Abstract
Madariaga virus (MADV), the new species designation for the South American isolates of eastern equine encephalitis virus (EEEV) is genetically divergent, and substantially different in ecology and pathogenesis from North American EEEV strains. We isolated and characterized a MADV isolate obtained from a horse in Brazil. Our results support previous phylogenetic studies showing there are 3 genetically distinct MADV lineages. The MADV isolate from Paraíba State belongs to the South American lineage III and is closely related to Peruvian, Colombian and Venezuelan isolates.
Several equine encephalitis viruses belong to the family Togaviridae, genus Alphavirus, including western (WEEV), Venezuelan (VEEV) and eastern equine encephalitis viruses (EEEV) (1). EEEV was previously classified into four distinct lineages/subtypes (I to IV) (2). Indeed, the South American (SA) strains (lineages II-IV), which include the isolates from South and Central America, are genetically diverse in contrast to the conserved North American (NA) strains (lineage I)(3). Because of the genetic divergence and significant differences in ecology and pathogenesis, the SA isolates were recently classified as a distinct species, Madariaga virus (MADV) (3, 4).
Previously in South America, only three human EEE cases have been reported, detected in Brazil and Trinidad (5, 6). However, an outbreak of encephalitis caused by MADV was confirmed in Panama in 2010, with 13 human and 50 equine positive cases (8). Unlike the epidemiological profile in South America, in North America an average of eight human cases of EEE neuroinvasive disease has been reported annually between 2004-2013 (7).
In Brazil, few studies have reported the isolation and molecular characterization of MADV from horses (9). Most information on MADV distribution is based on serological surveys in horses, humans and wild vertebrates, or virus isolation only from mosquito vectors, as previously reported in the states of Amazonas, Pará, Rondônia, Tocantins, Ceará, Paraíba, Pernambuco, Bahia, Mato Grosso, Mato Grosso do Sul, Minas Gerais, Rio de Janeiro and São Paulo (10-16).
Here, we describe the complete genomic sequence of a MADV isolate from a horse in Paraíba State, Brazil, in 2009. We analyzed samples from a five-year-old crossbred horse, from the city of São José do Rio do Peixe, Paraíba State, in the northeast of Brazil. The horse presented with signs of encephalitis for three days and tested negative for rabies. While under veterinary care, it showed blindness with depression, anorexia, severe ataxia, head pressing, circular walking while occasionally collapsing. On the second day, it presented with lip ptosis and lateral recumbence, and on the third day, the animal was euthanized. At necropsy, corneal opacity, scarifications of the head tissue and congestion in the leptomeninges and brain were observed.
As part of a histological examination, samples from various organs and the central nervous system (CNS) were fixed in 10% buffered formalin, cut transversely into 3-5 mm thick sections and then stained with hematoxylin-eosin. We observed mainly in the frontal cortex a diffuse multifocal non-suppurative lymphoplasmocytic encephalomyelitis with perivascular infiltration by lymphocytes, plasma cells, macrophages and some neurophils, vasculitis with endothelial cells swelling, neuronal death, satellitosis, neuronophagia, hemorrhages, edema and multifocal lymphocytic infiltration of the neurophil. Moderate lesions were also observed in the brain stem affecting mainly the grey matter, and mild lesions were observed in the cerebellum and cervical spinal cord. No lesions were observed in the dorsal and lumbar spinal cord.
Virus isolation was performed in vitro and in vivo. The brain tissue (frontal cortex, parietal cortex, basal nuclei, thalamus) was homogenized and the suspension was inoculated into C6/36 (Aedes albopictus) cells and in newborn albino mice (CH3– Rockefeller strain) intracerebrally. Mice were observed daily for 30 days for neurologic signs or death.
The 5 day-old mice presented signs of illness 24 hours post-inoculation including isolation from the others in the litter, failure to nurse, difficulty breathing, and paresis progressing to flaccid paralysis.
