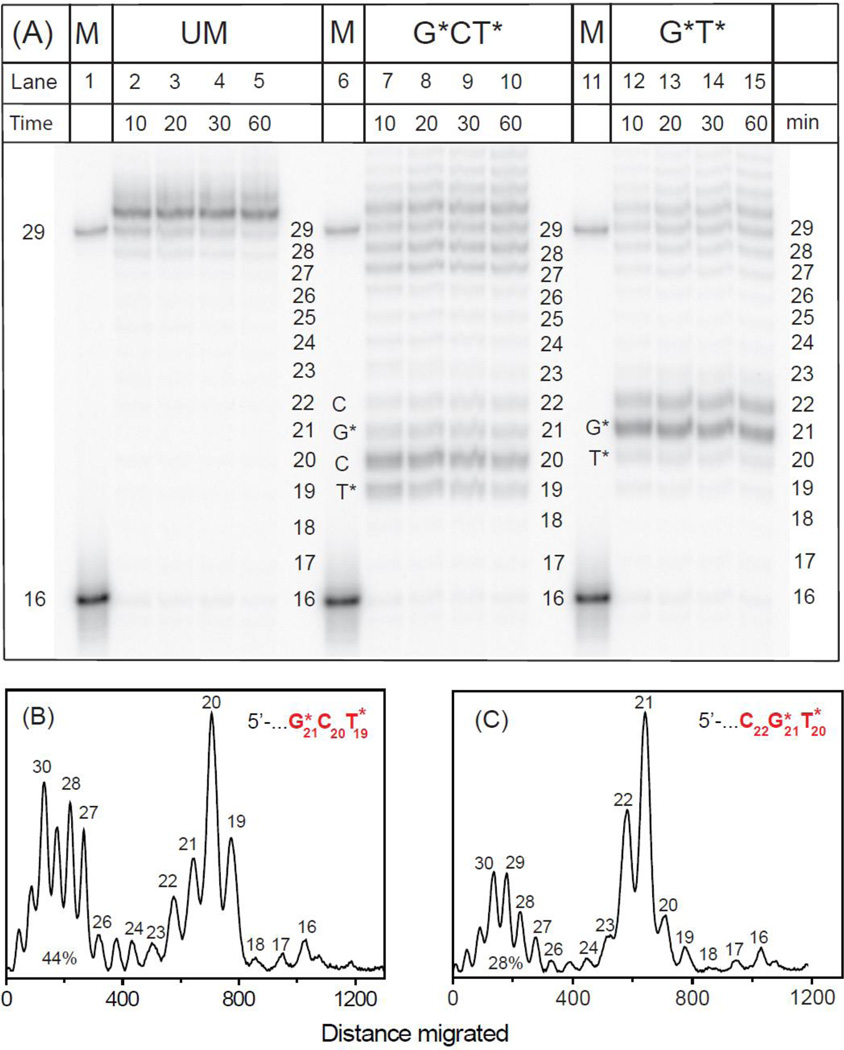
The TLS of both types of IntraCL catalyzed by pol η at 30 °C is significantly more efficient than in the case of BF and DpO4. As in the case of Dpo4, there is an accumulation of 19 – 20-mer primer extension products after the slow insertion of nucleotides at the cross-linked T19* → C20 step, a prominent accumulation of C20 extension products, followed by a slower C20 → G21* extension step. Smaller product accumulations after nucleotide insertions opposite G21* and C22 are also observed (Fig. 4A,B). Primer extension is markedly distributive with prominent amounts of 27 – 30-mer extension products formed (28–44%), including several blunt-end extension products more than 29 nucleotides in lengths (~10%); similar distributive and blunt-end primer extension products have been reported in the literature.12 The histograms derived from the denaturing gel autoradiographs show that in the case of G*T* primer, the extension patterns are similar, although the yields of products > 27 nucleotides in lengths (~28%, Fig. 4C) are lower than in the case of the G*CT* templates (~44%, Fig. 4B). The T20 → G21* step is the slowest, and the G21* → C22 step is also slow. Extension beyond the cross-linked C20 is the slowest, rate-determining step (Fig. 4C), as it is in the case of the G*CT* template (Fig. 3B). Similar in vitro full-length primer extension patterns catalyzed by pol η past G[8-5m]T and G[8-5]C cross-linked lesions were also observed by Colis et al.14 and Gu and Wang,12 respectively.
