Abstract
Lysosomes are membrane-bound intracellular organelles that receive macromolecules delivered by endocytosis, phagocytosis, and autophagy for degradation and recycling. Over the last decade, advances in lysosome research have established a broad role for the lysosome in the pathophysiology of disease. In this review, we highlight the recent discoveries in lysosome biology, with an emphasis on their implications for cancer therapy. We focus on targeting the lysosome in cancer by exploring lysosomal biogenesis and its role in the crosstalk between apoptosis and autophagy. We also discuss how lysosomal inhibition could emerge as a new therapeutic strategy to overcome drug resistance in cancer.
Keywords: lysosome, cell death, autophagy, apoptosis, cancer therapy, drug resistance
Introduction
Over the past several years, there have been exciting developments in cancer therapy. However, the acquisition of resistance to most cancer therapeutics is almost inevitable and ultimately limits the successful treatment of advanced malignancies. Tumor cells contain a very high degree of molecular heterogeneity within their microenvironment; drug resistance can gradually emerge through therapy-induced selection of a resistant subpopulation of cells in the original tumor.1 Therefore, personalized orthogonal therapies that target totally different pathways may be necessary. The application of genomic, postgenomic, proteomic, and other modern analytical techniques, especially the use of high-throughput techniques in combination with the methodologies of bioinformatics and systems biology, have resulted in significant progress in identifying novel targets that are involved in drug resistance. Among them, targeting the lysosome in cancer emerges as an attractive approach to overcome drug resistance. Over the past few decades, research on the molecular regulation of lysosomes has advanced quickly. It is now recognized that lysosomes play a critical role in fundamental cellular processes, such as protein secretion, endocytic receptor recycling, energy metabolism, and cell signaling.2 In addition, the critical role of the lysosome in autophagy has important practical implications for developing autophagy inhibitors as cancer therapeutics. The autophagy–lysosome pathway is intimately associated with the hallmarks of cancer, such as escaping cell death pathways, evading immune surveillance, and deregulating metabolism.1 Targeting lysosomes therefore has great therapeutic potential in cancer, because it not only triggers apoptotic and lysosomal cell death pathways but also inhibits cytoprotective autophagy. Furthermore, lysosomes play an important role in cancer drug resistance by sequestering cancer drugs in their acidic environment, resulting in a blunting of the drugs’ effects.3 Therefore, the lysosome could be considered an Achilles’ heel of cancer cells.
Lysosomal proteins in cancer
The term lysosome is derived from the Greek word for digestive body and describes degradative organelles present in all eukaryotic cells. Lysosomes are surrounded by a lipid protein membrane and contain different hydrolases. The basic function of lysosomes is to digest extracellular material that has been internalized by endocytosis and intracellular components that have been sequestered by autophagy. They recycle the unwanted cellular material as energy, providing a nutrient source for maintaining cellular homeostasis. Lysosomes were first discovered in 1955 by Christian de Duve, who later won the Nobel Prize in Physiology or Medicine for his discovery. Since then, lysosomes have mainly been considered to be digestive sacs made of a lipoprotein membrane and filled with various hydrolases for carrying out their basic digestive function. Although its digestive role has been appreciated for more than 50 years, the lysosome is now increasingly recognized as a highly advanced organelle with more complex functions. Lysosomes not only receive extracellular material through the endocytic pathway or intracellular material via autophagy but also secrete their contents by fusing with the plasma membrane.4 It is this two-way trafficking that makes them highly dynamic in many fundamental cellular processes, such as cell death, signaling, immunity, and stress responses. Lysosomes contain two classes of proteins that are essential for their functions: soluble hydrolases and integral lysosomal membrane proteins. There are over 60 hydrolases that have been identified and characterized,5 which are produced in the endoplasmic reticulum (ER) and are transported to the Golgi apparatus, where they receive a mannose-6-phosphate tag that targets them for the lysosome.4 Lysosomal hydrolases are also referred to as acid hydrolases because they have optimal activity at the acidic pH 4–5 in the lysosome. But, in some cases, they can also function at a neutral pH outside of the lysosomes. For instance, some cathepsins can function in the cytosol to initiate cell death following lysosomal membrane permeabilization (LMP).6
The best studied lysosomal hydrolases are the cathepsin proteases, subdivided into three subgroups on the basis of the active site of amino acids, which confers the catalytic activity: cysteine cathepsins, serine cathepsins, and aspartic cathepsins. It has been suggested that, depending on the location of the cathepsins, they can either suppress or promote tumor growth. Specifically, the cytosolic cathepsins can suppress tumor growth through activation of the intrinsic apoptotic pathway. In contrast, the extracellular cathepsins can promote tumor growth through their ability to break down basement membrane and activate other protumorigenic proteins.7 In fact, cathepsins B, S, and E are all associated with cancer progression and metastasis in various cancer types.8–10
Though many research groups focus on the luminal lysosomal hydrolases, lysosomal membrane proteins have been found to have indispensable functions and thus may also have potential as targets for cancer therapeutics. More than 25 lysosomal membrane proteins have been identified,11 with the most abundant lysosomal membrane proteins being lysosome-associated membrane protein 1 (LAMP-1) and LAMP-2, which represent 50% of all membrane proteins of the lysosomal membrane.12 These proteins are essential for lysosomal biogenesis, lysosomal acidification, transportation of metabolites, as well as chaperone-mediated autophagy (CMA).11 LAMP-2A functions as a receptor of CMA in the lysosomal membrane that can be increased by reduced degradation and/or redistribution from the lysosomal lumen to the lysosomal membrane.13 LAMP-1 has been found on the cell surface of highly metastatic tumor cells, especially in metastatic colon cancer cells, suggesting a role for this protein in cell–cell adhesion and migration.14
Another important lysosomal membrane protein is vacuolar H+-ATPase (V-ATPase), a proton pump that uses the energy from ATP hydrolysis to pump hydrogen ions into the lumen, thereby creating the acidic pH of lysosomes. It was found that V-ATPase is an important regulator of endocytotic trafficking and affects the tumor microenvironment by proton extrusion into the extracellular medium.15 The acid environment favors tissue damage because of the activation of destructive enzymes in the extracellular matrix. The application of specific inhibitors of V-ATPase can decrease the acidity of tumors and may allow the reduction of tumor metastasis and prevent tumor progression.16 A recent study shows that V-ATPase also functions as a master effector of E2F1-mediated lysosomal trafficking, mTORC1 activation, and autophagy in tumor malignancy. These findings suggest that pharmacological intervention at the level of V-ATPase may be beneficial for the treatment of metastatic tumors overexpressing E2F1.17
Recently, acid sphingomyelinase (ASM) has emerged as another extensively studied hydrolase within the lysosome. ASM breaks down the sphingomyelin into the ceramide, which serves as a substrate for the generation of other sphingolipids, such as sphingomyelin and sphingosine 1-phosphate (S1P).18 Abnormal sphingolipid metabolism is involved in many cancers, with either decreased levels of ceramide, which functions as a proapoptotic lipid, or with increased levels of S1P, which functions as a proliferative lipid.19 As a result, many investigators have focused their efforts on finding agents that promote ceramide-mediated cell death.