Viral RNA from the CNS of the horse and mice were extracted using the RNeasy mini kit (Qiagen Inc., Venlo, Netherlands) following the manufacturer's instruction. RT-PCRs for several flaviviruses and alphaviruses were performed as described by Bronzoni (17) to confirm the presence of MADV, followed by complete genomic sequencing of the MADV isolate. Complementary DNA (cDNA) was prepared using the RNA High Capacity cDNA Reverse Transcription Kit© with random primers (Thermo Fisher Scientific Inc., Waltham, MA). Nextera-XT DNA Library Prep Kit (©Illumina, San Diego, CA) was used to prepare the dsDNA library that was sequenced using the MiSeq Reagent Kit© v3, 300 cycles (Illumina Inc., San Diego, CA). Geneious v7.1.4 (Biomatters Inc., San Francisco, CA) was used to assemble the sequence using MADV complete genome strain PE-0.0155 as a reference (DQ241304 GenBank accession number). The derived sequence was submitted to GenBank under the accession number KR132531. The polyprotein sequences available in GenBank were aligned using ClustalW, implemented in MEGA 6 (9). A set of 44 sequences of 3,729 nt was used for analysis. Maximum likelihood (ML) phylogenetic trees were constructed using the General Time Reversible model (GTR+Γ4+I), which was identified as the best-fit model of nucleotide substitution using MODELTEST version 3.7 (18). Bootstrapping was performed to assess the robustness of tree topologies using 1,000 replicate neighbor-joining (NJ) trees under the ML substitution model. Analyses were performed with PAUP* version 4.0b (Sinauer Associates Inc., Sunderland, MA).
Divergence estimates between the Paraíba strain (MADY/Equus ferus/BR/PB/01/2009) and the nearest sister, MADV/Equus ferus caballus/COL/980150/2002 (GenBank accession number KJ469590), was conducted using a total of 11,217nt positions in the two open reading frames, using the Tamura-Nei model. Evolutionary analyses were conducted in MEGA 6 (19).
Previous phylogenetic studies based on partial structural polyprotein gene sequences indicate that EEEV and MADV are distinct species. The EEEV complex (lineage I) contains strains from North America and the Caribbean while the MADV (lineages II-IV) includes isolates collected throughout South and Central America (3). Our ML phylogeny (Figure 1) further supports the previously defined lineages (3). Our isolate also grouped most closely with recent strains from Peru, Colombia and Venezuela within EEE complex lineage III, with strong bootstrap support (100%). The MADV/Equus ferus/BR/PB/01/2009 strain was genetically distinct from the other isolates in this phylogeny and showed highest sequence identity with isolate MADV/Equus ferus caballus/COL/980150/2002, with overall divergence of 0.02 substitutions per site.
Figure 1.
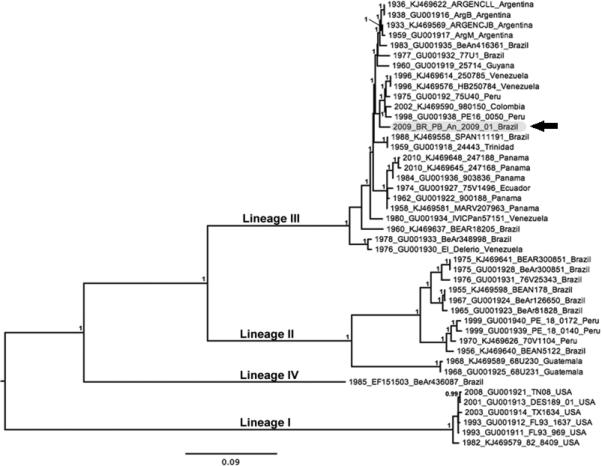
Maximum likelihood (ML) phylogeny tree of 44 strains of EEEV and MADV inferred by 3,729 nt of structural polyprotein gene sequences. Numbers at nodes indicate posterior probabilities ≥0.95. Taxon/tip labels include years of isolation, accession numbers, strain names and isolation countries. Scale bar shows percent nucleotide sequence divergence, 0.09 nucleotide substitutions/per site. Paraíba strain (MADY/Equus ferus/BR/PB/01/2009) is highlighted in gray and indicated with a black arrow.
In conclusion, our data suggest ongoing enzootic transmission of equine-virulent MADV in Paraíba State, Brazil. The etiologic agent of this case of equine encephalitis was a South American lineage III strain.
Acknowledgments
This work was supported by a FAPESP grant to MLN (2013/21719-3) and National Institutes of Health Contract HHSN272201000040I/HHSN27200004/D04.AJA is funded by the James W. McLaughlin Endowment Fund.