Fig. 4.
Panel A: Running-start primer extension on 29-mer unmodified or modified templates with G*CT* and G*T* lesions catalyzed by pol η polymerase. In the presence of all four dNTPs 5’-32P-labeled 16-mer primer (Fig. 1) was extended by pol η polymerase (see text for experimental conditions) at 30 °C. GCT unmodified template (UM): lanes 2 – 5, incubation time 10, 20, 30 and 60 min. G*CT* template: lanes 7 – 10, incubation time 10, 20, 30 and 60 min. G*T* template: lanes 12 – 15, incubation time 10, 20, 30 and 60 min. The 29-mer would constitute a fully extended primer and its position together with the unextended 16-mer are shown in the lanes labeled ctrl. Panel B and C: Histograms of the denaturing gel autoradiograph shown in Panel A for extension of 16-mer primers on the G*CT* (B) and G*T* (C) templates catalyzed by pol η polymerase. In each case, the profiles are derived from the respective lanes for 30 min incubation time, and the percent lesion bypass (defined as the sum of all primer extension products 27 – 32 nucleotides long is also shown). The averages of three trials were 45±5% and 28±4% for the G*CT* and G*T* template strand sequences, respectively.
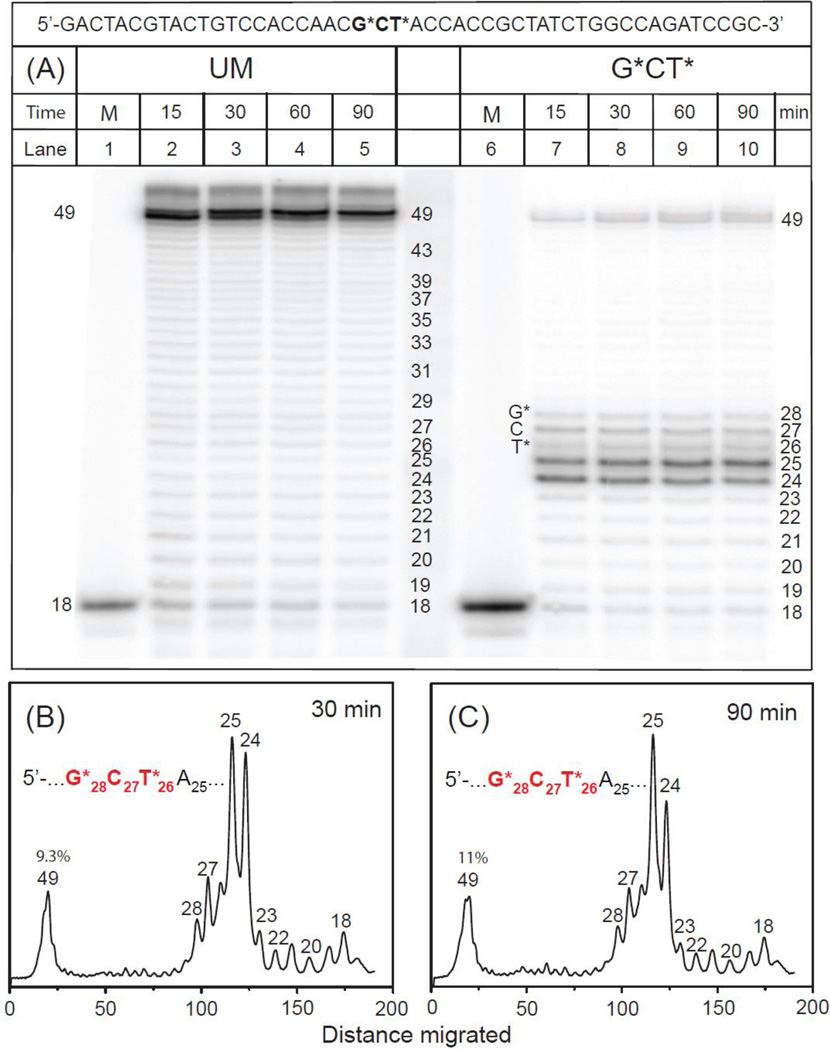
We also investigated TLS catalyzed by pol κ, an ortholog of the archeal polymerase Dpo4.30 The primer extension patterns catalyzed by pol κ with the same central 17-mer 3’-…CCACCAACG*CT*ACCACC… sequence used in the other experiments, but embedded in a longer 49-mer template – 18-mer primer strand duplex was also explored (the longer sequence became available from another project; we surmised that the TLS activity should not be affected significantly by the greater length, relative to the lengths of the 29-mer templates because pol κ grips only 9 – 10 base pairs at a time).39 The overall pattern of translesion bypass of the lesion-containing segment of the template strand 3’-…C29G28*C27T26*A25C24… catalyzed by pol κ exhibited major stalling sites at the unmodified template bases C24 and A25, just before the cross-linked template base T26*(Fig. 5). While the overall efficiency of full primer extension is similar in the case of pol κ and Dpo4 (Fig. 3B and 5B), the stalling patterns are different. In the case of Dpo4 translesion synthesis, stalled products accumulate after nucleotide insertions opposite T19* and C20, with lesser accumulations after insertion opposite A18 in the G21*C20T19*A18 sequence context (Fig. 3B). Finally, in the case of pol κ, the 25-mer/24-mer primer extension product ratio is slightly greater after a 90 min (Fig. 5C) than after a 30 min (Fig. 5B) incubation time, thus indicating that slow nucleotidyl transfer reactions continue to occur, albeit slowly, beyond 30 min.