Advantages of provoking LMP
LMP results in the release of lysosomal enzymes, especially cathepsins, into the cytosol and can give rise to necroptosis, autophagy, and apoptosis.20 Despite this advanced knowledge of the consequences of LMP, rigorous methods to track and measure LMP have only recently been reported.21 LMP is a complex process with different outcomes, depending on the level of permeabilization. It has been proposed that limited lysosomal enzyme release can result in lysosomal cell death and apoptosis, whereas extensive lysosomal enzyme release can result in necrosis. This necrotic type of cell death and lysosomal cell death are of major interest in cancer therapy, because cancer cells often acquire mutations in the apoptotic machinery protecting them from cell death by the classical apoptotic pathways.
Many stimuli can cause the release of cathepsins from the lysosomal lumen into the cytosol, which could have potential applications for targeting the lysosome in cancer. Among these stimuli, reactive oxygen species (ROS) may be the most commonly encountered within the tumor microenvironment and can induce LMP through activation of phospholipases A2, causing alteration of lysosome membranes by degradation of membrane phospholipids.22 Another important stimulus is sphingomyelin, which can be converted to ceramide and further to sphingosine by ASM and ceramidase.23 Accumulation of sphingosine, as well as ceramide, in lysosomes can induce LMP.24
There are numerous methods for quantifying LMP, such as detecting the enzymatic activity of lysosomal hydrolases in cytosol, visualizing LMP by tracking the imaging of lysosomes with fluorescent dextran, or, most recently, by monitoring LMP using fluorescent antibody probes against galcetins. Since LMP is a highly dynamic process and each assay has its own limitations with some intrinsic technical problems, accurate quantification of LMP is both biologically and technically challenging. Therefore, in order to obtain reliable results, carefully selecting an assay and/or using combined complementary methods is critical to correct interpretation of the results.20,21,25
In contrast, there are some molecules known to protect lysosomal membranes against permeabilization, such as the heat shock protein 70 (HSP70), LAMP-1, and LAMP-2.26 Among these, HSP70 has attracted the most attention and belongs to the HSP70 family, which contains both heat-inducible and constitutively expressed proteins. The latter are called heat-shock cognate (HSC) proteins and include HSC70, which can interact with LAMP-1 for CMA.27 Here, HSP70 is referred to as a heat-inducible protein, an evolutionarily conserved chaperone protein, which modulates many lysosomal proteins. HSP70 is expressed in many tumor types and can specifically bind to bis monoacylglycero phosphate in the lysosome lumen. This binding activates ASM, which breaks down the lipid sphingomyelin. It has been suggested that increased ASM activity supports lysosomal integrity; thus inhibition of ASM in cancer cells would increase lysosomal LMP and lead to cell death.28 An example of a novel HSP70 inhibitor is the small-molecule 2-phenylethyenesulfonamide (PES), which inhibits lysosomal function, inhibits autophagy, and promotes tumor cell death.29
The endosome–lysosome pathway and lysosome formation
The endosome–lysosome pathway can be viewed as the main means of trafficking and exchanging organelles and membrane components within the cell. Two sources of cell components via three routes are delivered to lysosomes for degradation. From inside the cell, intracellular components for lysosomal degradation are delivered to the lysosomes via autophagy, and from outside the cell, exogenous materials are delivered to the lysosomes via endocytosis and phagocytosis, which includes processes such as micropinocytosis or LC3-associated phagocytosis.4,30 Structurally, the trafficking vesicles can be categorized as early endosomes, with an intraluminal pH of 6.0–6.6; late endosomes, with a more acidic pH of 5; or lysosomes with the most acidic pH of 4.5. Lysosomes serve as the terminal station for the endosome–lysosome pathway. The cargo for endocytosis starts with the delivery of cellular components to the early endosome, where they are sorted. The majority of cargo and receptors are delivered back to the plasma membrane. In contrast, the cargo destined for degradation in lysosomes is retained in the early endosome and then converted into a late endosome, which has a multivesicular appearance, also known as multivesicular bodies, and finally fuses with a lysosome.31
Lysosome formation involves vesicular trafficking through the entire endolysosomal system and occurs by the fusion of vesicles from the trans-Golgi and plasma membrane via endocytosis. Lysosomes receive not only cargo from late endosomes but also new lysosomal hydrolases and membrane proteins, and growth factors from the trans-Golgi network (TGN).32 Trafficking between the TGN and endosomes is a highly dynamic process, in which newly synthesized enzymes are delivered to the endolysosomal compartment from the TGN, and some of them are returned to the TGN for recycling. Because of the complexity of the endocytic network, it is difficult to distinguish the lysosomes from other related vesicles, such as early and late endosomes. Morphologically, the early endosome is shown by electron microscopy as a central vacuole with multiple tubules, whereas the late endosomes/lysosome appear as globular vacuoles.33 Biochemically, the early endosome may be identified by a marker of Rab5, a protein that in humans is encoded by the gene RAB5A. Rab5 localizes to early endosomes, where it regulates intracellular membrane trafficking and is involved in the maturation of cell compartments to late endosomes.34 The late endosome may be identified by a marker of mannose 6-phosphate receptors (M6PRs),35 whereas, the lysosome may be distinguished from others by an absence of M6PRs and their internal acidity, which is usually less than pH 5.2 As such, the highly dynamic changes of the compartments of the endocytic–lysosomal system pose a problem for the development of drugs targeting the lysosome with high specificity.
