Abstract
Attention-deficit/hyperactivity disorder (ADHD) is a heritable, chronic, neurodevelopmental disorder with serious long-term repercussions. Despite being one of the most common cognitive disorders, the clinical diagnosis of ADHD is based on subjective assessments of perceived behaviors. Endophenotypes (neurobiological markers that cosegregate and are associated with an illness) are thought to provide a more powerful and objective framework for revealing the underlying neurobiology than syndromic psychiatric classification. Here, we present the results of applying genetic linkage and association analyses to neuropsychological endophenotypes using microsatellite and single nucleotide polymorphisms. We found several new genetic regions linked and/or associated with these endophenotypes, and others previously associated to ADHD, for example, loci harbored in the LPHN3, FGF1, POLR2A, CHRNA4 and ANKFY1 genes. These findings, when compared with those linked and/or associated to ADHD, suggest that these endophenotypes lie on shared pathways. The genetic information provided by this study offers a novel and complementary method of assessing the genetic causes underpinning the susceptibility to behavioral conditions and may offer new insights on the neurobiology of the disorder.
Introduction
Attention-deficit/hyperactivity disorder (ADHD) is one of the most common neurodevelopmental behavioral disorders, affecting ~5.3% of children and adolescents worldwide.1 The etiology and pathophysiology of ADHD are still not completely defined, but twin, adoption and family-based studies indicate a strong genetic component, particularly because first- and second-degree relatives of patients with ADHD have markedly higher prevalence of the illness.2 Multiple studies, based on twin concordance comparisons and complex segregation analyses of pedigrees, have shown that ADHD is highly heritable;2 the additive variance of the phenotype attributed to genetic factors is approximately 76%.2
Though genetic factors have been broadly linked to the susceptibility to develop ADHD and some susceptibility genes have been identified, functional mutations harbored at these loci, such as the precise differences in base pairs, remain undefined.3 Given that ADHD has a highly variable clinical manifestation with a complex syndromic clinical definition, it has been suggested that quantitative phenotypes, that is, endophenotypes, could be useful for dissecting the genetic basis of ADHD. As hypothesized intermediates between genes and disease outcomes, endophenotypes are thought to be directly influenced by fewer genes than disease phenotypes.4
A previous study conducted on 288 individuals affected and unaffected with ADHD from the Paisa community, a population exhibiting features of genetic isolation from Colombia, South America, found a number of neuropsychological tests that met the criteria of endophenotypes.5 The tests were aimed at ascertaining neuropsychological impairments frequently observed in patients with ADHD such as visual-motor functioning, executive function and intelligence. For executive function and intelligence, the following neuropsychological tests were performed: the Wechsler Intelligence Scale for Children (WISC) Block Design, performance intelligence quotient (PIQ) and full scale intelligence quotient (FSIQ). Correct responses and omissions on the ‘A'-cancelation and vigilance test (ACVT) were used to assess sustained attention, that is, vigilance. Finally, the Rey-Osterrieth complex figure test (ROCFT), standardized for Colombian children, was used to test visual-motor skills and immediate visual-motor memory recall.5
Starting with the above-mentioned neuropsychological endophenotypes, we hypothesized that applying genetic linkage and association analyses would implicate new genetic regions. To this end, we performed such analyses between our putative ADHD endophenotypes and genomic polymorphisms, that is, microsatellites and single nucleotide polymorphisms (SNPs). Our results provide novel findings that extend previous approaches and may offer new insights on the etiological components of this common behavioral condition, if replicated.