Fig. 5.
Panel A: One-base incorporation opposite the adduct site catalyzed by pol κ polymerase. In the presence of all four dNTPs 5’-32P-labeled 18-mer primer 5’-GCGGATCTGGCCAGATAG was extended by pol κ polymerase (see text for experimental conditions) at 30 °C. Unmodified template (UM): lanes 2 – 5, incubation time 15, 30, 60 and 90 min. G*CT* template: lanes 7 – 10, incubation time 15, 30, 60 and 90 min. The 49-mer constitutes a fully extended primer. Positions of the unextended 18-mer are shown in the lanes 1 and 6. Panel B and C: Histograms of the denaturing gel autoradiograph (Panel A) for extension of 18-mer primers on the G*CT* template catalyzed by pol κ polymerase. In each case, the profiles are derived from the respective lanes for 30 min (B), 10 ± 2% average full extensions, and for 90 min (Panel C).
4 Discussion
4.1 Replicative Polymerases
Replicative polymerases are known to be strongly blocked by the IntraCL G[8-5]C and G[8-5m]T lesions that are formed by gamma irradiation pathways.12, 13, 15, 17, 18 The oxidatively generated and isomeric IntraCL G[8-3]T lesions strongly block the replicative polymerase BF; full bypass is completely inhibited in the case of G*T*, and is only ~ 1 – 2% in the case of the G*CT* sequence (Fig. 2). This is consistent with the results obtained with the G[8-5]C12, 17 and G[8-5m]T13, 18 IntraCL lesions that strongly inhibit a series of other replicative A-family polymerases.
4.2 The Y-family polymerases Dpo4 and pol κ
Dpo4 is the archaeal ortholog of human pol κ30 that has a flexible, spacious and solvent-exposed active site.29 In the case of our G[8-3]T lesion in the G21*T20* sequence context, primer extension is slowed significantly from T20* → G21*, and even more strongly beyond G21*, thus leading to the low (~ 8%) observed full primer extension (Fig. 3). In the case of the G[8-3]T lesion positioned in the G21*C20T19* sequence context, there is an accumulation of primer extension products after nucleotide insertions opposite T19* and especially opposite the unmodified C20. This indicates that the primer extension step C20 → G21* is the slowest one. In the case of the G21*T20* lesion, nucleotide insertion is more facile opposite T20* than opposite G21*, while primer extension beyond G21* is the slowest step at 37 °C (Fig. 3C).