Lysosomes and cancer cell death pathways
Cancer cell death can be classified as type I (apoptosis), type II (autophagy), or type III cell death (necrosis),which does not involve digestion.36 Lysosomes can play a role in each of these types of cell death.
Apoptosis is divided into intrinsic and extrinsic pathways. The intrinsic pathway is activated by permeabilization of the mitochondrial outer membrane and the release of cytochrome c into the cytoplasm, which initiates activation of the caspase cascade, while the extrinsic apoptotic pathway is initiated by cell death receptors, which leads to activation of the caspase cascade.37 Caspase activation is regulated by a family of intracellular proteins known as B cell lymphoma 2 (Bcl-2),which governs mitochondrial outer membrane permeabilization. Bcl-2 proteins can be either antiapoptotic (e.g., Bcl-2 and Bcl-xL) or proapoptotic (e.g., Bax and Bid). LMP can contribute to or accelerate apoptosis through cathepsin cleavage of Bid, which induces Bax-mediated release of cytochrome c.
Autophagy is another major cell death pathway with dual roles in cancer therapy and is an evolutionarily conserved process that allows the cell to digest and recycle cellular components. In the setting of advanced cancers treated with cancer therapeutics, while very high doses of certain drugs can induce an apoptosis-independent, autophagy-dependent cell death, in most cases, a secondary death process, such as necrosis, is the ultimate cause of death. On the other hand, because of its recycling capacity, multiple experiments have demonstrated that therapy-induced autophagy mainly serves as a tumor survival pathway. Autophagy can be divided into noncanonical and canonical pathways, with canonical pathways having three different forms: macroautophagy, microautophagy, and CMA. Both microautophagy and CMA directly deliver cellular components to the lysosome. In contrast, macroautophagy delivers unwanted cellular materials to the lysosome through double-membraned vacuoles, called autophagosomes, which sequester cytoplasmic materials.38 Autophagy was previously regarded as a nonselective process; however, it has been recognized in recent years that autophagy can also selectively degrade specific cellular constituents in the lysosome (such as mitochondria), referred to as mitophagy,39 and in the nucleus, referred to as nucleophagy,40 as well as in damaged lysosomes, a process known as lysophagy.41 Regardless of the mechanisms underlying different kinds of autophagy and other endocytic compartments, all endocytic and autophagic pathways ultimately rely on the lysosome for completing their catabolic functions. Simply put, autophagy is a lysosome-dependent pathway. This provides a strong argument for targeting the lysosome to inhibit autophagy.42
Among cell death pathways, both apoptosis and autophagy are forms of programmed cell death, which is cell death mediated by a genetically-defined program. In contrast, necrosis was thought to be a nonprogrammed cellular catastrophe that involved membrane explosion. Necrosis was thought to be a nonphysiological process that occurs as a result of trauma, infection, or injury to the cell. A form of programmed necrosis, called necroptosis, has recently been recognized as an alternate form of programmed cell death, which can serve as a cell death backup to apoptosis when apoptosis signaling is blocked by endogenous or exogenous factors, such as viruses or mutations.43 Most recently, the receptor-interacting serine/threonine protein kinase 1 (RIPK1) and RIPK3 have been suggested to regulate both apoptosis and necroptosis, which involves lysosomal function. When caspase activation in apoptosis is interrupted, RIPK1 and RIPK3 can induce necroptosis through sphingomyelinase-mediated LMP.44 In human renal cell carcinoma cells, inhibition of mTOR with CCI-779 induces autophagy and suppresses RIPKs, and autophagy inhibition stimulates RIPK- and ROS-dependent necroptosis in vitro and suppresses xenograft growth that may enhance cancer treatment efficacy.45