REFERENCES
- 1.Griffin DE. Alphaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 6 ed. LIPPINCOTT WILLIAMS & WILKINS; Philadelphia: 2012. [Google Scholar]
- 2.Brault AC, Powers AM, Chavez CL, Lopez RN, Cachon MF, Gutierrez LF, et al. Genetic and antigenic diversity among eastern equine encephalitis viruses from North, Central, and South America. Am J Trop Med Hyg. 1999 Oct;61(4):579–86. doi: 10.4269/ajtmh.1999.61.579. [DOI] [PubMed] [Google Scholar]
- 3.Arrigo NC, Adams AP, Weaver SC. Evolutionary patterns of eastern equine encephalitis virus in North versus South America suggest ecological differences and taxonomic revision. J Virol. 2010 Jan;84(2):1014–25. doi: 10.1128/JVI.01586-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mota MTO, Ribeiro MR, Vedovello D, Nogueira ML. Mayaro virus: a neglected arbovirus of the Americas. Future virology. 2015 [Google Scholar]
- 5.Alice F. Infecção humana pelo vírus “Leste” de Encefalite Eqüina. Bol Inst Biol, Bahia. 1956;3:3–9. [Google Scholar]
- 6.Corniou B, Ardoin P, Bartholomew C, Ince W, Massiah V. First isolation of a South American strain of Eastern Equine virus from a case of encephalitis in Trinidad. Tropical and geographical medicine. 1972 Jun;24(2):162–7. [PubMed] [Google Scholar]
- 7.CDC . Eastern Equine Encephalitis: Epidemiology & Geographic Distribution. Prevention CfDCa; [Google Scholar]
- 8.Carrera JP, Forrester N, Wang E, Vittor AY, Haddow AD, Lopez-Verges S, et al. Eastern equine encephalitis in Latin America. N Engl J Med. 2013 Aug 22;369(8):732–44. doi: 10.1056/NEJMoa1212628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silva ML, Galiza GJ, Dantas AF, Oliveira RN, Iamamoto K, Achkar SM, et al. Outbreaks of Eastern equine encephalitis in northeastern Brazil. J Vet Diagn Invest. 2011 May;23(3):570–5. doi: 10.1177/1040638711403414. [DOI] [PubMed] [Google Scholar]
- 10.Aguiar DM, Cavalcante GT, Lara MCCSH, Villalobos EMC, Cunha EMS, Okuda LH, et al. The antibody prevalence of viral and bacterial diseases in equids in the municipality of Monte Negro, Rondonia, Amazônia Ocidental Brasileira. Braz J Vet Res Anim Sci. 2008;45:269–76. [Google Scholar]
- 11.Fernandez Z, Richartz R, Travassos da Rosa A, Soccol VT. [Identification of the encephalitis equine virus, Parana, Brazil]. Rev Saude Publica. 2000 Jun;34(3):232–5. doi: 10.1590/s0034-89102000000300004. [DOI] [PubMed] [Google Scholar]
- 12.Ferreira IB, Pereira LE, Rocco IM, Marti AT, Souza LTM, Iversson LB. Surveillance of arbovirus infection in the Atlantic Forest Region, State of São Paulo, Brazil. I. Detection of hemagglutination-inhibiting antibodies in wild birds between 1978 and 1990. Revista do Instituto de Medicina Tropical de São Paulo. 1994;6(3):265–74. doi: 10.1590/s0036-46651994000300011. [DOI] [PubMed] [Google Scholar]
- 13.Heinemann MB, Souza MC, Cortez A, Ferreira F, Homem VSF, Ferreira-Neto JS, et al. Seroprevalence of eastern and western equine encephalomyelitis virus in Uruará, PA, Brazil. Braz J Vet Res Anim Sci. 2006;43:137–9. [Google Scholar]
- 14.Kotait I, Peixoto ZMP, Coimbra TLM, Cunha EMS, Queiroz LH, Macruz R, et al. Isolamento e identificação do Vírus da Encefalomielite eqüina, tipo Leste, em eqüinos do Estado de São Paulo, Brazil. Arq Inst Biol. 1992;59(1/2):37–41. [Google Scholar]
- 15.Vasconcelos PF, Da Rosa JF, Da Rosa AP, Degallier N, Pinheiro Fde P, Sa Filho GC. [Epidemiology of encephalitis caused by arbovirus in the Brazilian Amazonia]. Rev Inst Med Trop Sao Paulo. 1991 Nov-Dec;33(6):465–76. [PubMed] [Google Scholar]
- 16.Villari P, Spielman A, Komar N, McDowell M, Timperi RJ. The economic burden imposed by a residual case of eastern encephalitis. Am J Trop Med Hyg. 1995 Jan;52(1):8–13. doi: 10.4269/ajtmh.1995.52.8. [DOI] [PubMed] [Google Scholar]
- 17.de Morais Bronzoni RV, Baleotti FG, Ribeiro Nogueira RM, Nunes M, Moraes Figueiredo LT. Duplex reverse transcription-PCR followed by nested PCR assays for detection and identification of Brazilian alphaviruses and flaviviruses. J Clin Microbiol. 2005 Feb;43(2):696–702. doi: 10.1128/JCM.43.2.696-702.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–8. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 19.Tamura K, Stecher G, Peterson D, Filipski A, S K. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]