Dpo4 has the ability to bypass a variety of other DNA lesions,40 including the intrastrand cisplatin-derived cis-Pt-G*G* lesions.41, 42 While the bypass of the cis-Pt-G*G* lesions was significant, prominent pause sites were observed after the successful insertion of nucleotides opposite the 5’-template G*, and to a lesser extent opposite the upstream 3’-G*. Similar to the results observed with the G*T* lesion, in the case of cis-Pt-G*G*, nucleotide insertion opposite the first cross-linked 5’-G* was more efficient than opposite the second cross-linked G* base.41, 42
Another extensively studied IntraCL is the cis-syn cyclobutane thymine-thyme dimer T*T*, which is bypassed inefficiently by Dpo4 because of the slow insertion efficiencies across the first cross-linked T* encountered by the polymerase.43
Pol κ is well known for bypassing primarily N2-guanine lesions,31, 44 and is also known to be involved in the bypass of interstrand cross-linked lesions.3, 45 In the case of the G[8-3]T lesion in the G28*C27T26*A25C24 sequence context, significant amounts of extension products accumulate with the 3’-end nucleotide of the primer strand opposite C24 and A25, indicating that the A25 → T26* is the slowest step. The overall full-length bypass is in the range of 9 – 11% under the specified reaction conditions (Fig. 5). Similarly, the cis-syn T*T* thymine dimers are not bypassed by pol κ in vitro,31 and it has been proposed that pol κ is involved only in the extension step beyond the cross-linked thymines.46
4.3. Pol η bypasses G[8,3]T, G[8,5]T and other intrastrand cross-linked DNA lesions
Pol η was the most efficient polymerase in bypassing the G[8-3]T lesions in both sequence contexts (Fig. 4). It is interesting to compare the human pol η TLS patterns obtained with the G[8-3]T lesion in the G*T* sequence context with those observed in the case of the related G[8-5m]T IntraCL generated by gamma irradiation; in the case of the G[8-5m]T and G[8-5]C lesions, significant accumulations of partially extended primer strands are observed after the insertion of nucleotides opposite the cross-linked T* and G* template bases.12–15 These results indicate that the insertion of nucleotides opposite T* by human (and yeast) pol η is more efficient than opposite G*, while primer extension beyond the cross-linked G* is the slowest step. To summarize, both of the IntraCLs, G[8-3]T and G[8-5m]T are bypassed by pol η, although less efficiently than the unmodified control sequences, and in both cases a significant fraction of partially extended products are observed, including blunt end extension products.
Pol η is best known for efficiently bypassing the IntraCL cis-syn CPD thymine dimers in an error-free manner,34, 43, 47, 48 but is stalled by the 6-4 pyrimidine-pyrimidone thymine-thymine photoproduct.48 On the other hand, the cisplatin-derived IntraCL product Pt-G*G* is partially bypassed by pol η.48, 49 It has been proposed that the bypass of different IntraCL lesions by pol η depends strongly on the abilities of the cross-linked nucleotides to form proper hydrogen bonds with the incoming dNTPs, the structural features of the primer-template junction at the polymerase active site, and the positions of the cross-linked bases.43, 49, 50
It is of interest to consider the structural differences between the G[8,3]T and cisplatin derived IntraCL that may account for the different bypass efficiencies catalyzed by pol η. The Pt-G*TG* lesion is more poorly bypassed by pol η than the Pt-G*G* lesion, because pol η fails to insert nucleotides either across the first cross-linked 3’-side G* and across the following T.51 These results suggest that the rigid and square-planar Pt coordination structure displaces the intervening thymine as well as the 3’-G* that is connected to the 5’-G* from their normal positions at the active site of the polymerase. These unfavorable alignments lower the overall bypass of the cisplatin-derived Pt-G*TG* lesions,51 while in the Pt-G*G* case the absence of the T may allow for a more favorable alignment of the rigid cis-Pt-G*G* structure for translesion bypass. In the case of the G*CT* and G*T* lesions, the absence of a bulk substituent like cis-Pt is more favorable for primer extension. In contrast to the cisplatin modified cis-Pt-G*TG*/-G*G* sequences, the G[5,3]T lesion in the G*CT* sequence context is better bypassed by pol η than in the G*T* sequence (Fig.4), as discussed in the following section.