Lysosomal cell death is referred to as a process involving LMP and leakage of the lysosomal proteases, namely cathepsins, into the cytosol. Cytosolic cathepsins can initiate caspase-dependent and/or-independent cell death, as well as necroptosis.25,44 The original concept of lysosomal cell death was defined as necrotic cell death and was first presented in 1974 by Christian de Duve, who described the lysosomes as “suicide bags” that can cause cell autolysis upon rupture. Lysosome-mediated autolysis usually occurs in injured cells, such as those infected with virus or bacteria or those with nutrient depletion. It also occurs when cell organelles, such as mitochondria and peroxisomes, become old or inactive, leading to release of digestive enzymes out of lysosomal membranes in order to digest themselves.46 However, recent studies on LMP reveal that this form of cell death functions as more than “suicide bags” and can have both necrotic and apoptotic features as well as the features of functional impairment of autophagy.47
Transcriptional regulation of lysosomal biogenesis
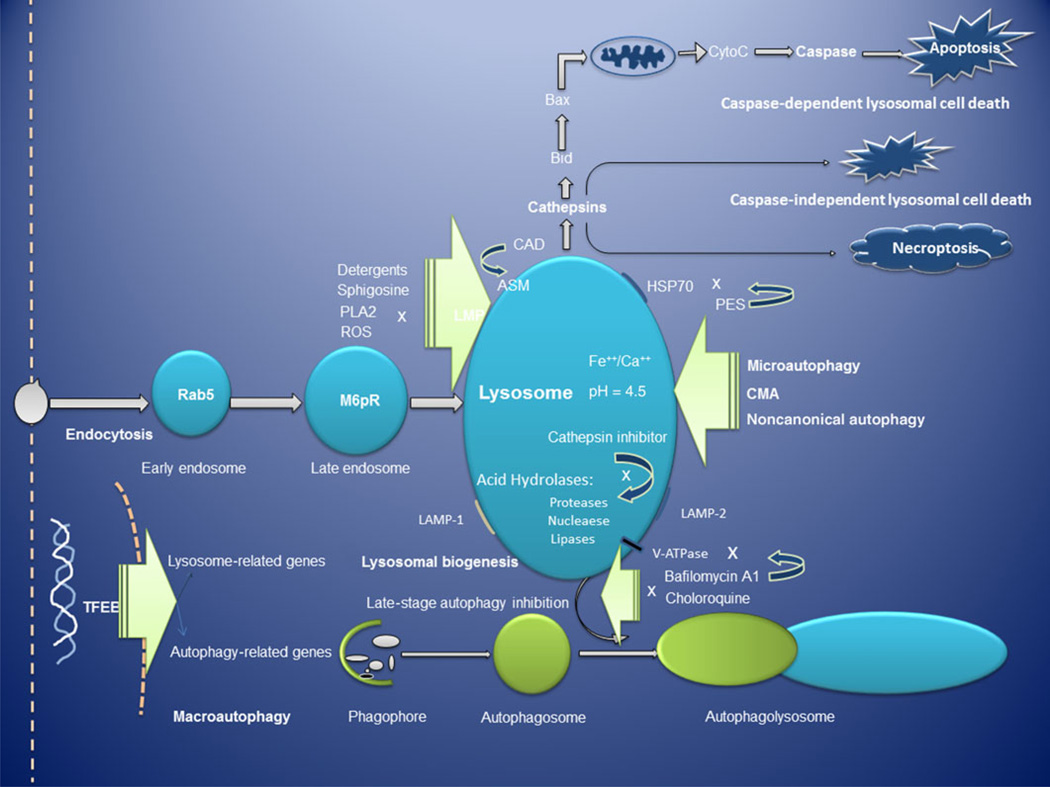
The molecular machinery that regulates lysosome formation and biogenesis was thought to be fully explained by the highly dynamic process of vesicular trafficking from the ER, Golgi apparatus, endosomes, and lysosomes. However, recent work48 has demonstrated that lysosomal biogenesis is fine-tuned at the transcriptional level by a specific gene network named CLEAR (coordinated lysosomal expression and regulation).49 Most recently, research has shown that transcriptional regulation of lysosome biogenesis and autophagy is mediated by transcription factors of the MiT/TFE family, which have been shown to reprogram the metabolism of pancreatic ductal adenocarcinoma (PDA). These findings demonstrate that the transcriptional activation of the clearance pathways that converge on the lysosome plays a crucial role in the pathogenesis of PDA.50 Many genes encoding lysosomal proteins harbor a CLEAR sequence near the transcription start site. This gene network is coordinated by the transcription factor EB (TFEB), which can enter the nucleus and bind to the CLEAR elements, inducing gene transcription. TFEB is regarded as a master regulator of stress responses in the process of lysosomal biogenesis and autophagy,51 and there are 500–800 direct TFEB target genes, many of which involve lysosomal biogenesis and autophagy.48 These genes may represent potential targets for drug development in cancer, either as direct targets or as reporters for assays to identify lysosomal inhibitors (Fig. 1).
Figure 1.
Overview of the lysosomal–autophagy pathway. TFEB regulates both autophagy and lysosome biogenesis.51 After LMP, cathepsin initiates both caspase-dependent and -independent lysosomal cell death, as well as necroptosis.44 The lysosome is the final destination of the lysosomal–autophagy pathway, and drugs targeting the lysosome in cancer not only disable all pathways of autophagy but also block both apoptotic and lysosomal cell death.42 Abbreviations: M6pR, mannose 6-phosphate receptor; PLA2, phospholipases A2; ROS, reactive oxygen species; LMP, lysosomal membrane permeabilization; HSP70, heat shock protein 70; CMA, chaperone-mediated autophagy; TFEB, transcription factor EB; PES, 2-phenylethynesulfonamide; LMP-1 and LMP-2, lysosome-associated membrane protein-1 and -2; ASM, acid sphingomyelinase; CAD, cationic amphiphilic drug.
Lysosomes promote tumor metastasis via lysosomal changes
Advanced cancer cells are highly dependent on effective lysosomal function. As a result, cancer progression and metastasis are associated with striking changes in lysosomal compartments, as compared with normal cells, including lysosome volume, composition, cellular distribution, and lysosomal enzyme activity.52,53 Most of those changes are closely correlated with invasive growth, angiogenesis, and drug resistance.54 Since cancer cells usually exhibit weaker lysosomal membranes as compared to noncancerous cells, they can be selectively sensitized to cell death.
Drugs targeting the lysosome in cancer
Currently, there are five major categories of agents that target the lysosome in cancer (Table 1). The majority of these agents are still being investigated in the preclinical research process, except hydroxychloroquine (HCQ), which is widely tested in many clinical trials combining other anticancer therapies.
Table 1.