Materials and methods
Sample population
We studied multigenerational and extended and nuclear pedigrees from the Paisa genetic isolate from the Medellin metropolitan area in the state of Antioquia, Colombia. Family ascertainment and sample size has been described in our previous studies.5, 6 Briefly, Paisa descent was considered as having all four grandparents originating from the Paisa region of Colombia. The original sample consisted of 1077 family members, 725 (67.3%) adults (17 years and older) and 352 (32.7%) children and adolescents (6–16 years old), from 141 nuclear and multigenerational families (126 nuclear and 15 extended and multigenerational families) from the Paisa genetic isolate5, 6, 7, 8 with an average family size of 38.5 (range 5–85 individuals) and an average of 3.62 generations (range 2–5). Upon obtaining written informed consent from participating subjects and/or their parents/legal guardians, pedigrees were built through a fixed sampling scheme from a parent or grandparent of an index proband as approved by the University of Antioquia Ethics Committee (Protocol: 11-13-342). This mentioned Committee also approved a subsequent collaboration with researchers from the US National Institutes of Health (Protocol: 00-HG-0058). Full details of the clinical, demographic and genetic ascertainment features, as well as the methodology of endophenotype characterization were published elsewhere.5, 6, 7, 8
Whole-genome scan non-parametric linkage
Genotyping of microsatellite markers was previously described.7 A total of 372 markers across the whole genome, with an average distance of 8.68 cM, were genotyped in the aforementioned study (Table 1). In the present effort, given the large and multigenerational unique nature of these pedigrees, we applied non-parametric linkage analyses, as implemented in MERLIN, to the genotyped data.9 Association analyses, using a variance component model to estimate an additive effect for each microsatellite, was carried out, in addition to non-parametric linkage analysis. Before association evaluation, missing genotypes were estimated to increase statistical power. In addition to the non-parametric linkage analysis, we applied family-based association tests to the whole-genome scan data, under the null hypothesis of presence of no linkage and no association. In the case of previously linked regions, we applied family-based association tests under the null hypothesis of linkage and no association for single marker evaluation. In the case of haplotype analyses (shown in Supplementary Materials), the null hypothesis of no linkage and no association was tested. Five equidistant steps were arbitrarily defined between markers to estimate multipoint linkage statistics. Despite attempts to improve MERLIN's performance and resource use, it was computationally intensive and unable to run optimally because of the large pedigree sizes. To resolve this, we pruned some branches to decrease pedigree sizes. The pruning criteria were based on the availability of genotype and endophenotype information as well as family structure. The linkage analysis was carried out on six different endophenotypes (WISC Block Design, WISC PIQ, WISC FSIQ, ACVT Correct Responses, ACVT Omissions and ROCFT Copy scores) using age, sex, school grade and ADHD status as covariates to control for potential confounding factors.
Table 1. Summary for linkage and association analysis of peak LOD scores, P-values, position of peak and position relative to the closest marker.
| Endophenotype | Chromosome | Peak LOD | P-value | Peak LOD position (cM) | Closest marker | Distance from marker (cM) |
|---|---|---|---|---|---|---|
| WISC Block Design | 2 | 2.51 | 0.00034 | 31.333 | D2S1360 | 3.333 |
| 3 | 1.25 | 0.0082 | 216 | D3S1311 | 0 | |
| 5 | 1.11 | 0.0118 | 171 | D5S1471 | 1 | |
| 9 | 1.33 | 0.0067 | 104 | D9S938 | 0 | |
| 10 | 1.73 | 0.0024 | 61.334 | D10S1221 | 1.666 | |
| 11 | 1.41 | 0.0054 | 109.667 | D11S4464 | 3.333 | |
| 12 | 1.86 | 0.0017 | 56 | D12S398 | 0 | |
| 13 | 1.19 | 0.0097 | 36 | D13S325 | 3 | |
| 14 | 1.75 | 0.0023 | 109.5 | D14S1434 | 3.5 | |
| 15 | 1.42 | 0.0053 | 58.667 | D15S131 | 1.333 | |
| 16 | 1.02 | 0.015 | 27.5 | D16S3103 | 4.5 | |
| 18 | 1.05 | 0.0141 | 28 | D18S542 | 0 | |
| 19 | 1.41 | 0.0054 | 10 | D19S1034 | 0 | |
| WISC PIQ | 2 | 1.68 | 0.0027 | 38 | D2S405 | 0 |
| 3 | 1.34 | 0.0064 | 210.167 | D3S2418 | 1.167 | |
| 4 | 1.28 | 0.0076 | 33 | D4S391 | 0 | |
| 5 | 1.61 | 0.0032 | 172 | D5S1471 | 0 | |
| 9 | 1.21 | 0.0091 | 104 | D9S938 | 0 | |
| 12 | 1.78 | 0.0021 | 51.333 | C12S916 | 2.333 | |
| 13 | 2.01 | 0.00118 | 56 | D13S317 | 0 | |
| 15 | 2.06 | 0.00103 | 60 | D15S131 | 0 | |
| 18 | 1.07 | 0.0132 | 67.667 | D18S851 | 3.667 | |
| 19 | 1.33 | 0.0067 | 3.333 | D19S591 | 3.333 | |
| WISC FSIQ | 2 | 1 | 0.016 | 84.833 | D2S1394 | 2.167 |
| 3 | 1.06 | 0.0137 | 182 | D3S2427 | 0 | |
| 12 | 2.05 | 0.00106 | 36 | D12S1042 | 0 | |
| 19 | 1.12 | 0.0116 | 0 | D19S591 | 0 |
Abbreviations: FSIQ, full scale intelligence quotient; LOD, logarithm of odds; PIQ, performance intelligence quotient; WISC, Wechsler Intelligence Scale for Children.