4.4 More efficient bypass of G[8,3]T lesions in the G*CT* than the G*T* sequence context
The overall bypass catalyzed by pol η or Dpo4 of the G[8-3]T IntraCL lesion in the G*CT* sequence is more efficient than in the case of the G*T* template sequences (Figs. 3 and 4). This suggests that an unmodified C positioned between the G* and T* cross-linked bases facilitates the overall bypass of these oxidatively generated cross-linked lesions.
Here we examine the structural features of these lesions that might account for these observations. The sterically and energetically feasible conformations of the G*T* and G*CT* were analyzed by molecular modeling, free energy calculations, and molecular dynamic simulations of G*T* and G*CT* in double-stranded DNA by Ding et al.32 In the case of G*CT*, eight energetically feasible conformations were identified, while in the case of G*T* there were only two. It was concluded from these findings that the hindered rotation about the G*-T* cross-linked bond limits the conformational possibilities in the case of G*T*, while in the case of G*CT* more flexibility is permitted in the DNA backbone because of the intervening deoxycytidine nucleotide in G*CT*. These are intrinsic properties of the G*T* and G*CT* lesions that occur in the absence of the secondary structure of the DNA. In the case of G*CT* lesions, multiple configurations at the active sites are possible, some being favorable for TLS and some not. An example of a lack of flexibility in TLS is the crystallographic structure of the cis-Pt-G*G* lesions positioned at the active site of pol η published by Alt et al;49 two structures were identified, one of these structures has the correct alignment of residues for primer extension, while the other does not. In an analogous manner, we propose the hypothesis that the G*CT* can assume more different configurations than G*T*, including some that are favorable for TLS, thus allowing for an overall higher yield of TLS primer extension products.
5 Conclusions
The intra-strand cross-linked G[8,3]T lesions in two different sequence contexts G*CT* and G*T* represent strong blocks of bypass catalyzed by the replicative thermophilic polymerase BF at 37 or 55 °C in vitro. The archaeal polymerase Dpo4 and its human ortholog pol κ are also inhibited by the cross-linked G* and T* lesions, but a small amount of full primer extension is nevertheless observed in the range of ~ 7 – 11% under similar incubation conditions. By contrast, a much stronger but highly distributive pattern of translesion bypass catalyzed by the Y-family polymerase pol η is observed; the sum of primer extension products varying in lengths from two nuclotides shorter than the fully extended primer, to three nucleotide-longer blunt-end extension products, varied from 28±4% in the case of the G*T* to 42 ± 5% in the case of the G*CT* sequences. The translesion bypass is consistently lower in the case of the G*T* lesion. It is proposed that this difference is associated with the rigidity of the sterically hindered G*T* cross-link, while the intervening unmodified cytosine base imparts some flexibility to the G*CT* lesions that can assume multiple configurations,32 some of which are favorable for lesion bypass.
Acknowledgments
This work was supported by the National Institute of Environmental Health Sciences Grant R01 ES 011589. Components of this work were conducted in the Shared Instrumentation Facility at NYU that was constructed with support from a Research Facilities Improvement Grant (C06 RR-16572) from the National Center for Research Resources, National Institutes of Health. The acquisition of the MALDI-TOF mass spectrometer was supported by the National Science Foundation (CHE-0958457).
Footnotes
Author’s contribution
YAL carried out the running start primer extension assays, YCL performed the one-base nucleotide opposite the adduct site experiments, and VS and NEG supervised the experimental work and wrote the manuscript. All authors approved the final version of the manuscript.