Examples of drugs targeting the lysosome in cancer
| Target | Compound | Mode of action | Clinical potential | Reference |
|---|---|---|---|---|
| End stage of autophagy | HCQ | Autophagolysosome formation | Autophagy inhibition | 42, 55 |
| Lyso 5 | Autophagolysosome formation | Autophagy inhibition | 57 | |
| Vacuolar H+-ATPase | Bafilomycin A1 | Deacidification | H+-ATPase inhibition | 62 |
| Archazolid | Deacidification | H+-ATPase inhibition | 64 | |
| Cleistanthin A | Deacidification | H+-ATPase inhibition | 65 | |
| Manzamine A | Deacidification | H+-ATPase inhibition | 66 | |
| ASM | Cationic amphiphilic drugs | Modulate ASM metabolism | ASM modulator | 77 |
| Lysosomal hydrolases | Pepstatin | Inhibition of cathepsin D | Acid protease inhibitor | 68 |
| CA-074 | Inhibition of cathepsin B | Acid protease inhibitor | 69 | |
| Z-FF-FMK | Inhibition of cathepsin L | Acid protease inhibitor | 72 | |
| Cat S/6n | Inhibition of cathepsin S | Acid protease inhibitor | 71 | |
| HPS70 | PES | ASM metabolism, LMP | ASM modulator | 76 |
Choloroquine derivatives
Chloroquine (CQ), a well-established drug used in the treatment or prevention of malaria, is a lysosomotropic agent that is presumed to accumulate in, and prevent and/or impair, endosomal acidification, thereby impairing lysosomal enzymatic function and blocking autophagy.55 Little is known about the mechanism of action of CQ derivatives within lysosomes in mammalian cells. CQ accumulates inside of endosomes and lysosomes; however, many cancer drugs are weak bases that accumulate in lysosomes without the effect of autophagy inhibition.56 One explanation for this is that HCQ can displace other combined drugs from the lysosome to the cytosol or nucleus, improving their intracellular bioavailability.56 An alternative hypothesis is that there is a protein or nonprotein target of CQ within the lysosome. There are currently over 40 clinical trials involving HCQ as a potential autophagy inhibitor worldwide, and the promising results of six trials, five in humans and one in dogs, were recently published.42 These studies demonstrated that high doses of HCQ were required to block autophagy in human tissues, including in tumors. Some drug combinations involving HCQ were safe enough where relatively high doses of HCQ could be reached, whereas other combinations produced dose-limiting toxicities such that dose escalation of HCQ was halted. Therefore, while the safety and feasibility of autophagy inhibition was clearly established, the required potency for effective autophagy inhibition is still unknown. The search for more potent autophagy inhibitors has continued. One example of a more potent CQ derivative that we reported on is Lys05, a novel dimeric derivative of CQ,57 which has shown to have significant in vivo activity both as a single agent58 and in combination with a BRAF (B-Raf proto-oncogene serine/threonine protein kinase) inhibitor.42 A number of other more potent novel CQ derivatives or chemical analogs for cancer have recently been reported, such as EAD1,59 VR23,60 and quinacrine (QN) analogs.61 EAD1 is a CQ derivative that retained the 4-aminoquinoline subunit of CQ but incorporated different triazoles into the structure. This compound was tested in human lung cancer and pancreatic cancer cells and was found to be eightfold more potent than CQ and HCQ.59 VR 23 is another CQ derivative that was developed using a hybrid pharmacophore approach. This compound was tested in an in vitro model of cancer cells and found to cause massive cell death in a variety of cancer cell types.60 QN is an acridine derivative with a similar chemical structure to CQ and has a 60-fold higher potency than CQ in autophagy inhibition but has high cytotoxicity. Newly designed QN analogs have reduced cytotoxicity, yet strong autophagy inhibition effects; these improved QN analogs may be used as anticancer agents.61
V-ATPase inhibitors
Most V-ATPase inhibitors are natural compounds of microbial origin, such as cleistanthin A, manzamine A, archazolid, and bafilomycin A1. Bafilomycin A1 is considered to be the prototypical V-ATPase inhibitor at low nanomolar concentrations and is therefore often used to block distal autophagic flux in a key autophagic flux assay known as the bafilomycin clamp. However, caution should be exercised when using bafilomycin, as other mechanisms of action have now been proposed. Bafilomycin A may target both early and late stages of the autophagy pathway by activating mammalian target of rapamycin signaling and by disassociating the Beclin 1–Vps34 complex, as well as by inhibiting the formation of autolysosomes.62 In addition, bafilomycin A1 induces the binding of Beclin 1 to Bcl-2, which further inhibits autophagy and promotes apoptotic cell death. Bafilomycin A1 also targets mitochondria and induces caspase-independent apoptosis by inducing the translocation of apoptosis-inducing factor from mitochondria to the nucleus.63
Another well studied inhibitor of V-ATPase is archazolid, a myxobacterial agent. Research shows that inhibition of V-ATPase by archazolid reduces the activity of proteases such as cathepsin B in vitro and in vivo.64 Other inhibitors of V-ATPase include cleistanthin-A, which is a diphyllin glycoside isolated from a tropical plant and has shown cytotoxicity in several tumor cell lines.65 Manzamine A was isolated from marine sponges of the genus Haliclona, which has shown activity against pancreatic cancer cells through inhibition of V-ATPase and autophagy.66
ASM modulators
ASM is mainly located within the lysosome, where it cleaves sphingomyelin to ceramide and sphingosine in the cellular response to stress. Cancer cells show high susceptibility to ASM targeting, because ASM activity is usually lower in cancer cells than in normal cells, leading to higher sphingomyelin levels. Further inhibiting ASM activity results in even higher levels of sphingomyelin, which interferes with the normal function of the lysosomal membrane.67 An example of ASM modulators are cationic amphiphilic drugs, such as clorpromazine, CQ, and amiodarone, which can reduce ASM activity by displacing ASM from vesicular membranes in the lysosome. This elicits LMP and an induction of tumor cell death.28
Cathepsin inhibitors
Subcellular localization of cathepsins is a crucial factor in determining the role of cathepsins in cancer. The intercellular release of cathepsins into the extracellular space is of oncogenic character, which has been associated with tumor progression and metastasis in various cancer types. As such, cathepsin inhibitors, including the inhibitors of cathepsins D,68 B,69 K,70 E,68 S,71 and L,72 have been developed for cancer therapy. Although many studies have indicated that cathepsins would be effective targets for cancer drugs, there are currently no cathepsin inhibitors that have advanced to clinical trials in cancer. The main focus of research on the development of cathepsin inhibitors is on osteoporosis73 and rheumatoid arthritis.74
HSP70 inhibitors
HSP70 protects lysosomal membrane integrity and promotes tumor cell metastasis, conferring a survival advantage to cancer cells. Elevated expression of HSP70 is common and correlates with poor prognosis in many cancers. PES is a recently developed HSP70 modulator,75 which has been shown to have anticancer activities by disrupting protein–protein interactions between HSP70 and p53. It triggers apoptosis and blocks autophagy at the level of the lysosome, leading to massive accumulation of autophagosomes with undigested cargo.76
Other potential lysosomal targets
The recent discovery of the TFEB, linked to a novel mechanism that controls the lysosomal–autophagic pathway, suggests that the 500–800 TFEB-regulated genes may include new targets for drug development in cancer. Most recently, studies have also indicated that lysosomes may tightly regulate the cell signaling of apoptosis and autophagy through their critical roles in the metabolism of iron, heme,78 and calcium.79 As a result, targeting nonprotein constituents of the lysosome, such as high calcium, iron, and heme content, may provide another alternative therapeutic strategy for drugs targeting the lysosome in cancer.