Association analyses to loci linked to ADHD
Fine-scale targeted genetic association with a resolution of ~68 Kilo-base pairs (kb) was conducted to SNP markers spanning regions linked to ADHD, that is, 4q, 5q, 11p, 17p, 20q from the Paisa genetic sample3 by using an approach similar to that previously described.10 The 11p region refers to the linkage and association of variants of DRD4 to ADHD.11 DRD4 is harbored in 11p. Unfortunately, we did not have enough data of the 11q-linked region in the set of paisa families. In the present studies, the data obtained were imported to SNP and Variation Suite (SVS) 7.6.7 (Golden Helix, Bozeman, MT, USA; http://www.goldenhelix.com) for association analyses.12, 13, 14 The Golden Helix SVS 7.6.7 is an integrated collection of analytic tools for managing, analyzing and visualizing multifaceted genomic and phenotypic data. Parameters for excluding markers from analyses included: (i) deviations from Hardy–Weinberg equilibrium, (ii) a minimum genotype call rate of 70%, (iii) the presence of more than two alleles and (iv) monoallelic markers. Genotype and allelic frequencies were estimated by maximum likelihood. Family-based association tests as implemented in SVS 7.6.7 were applied to the whole set of markers that passed quality control. Genetic analysis, using the dominant model, and allelic tests of association were applied as implemented in Golden Helix's SVS 7.6.7. Each endophenotype (WISC Block Design, WISC PIQ, WISC FSIQ, ACVT Correct Responses, ACVT Omissions and ROCFT Copy scores) was independently analyzed while age, sex and school grade were considered as covariates of interest. ADHD status was considered as an interacting variable. Multiple test correction to determine significance was performed using the false discovery rate (FDR) approach. Haplotype analyses were also applied to contrast with marker-wise results (described in detail in Supplementary Materials).
Results
Sample population—inclusion/exclusion criteria
From the 352 children and adolescents, 16 were excluded; 10 had a diagnosis of probably affected with ADHD and 6 were excluded because of incomplete clinical information. This left 336 young subjects, including 228 affected and 108 unaffected with ADHD in whom FSIQ was assessed. Only children and adolescents with FSIQ⩾81 and with regular school grades corresponding to their age were included in subsequent analyses to exclude participants potentially affected with generalized learning disorders. After applying this exclusion criterion, a final sample of 288 children and adolescents remained, including 194 (67.4%) affected with ADHD and 94 (32.6%) unaffected. The proportion of excluded children and adolescents with FSIQ⩽80 and academic problems did not differ statistically between affected (34/228; 14.9%) and unaffected children (14/108; 13.0%), (odds ratio=1.17, 95% confidence interval: 0.6–2.3, chi-square=0.2274, P=0.63). We observed expected significant differences between ADHD affected and unaffected individuals on demographic covariates: sex (P<0.00001), age (P<0.0001) and school grade (P<0.0001).