References
- 1.Wang Y. Chem. Res. Toxicol. 2008;21:276–281. doi: 10.1021/tx700411g. [DOI] [PubMed] [Google Scholar]
- 2.Cadet J, Ravanat JL, TavernaPorro M, Menoni H, Angelov D. Cancer Lett. 2012;327:5–15. doi: 10.1016/j.canlet.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Ho TV, Scharer OD. Environ. Mol. Mutagen. 2010;51:552–566. doi: 10.1002/em.20573. [DOI] [PubMed] [Google Scholar]
- 4.Jung Y, Lippard SJ. Chem. Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 5.Friedberg EC, Graham WC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis, Second Edition. Washington, DC: ASM Press; 2006. [Google Scholar]
- 6.Dizdaroglu M. Biochem. J. 1986;238:247–254. doi: 10.1042/bj2380247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romieu A, Bellon S, Gasparutto D, Cadet J. Org. Lett. 2000;2:1085–1088. doi: 10.1021/ol005643y. [DOI] [PubMed] [Google Scholar]
- 8.Box HC, Budzinski EE, Dawidzik JB, Gobey JS, Freund HG. Free Radic. Biol. Med. 1997;23:1021–1030. doi: 10.1016/s0891-5849(97)00166-4. [DOI] [PubMed] [Google Scholar]
- 9.Bellon S, Ravanat JL, Gasparutto D, Cadet J. Chem. Res. Toxicol. 2002;15:598–606. doi: 10.1021/tx015594d. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Q, Wang Y. J. Am. Chem. Soc. 2003;125:12795–12802. doi: 10.1021/ja034866r. [DOI] [PubMed] [Google Scholar]
- 11.Box HC, Patrzyc HB, Dawidzik JB, Wallace JC, Freund HG, Iijima H, Budzinski EE. Radiat. Res. 2000;153:442–446. doi: 10.1667/0033-7587(2000)153[0442:dblidx]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 12.Gu C, Wang Y. Biochemistry. 2004;43:6745–6750. doi: 10.1021/bi0497749. [DOI] [PubMed] [Google Scholar]
- 13.Jiang Y, Hong H, Cao H, Wang Y. Biochemistry. 2007;46:12757–12763. doi: 10.1021/bi7012195. [DOI] [PubMed] [Google Scholar]
- 14.Colis LC, Raychaudhury P, Basu AK. Biochemistry. 2008;47:8070–8079. doi: 10.1021/bi800529f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raychaudhury P, Basu AK. J. Nucleic Acids. 2010;2010 doi: 10.4061/2010/101495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raychaudhury P, Basu AK. Biochemistry. 2011;50:2330–2338. doi: 10.1021/bi102064z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu C, Wang Y. Biochemistry. 2005;44:8883–8889. doi: 10.1021/bi050036+. [DOI] [PubMed] [Google Scholar]
- 18.Bellon S, Gasparutto D, Saint-Pierre C, Cadet J. Org. Biomol. Chem. 2006;4:3831–3837. doi: 10.1039/b609460k. [DOI] [PubMed] [Google Scholar]
- 19.Crean C, Uvaydov Y, Geacintov NE, Shafirovich V. Nucleic Acids Res. 2008;36:742–755. doi: 10.1093/nar/gkm1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crean C, Lee YA, Yun BH, Geacintov NE, Shafirovich V. Chem Bio Chem. 2008;9:1985–1991. doi: 10.1002/cbic.200800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crean C, Geacintov NE, Shafirovich V. Free Radic. Biol. Med. 2008;45:1125–1134. doi: 10.1016/j.freeradbiomed.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yun BH, Geacintov NE, Shafirovich V. Chem. Res. Toxicol. 2011;24:1144–1152. doi: 10.1021/tx200139c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rokhlenko Y, Geacintov NE, Shafirovich V. J. Am. Chem. Soc. 2012;134:4955–4962. doi: 10.1021/ja212186w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lonkar P, Dedon PC. Int. J. Cancer. 2011;128:1999–2009. doi: 10.1002/ijc.25815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cadet J, Wagner JR, Shafirovich V, Geacintov NE. Int. J. Radiat. Biol. 2014;90:423–432. doi: 10.3109/09553002.2013.877176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madugundu GS, Wagner JR, Cadet J, Kropachev K, Yun BH, Geacintov NE, Shafirovich V. Chem. Res. Toxicol. 2013;26:1031–1033. doi: 10.1021/tx400158g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiefer JR, Mao C, Braman JC, Beese LS. Nature. 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- 28.Kiefer JR, Mao C, Hansen CJ, Basehore SL, Hogrefe HH, Braman JC, Beese LS. Structure. 1997;5:95–108. doi: 10.1016/s0969-2126(97)00169-x. [DOI] [PubMed] [Google Scholar]