Perspective
The emerging role of the lysosome in cancer involves many complex processes of cancer biology, such as nutrient sensing, cell signaling, cell death, immune responses, and cell metabolism. The same processes and mechanisms that are used by cancer cells for their survival can be subverted to sensitize cancer cells to be killed by cancer therapy. What is currently known about lysosomes is still very limited, and many questions are yet to be answered. What is the exact role of the hundreds of genes involved in the network of lysosome biogenesis and autophagy? What is the exact molecular mechanism of lysosomal protein trafficking? What are the critical cell signaling modules that control lysosomal function? How can we effectively analyze the highly dynamic changes within the endosome–lysosome compartment in response to a variety of stimuli and therapeutic stresses? To better answer these questions, researchers need to move from gene to genome in exploring the role of lysosomes within the tumor microenvironment. Systematic and interdisciplinary approaches, with an emphasis on genomic and postgenomic analysis using high-throughput techniques such as transcriptomics, proteomics, and metabolomics to analyze patient-derived tissues, may lead to a deeper understanding of lysosomal variation. These new approaches may provide effective strategies to develop biomarkers that may detect a subset of human cancer that is more prone to lysosomal inhibition. Consequently, these biomarkers could be used to match emerging lysosome-targeted therapies that may be effective single agents and also to overcome cancer drug resistance.
Footnotes
Conflicts of interest
R.K.A. is a consultant for Presage Biosciences and Sprint Biosciences. He is an inventor on patents related to Lys05.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Appelqvist H, Waster P, Kagedal K, Ollinger K. The lysosome: from waste bag to potential therapeutic target. J. Mol. Cell Biol. 2013;5:214–226. doi: 10.1093/jmcb/mjt022. [DOI] [PubMed] [Google Scholar]
- 3.Gotink KJ, et al. Lysosomal sequestration of sunitinib: a novel mechanism of drug resistance. Clin. Cancer Res. 2011;17:7337–7346. doi: 10.1158/1078-0432.CCR-11-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Repnik U, Cesen MH, Turk B. The endolysosomal system in cell death and survival. Cold Spring Harb. Perspect. Biol. 2013;5:a008755. doi: 10.1101/cshperspect.a008755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schroder BA, Wrocklage C, Hasilik A, Saftig P. The proteome of lysosomes. Proteomics. 2010;10:4053–4076. doi: 10.1002/pmic.201000196. [DOI] [PubMed] [Google Scholar]
- 6.Kirkegaard T, Jaattela M. Lysosomal involvement in cell death and cancer. Biochim. Biophys. Acta. 2009;1793:746–754. doi: 10.1016/j.bbamcr.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Repnik U, Stoka V, Turk V, Turk B. Lysosomes and lysosomal cathepsins in cell death. Biochim. Biophys. Acta. 2012;1824:22–33. doi: 10.1016/j.bbapap.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 8.Withana NP, et al. Cathepsin B inhibition limits bone metastasis in breast cancer. Cancer Res. 2012;72:1199–1209. doi: 10.1158/0008-5472.CAN-11-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Small DM, et al. Cathepsin S from both tumor and tumor-associated cells promote cancer growth and neovascularization. Int. J. Cancer. 2013;133:2102–2112. doi: 10.1002/ijc.28238. [DOI] [PubMed] [Google Scholar]
- 10.Keliher EJ, et al. Targeting cathepsin E in pancreatic cancer by a small molecule allows in vivo detection. Neoplasia. 2013;15:684–693. doi: 10.1593/neo.13276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 2009;10:623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 12.Saftig P, Schroder B, Blanz J. Lysosomal membrane proteins: life between acid and neutral conditions. Biochem. Soc. Trans. 2010;38:1420–1423. doi: 10.1042/BST0381420. [DOI] [PubMed] [Google Scholar]
- 13.Dice JF. Chaperone-mediated autophagy. Autophagy. 2007;3:295–299. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- 14.Furuta K, et al. Expression of lysosome-associated membrane proteins in human colorectal neoplasms and inflammatory diseases. Am. J. Pathol. 2011;159:449–455. doi: 10.1016/S0002-9440(10)61716-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K. Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in. Arabidopsis. Plant Cell. 2006;18:715–730. doi: 10.1105/tpc.105.037978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fais S, DeMilito A, You H, Qin W. Targeting vacuolar H+ –ATPases as a new strategy against cancer. Cancer Res. 2007;67:10627–10630. doi: 10.1158/0008-5472.CAN-07-1805. [DOI] [PubMed] [Google Scholar]
- 17.Meo-Evoli N, et al. V-ATPase: a master effector of E2F1-mediated lysosomal trafficking, mTORC1 activation and autophagy. Oncotarget. 2015;6:28057–28070. doi: 10.18632/oncotarget.4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petersen NH, et al. Transformation-associated changes in sphingolipid metabolism sensitize cells to lysosomal cell death induced by inhibitors of acid sphingomyelinase. Cancer Cell. 2013;24:379–393. doi: 10.1016/j.ccr.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 19.Savic R, Schuchman EH. Use of acid sphingomyelinase for cancer therapy. Adv. Cancer Res. 2013;117:91–115. doi: 10.1016/B978-0-12-394274-6.00004-2. [DOI] [PubMed] [Google Scholar]
- 20.Boya P, Kroemer G. Lysosomal membrane permeabilization in cell death. Oncogene. 2008;27:6434–6451. doi: 10.1038/onc.2008.310. [DOI] [PubMed] [Google Scholar]