Whole-genome scan non-parametric linkage analyses
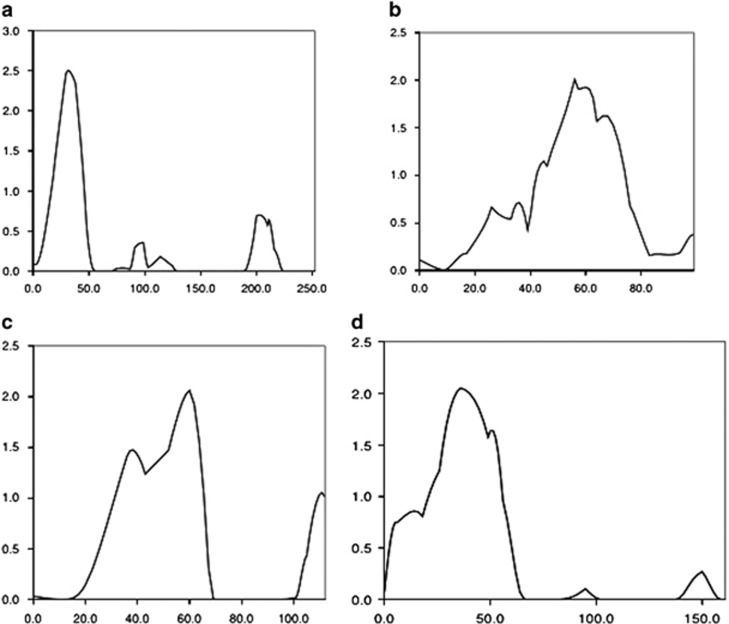
We found LOD scores >2.0 for WISC Block Design on chromosome 2, marker D2S1360 (LOD=2.51, P=0.00034); WISC PIQ on chromosome 15, marker D15S131 (LOD=2.06, P=0.00103; and at marker D13S317 (LOD=2.01, P=0.00118); and for WISC FSIQ on chromosome 12, marker D12S1042 (LOD=2.05, P=0.00106 (Figure 1 and Table 1). Nominal LOD scores >1.0 are presented in Table 1. Additional linkage results are presented in the Supplementary Materials.
Figure 1.
ADHD endophenotypes multipoint linkage chromosomal plots of non-parametric LOD scores >2.0. (a) WISC block design, chromosome 2p; (b) WISC PIQ, chromosome 13q; (c) the WISC PIQ, chromosome 15q; (d) WISC FSIQ chromosome 12p. LOD, logarithm of odds.
Association analysis to loci linked to ADHD
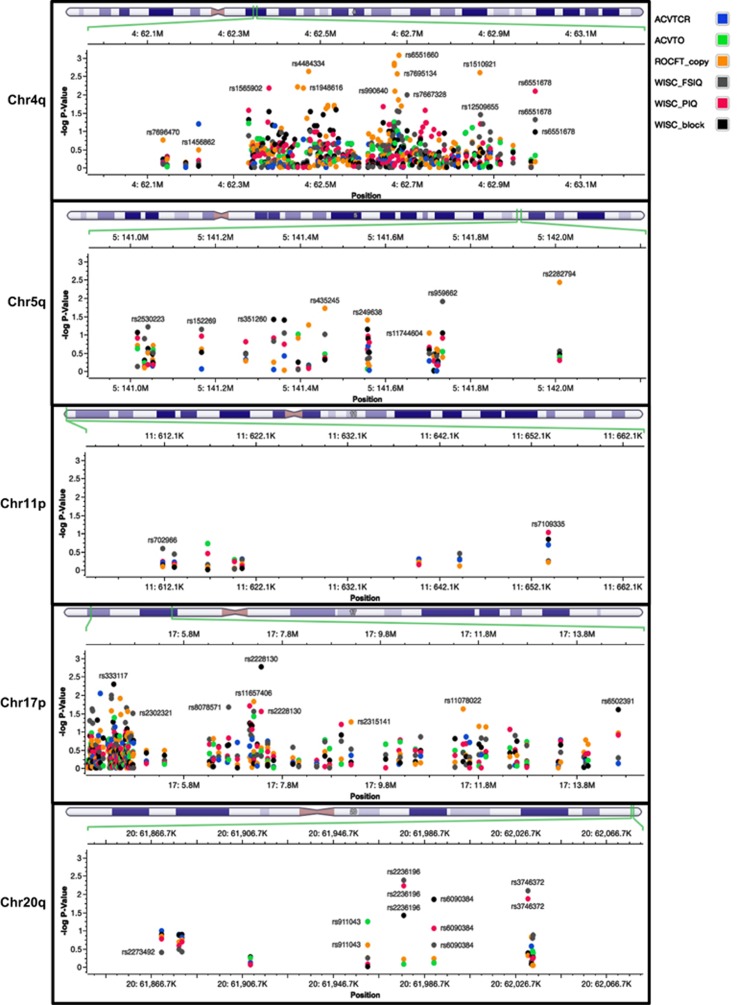
The targeted association analysis was carried out to SNP markers spanning regions previously described to be linked with ADHD, that is, 4q, 5q, 11p, 17p and 20q.3 Table 2 shows only the significant associations after FDR correction. The ROCFT endophenotype was associated to markers rs6551660 (G allele, Praw=0.0009, PFDR=0.0260) and rs2013374 (G allele, Praw=0.0014, PFDR=0.0260), both located within the Latrophilin 3 gene (LPHN3) (Figure 2). We also found that rs2282794, harbored in the Fibroblast growth factor 1 (FGF1) gene, was significantly associated with ROCFT. Two markers were found to be significantly associated with WISC Block Design, rs2228130 (A allele, Praw=0.0018, PFDR=0.0260) and rs333117 (C allele, Praw=0.0051, PFDR=0.0350) located within the Polymerase (RNA) II (DNA directed) polypeptide A (POLR2A) and between the Protein spinster homolog 3 (SPNS3) gene and the Protein spinster homolog 2 (SPNS2) gene, respectively. Two markers were found to be significantly associated with WISC FSIQ, rs2236196 and rs3746372 located within the Cholinergic receptor, nicotinic, alpha 4 (neuronal) (CHRNA4) gene and between the uncharacterized LOC100130152 and the Potassium voltage-gated channel subfamily KQT member 2 (KCNQ2) genes, respectively. Three markers were found to be significantly associated with WISC PIQ, the two