- 29.Boudsocq F, Iwai S, Hanaoka F, Woodgate R. Nucleic Acids Res. 2001;29:4607–4616. doi: 10.1093/nar/29.22.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sale JE, Lehmann AR, Woodgate R. Nat. Rev. Mol. Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Yuan F, Wu X, Wang M, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding S, Kropachev K, Cai Y, Kolbanovskiy M, Durandina SA, Liu Z, Shafirovich V, Broyde S, Geacintov NE. Nucleic Acids Res. 2012;40:2506–2517. doi: 10.1093/nar/gkr1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pages V, Fuchs RP. Oncogene. 2002;21:8957–8966. doi: 10.1038/sj.onc.1206006. [DOI] [PubMed] [Google Scholar]
- 34.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clark JM, Joyce CM, Beardsley GP. J. Mol. Biol. 1987;198:123–127. doi: 10.1016/0022-2836(87)90462-1. [DOI] [PubMed] [Google Scholar]
- 36.Zhuang P, Kolbanovskiy A, Amin S, Geacintov NE. Biochemistry. 2001;40:6660–6669. doi: 10.1021/bi010005o. [DOI] [PubMed] [Google Scholar]
- 37.Vyas R, Efthimiopoulos G, Tokarsky EJ, Malik CK, Basu AK, Suo Z. J. Am. Chem. Soc. 2015;137:12131–12142. doi: 10.1021/jacs.5b08027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pryor WA, Squadrito GL. Am. J. Physiol. 1995;268:L699–L722. doi: 10.1152/ajplung.1995.268.5.L699. [DOI] [PubMed] [Google Scholar]
- 39.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Mol. Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 40.Maxwell BA, Suo Z. Biochemistry. 2014;53:2804–2814. doi: 10.1021/bi5000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown JA, Newmister SA, Fiala KA, Suo Z. Nucleic Acids Res. 2008;36:3867–3878. doi: 10.1093/nar/gkn309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong JH, Brown JA, Suo Z, Blum P, Nohmi T, Ling H. EMBO J. 2010;29:2059–2069. doi: 10.1038/emboj.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson RE, Prakash L, Prakash S. Proc. Natl. Acad. Sci. U S A. 2005;102:12359–12364. doi: 10.1073/pnas.0504380102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H. Genes Dev. 2000;14:1589–1594. [PMC free article] [PubMed] [Google Scholar]
- 45.Minko IG, Harbut MB, Kozekov ID, Kozekova A, Jakobs PM, Olson SB, Moses RE, Harris TM, Rizzo CJ, Lloyd RS. J. Biol. Chem. 2008;283:17075–17082. doi: 10.1074/jbc.M801238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yagi Y, Ogawara D, Iwai S, Hanaoka F, Akiyama M, Maki H. DNA Repair (Amst) 2005;4:1252–1269. doi: 10.1016/j.dnarep.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 47.Johnson RE, Prakash S, Prakash L. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 48.Masutani C, Kusumoto R, Iwai S, Hanaoka F. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alt A, Lammens K, Chiocchini C, Lammens A, Pieck JC, Kuch D, Hopfner KP, Carell T. Science. 2007;318:967–970. doi: 10.1126/science.1148242. [DOI] [PubMed] [Google Scholar]
- 50.Ummat A, Rechkoblit O, Jain R, Roy Choudhury J, Johnson RE, Silverstein TD, Buku A, Lone S, Prakash L, Prakash S, Aggarwal AK. Nat. Struct. Mol. Biol. 2012;19:628–632. doi: 10.1038/nsmb.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chijiwa S, Masutani C, Hanaoka F, Iwai S, Kuraoka I. Carcinogenesis. 2010;31:388–393. doi: 10.1093/carcin/bgp316. [DOI] [PubMed] [Google Scholar]