- 21.Aits S, Jaattela M, Nylandsted J. Methods for the quantification of lysosomal membrane permeabilization: a hallmark of lysosomal cell death. Methods Cell Biol. 2015;126:261–285. doi: 10.1016/bs.mcb.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurz T, Terman A, Gustafsson B, Brunk UT. Lysosomes in iron metabolism, ageing and apoptosis. Histochem. Cell Biol. 2008;129:389–406. doi: 10.1007/s00418-008-0394-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeidan YH, Hannun YA. The acid sphingomyelinase/ ceramide pathway: biomedical significance and mechanisms of regulation. Curr. Mol. Med. 2010;10:454–466. doi: 10.2174/156652410791608225. [DOI] [PubMed] [Google Scholar]
- 24.LeGendre O, Breslin PAS, Foster DA. (−)-Oleocanthal rapidly and selectively induces cancer cell death via lysosomal membrane permeabilization. Mol. Cell. Oncol. 2015;2:e1006077. doi: 10.1080/23723556.2015.1006077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Repnik U, Hafner Cesen M, Turk B. Lysosomal membrane permeabilization in cell death: concepts and challenges. Mitochondrion. 2014;19(Pt A):49–57. doi: 10.1016/j.mito.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Hafner Česen M, Pegan K, Špes A, Turk B. Lysosomal pathways to cell death and their therapeutic applications. Exp. Cell Res. 2012;318:1245–1251. doi: 10.1016/j.yexcr.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 27.Cuervo AM, Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 2014;24:92–104. doi: 10.1038/cr.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saftig P, Sandhoff K. Cancer: killing from the inside. Nature. 2013;502:312–313. doi: 10.1038/nature12692. [DOI] [PubMed] [Google Scholar]
- 29.Leu JI, Pimkina J, Pandey P, et al. HSP70 inhibition by the small-molecule 2-phenylethynesulfonamide impairs protein clearance pathways in tumor cells. Mol. Cancer Res. 2011;9:936–947. doi: 10.1158/1541-7786.MCR-11-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romao S, Munz C. LC3-associated phagocytosis. Autophagy. 2014;10:526–528. doi: 10.4161/auto.27606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luzio JP, et al. Lysosome–endosome fusion and lysosome biogenesis. J. Cell Sci. 2000;113(Pt 9):1515–1524. doi: 10.1242/jcs.113.9.1515. [DOI] [PubMed] [Google Scholar]
- 33.Klumperman J, Raposo G. The complex ultra-structure of the endolysosomal system. Cold Spring Harb. Perspect. Biol. 2014;6:a016857. doi: 10.1101/cshperspect.a016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorvel JP, Chavrier P, Zerial M, Gruenberg J. rab5 controls early endosome fusion in vitro. Cell. 1991;64:915–925. doi: 10.1016/0092-8674(91)90316-q. [DOI] [PubMed] [Google Scholar]
- 35.Russell MR, Nickerson DP, Odorizzi G. Molecular mechanisms of late endosome morphology, identity and sorting. Curr. Opin. Cell Biol. 2006;18:422–428. doi: 10.1016/j.ceb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Duprez L, Wirawan E, Berghe TV, Vandenabeele P. Major cell death pathways at a glance. Microbes Infect. 2009;11:1050–1062. doi: 10.1016/j.micinf.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 37.Elmore S. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amaravadi RK, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin. Cancer Res. 2011;17:654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chourasia AH, Boland ML, Macleod KF. Mitophagy and cancer. Cancer Metab. 2015;3:4. doi: 10.1186/s40170-015-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mochida K, et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015;522:359–362. doi: 10.1038/nature14506. [DOI] [PubMed] [Google Scholar]
- 41.Hasegawa J, Maejima I, Iwamoto R, Yoshimori T. Selective autophagy: lysophagy. Methods. 2015;75:128–132. doi: 10.1016/j.ymeth.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 42.Rebecca VW, Amaravadi RK. Emerging strategies to effectively target autophagy in cancer. Oncogene. 2015 doi: 10.1038/onc.2015.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu X, Deng Q, Bode AM, et al. The role of necroptosis, an alternative form of cell death, in cancer therapy. Expert Rev. Anticancer Ther. 2013;13:883–893. doi: 10.1586/14737140.2013.811180. [DOI] [PubMed] [Google Scholar]
- 44.Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat. Rev. Mol. Cell Biol. 2010;11:700–714. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- 45.Bray K, et al. Autophagy suppresses RIP kinase-dependent necrosis enabling survival to mTOR inhibition. PLoS One. 2012;7:e41831. doi: 10.1371/journal.pone.0041831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Duve C. Lysosomes revisited. Eur. J. Biochem. 1983;137:391–397. doi: 10.1111/j.1432-1033.1983.tb07841.x. [DOI] [PubMed] [Google Scholar]
- 47.Kroemer G, Jaattela M. Lysosomes and autophagy in cell death control. Nat. Rev. Cancer. 2005;5:886–897. doi: 10.1038/nrc1738. [DOI] [PubMed] [Google Scholar]
- 48.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmieri M, et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 2011;20:3852–3866. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 50.Perera RM, et al. Transcriptional control of autophagy–lysosome function drives pancreatic cancer metabolism. Nature. 2015;524:361–365. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sardiello M, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 52.Nishimura Y, Sameni M, Sloane BF. Malignant transformation alters intracellular trafficking of lysosomal cathepsin D in human breast epithelial cells. Pathol. Oncol. Res. 1998;4:283–296. doi: 10.1007/BF02905219. [DOI] [PubMed] [Google Scholar]
- 53.Gocheva V, et al. Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Genes Dev. 2006;20:543–556. doi: 10.1101/gad.1407406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fehrenbacher N, Jaattela M. Lysosomes as targets for cancer therapy. Cancer Res. 2005;65:2993–2995. doi: 10.1158/0008-5472.CAN-05-0476. [DOI] [PubMed] [Google Scholar]
- 55.Steinman RM, Mellman IS, Muller WA, Cohn ZA. Endocytosis and the recycling of plasma membrane. J. Cell Biol. 1983;96:1–27. doi: 10.1083/jcb.96.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fu D, et al. Imaging the intracellular distribution of tyrosine kinase inhibitors in living cells with quantitative hyperspectral stimulated Raman scattering. Nat. Chem. 2014;6:614–622. doi: 10.1038/nchem.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amaravadi RK, Winkler JD. Lys05: a new lysosomal autophagy inhibitor. Autophagy. 2012;8:1383–1384. doi: 10.4161/auto.20958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McAfee Q, et al. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc. Natl. Acad. Sci. U.S.A. 2012;109:8253–8258. doi: 10.1073/pnas.1118193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nordstrom LU, et al. Discovery of autophagy inhibitors with antiproliferative activity in lung and pancreatic cancer cells. ACS Med. Chem. Lett. 2015;6:134–139. doi: 10.1021/ml500348p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vu H-YT, Pundir S, Solomon RV, Lee H. Abstract 4545: anticancer effects and mechanism of VR23, a novel chloroquine derivative. Cancer Res. 2014;74:4545. [Google Scholar]
- 61.Wang T, et al. Synthesis of improved lysomotropic autophagy inhibitors. J. Med. Chem. 2015;58:3025–3035. doi: 10.1021/jm501586m. [DOI] [PubMed] [Google Scholar]
- 62.Yamamoto A, et al. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- 63.Yuan N, et al. Bafilomycin A1 targets both autophagy and apoptosis pathways in pediatric B-cell acute lymphoblastic leukemia. Haematologica. 2015;100:345–356. doi: 10.3324/haematol.2014.113324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kubisch R, et al. V-ATPase inhibition by archazolid leads to lysosomal dysfunction resulting in impaired cathepsin B activation in vivo. Int. J. Cancer. 2014;134:2478–2488. doi: 10.1002/ijc.28562. [DOI] [PubMed] [Google Scholar]
- 65.Zhao Y, Lu Y, Ma J, Zhu L. Synthesis and evaluation of cleistanthin A derivatives as potent vacuolar H+-ATPase inhibitors. Chem. Biol. Drug Des. 2015;86:691–696. doi: 10.1111/cbdd.12538. [DOI] [PubMed] [Google Scholar]
- 66.Kallifatidis G, Hoepfner D, Jaeg T, et al. The marine natural product manzamine A targets vacuolar ATPases and inhibits autophagy in pancreatic cancer cells. Mar. Drugs. 2013;11:3500–3516. doi: 10.3390/md11093500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith EL, Schuchman EH. Acid sphingomyelinase overexpression enhances the antineoplastic effects of irradiation in vitro and in vivo. Mol. Ther. 2008;16:1565–1571. doi: 10.1038/mt.2008.145. [DOI] [PubMed] [Google Scholar]
- 68.Maynadier M, et al. Dipeptide mimic oligomer transporter mediates intracellular delivery of Cathepsin D inhibitors: a potential target for cancer therapy. J. Control. Release. 2013;171:251–257. doi: 10.1016/j.jconrel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 69.Kos J, Mitrovic A, Mirkovic B. The current stage of cathepsin B inhibitors as potential anticancer agents. Future Med. Chem. 2014;6:1355–1371. doi: 10.4155/fmc.14.73. [DOI] [PubMed] [Google Scholar]
- 70.Duong le T, Wesolowski GA, Leung P, et al. Efficacy of a cathepsin K inhibitor in a preclinical model for prevention and treatment of breast cancer bone metastasis. Mol. Cancer Ther. 2014;13:2898–2909. doi: 10.1158/1535-7163.MCT-14-0253. [DOI] [PubMed] [Google Scholar]
- 71.Tsai JY, et al. Effects of novel human cathepsin S inhibitors on cell migration in human cancer cells. J. Enzyme Inhib. Med. Chem. 2014;29:538–546. doi: 10.3109/14756366.2013.823957. [DOI] [PubMed] [Google Scholar]
- 72.Lankelma JM, et al. Cathepsin L, target in cancer treatment? Life Sci. 2010;86:225–233. doi: 10.1016/j.lfs.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 73.Bromme D, Lecaille F. Cathepsin K inhibitors for osteoporosis and potential off-target effects. Expert Opin. Investig. Drugs. 2009;18:585–600. doi: 10.1517/13543780902832661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang D, Bromme D. Drug delivery strategies for cathepsin inhibitors in joint diseases. Expert Opin. Drug Deliv. 2005;2:1015–1028. doi: 10.1517/17425247.2.6.1015. [DOI] [PubMed] [Google Scholar]
- 75.Wu Z, et al. Autophagy blockade sensitizes prostate cancer cells towards Src family kinase inhibitors. Genes Cancer. 2010;1:40–49. doi: 10.1177/1947601909358324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Granato M, et al. HSP70 inhibition by 2-phenylethynesulfonamide induces lysosomal cathepsin D release and immunogenic cell death in primary effusion lymphoma. Cell Death Dis. 2013;4:e730. doi: 10.1038/cddis.2013.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Petersen NH, et al. Transformation-associated changes in sphingolipid metabolism sensitize cells to lysosomal cell death induced by inhibitors of acid sphingomyelinase. Cancer Cell. 2013;24:379–393. doi: 10.1016/j.ccr.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 78.Dong C, et al. Heme oxygenase-1 enhances autophagy in podocytes as a protective mechanism against high glucose-induced apoptosis. Exp. Cell Res. 2015;337:146–149. doi: 10.1016/j.yexcr.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 79.Medina DL, et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 2015;17:288–299. doi: 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]