markers with the highest association being rs2236196 and rs1565902, which are located within the CHRNA4 and LPHN3 genes, respectively. Marker rs1982177, located within the Ankyrin repeat and FYVE domain-containing protein 1 gene (ANKFY1) was found to be associated to ACVT Omissions and ACVT Correct Responses. Haplotype results are presented in the Supplementary Materials section.
Table 2. Top 20 regions of association for endophenotype and SNP markers, chromosome position, allele and frequency, closest gene, P-values with and without FDR correction.
| Endophenotype | Marker | Chromosome | Position | Allele (Frequency) | Closest gene(s) | P-value |
|
|---|---|---|---|---|---|---|---|
| Raw | FDR-corrected | ||||||
| ROCFT | rs6551660 | 4 | 62708149 | G (0.43) | LPHN3 | 0.0009 | 0.0260 |
| rs2013374 | 4 | 62697759 | G (0.37) | LPHN3 | 0.0014 | 0.0260 | |
| rs2122642 | 4 | 62698263 | C (0.37) | LPHN3 | 0.0016 | 0.0260 | |
| rs2345041 | 4 | 62698356 | C (0.37) | LPHN3 | 0.0016 | 0.0260 | |
| WISC block design | rs2228130 | 17 | 7404990 | A (0.03) | POLR2A | 0.0018 | 0.0260 |
| ROCFT | rs4484334 | 4 | 62499840 | T (0.49) | LPHN3 | 0.0024 | 0.0260 |
| rs1510921 | 4 | 62895591 | T (0.19) | LPHN3 | 0.0026 | 0.0260 | |
| rs7695134 | 4 | 62704851 | T (0.42) | LPHN3 | 0.0027 | 0.0260 | |
| rs2282794 | 5 | 141981708 | A (0.11) | FGF1 | 0.0037 | 0.0317 | |
| WISC FSIQ | rs2236196 | 20 | 61977555 | G (0.26) | CHRNA4 | 0.0043 | 0.0331 |
| WISC block design | rs333117 | 17 | 4395169 | C (0.45) | SPNS3/SPNS2 | 0.0051 | 0.0350 |
| WISC PIQ | rs2236196 | 20 | 61977555 | G (0.26) | CHRNA4 | 0.0061 | 0.0331 |
| ROCFT | rs10001410 | 4 | 62474228 | C (0.48) | LPHN3 | 0.0062 | 0.0350 |
| rs1948616 | 4 | 62487687 | T (0.48) | LPHN3 | 0.0067 | 0.0350 | |
| WISC PIQ | rs1565902 | 4 | 62408619 | C (0.37) | LPHN3 | 0.0069 | 0.0350 |
| rs6551678 | 4 | 63023050 | G (0.31) | LOC391656/LOC100131441 | 0.0082 | 0.0350 | |
| WISC FSIQ | rs3746372 | 20 | 62032034 | G (0.27) | LOC100130152/KCNQ2 | 0.0083 | 0.0350 |
| ROCFT | rs990640 | 4 | 62698936 | T (0.37) | LPHN3 | 0.0083 | 0.0350 |
| ACVT-O | rs1982177 | 17 | 4119993 | C (0.45) | ANKFY1 | 0.0091 | 0.0350 |
| ACVTCR | rs1982177 | 17 | 4119993 | C (0.45) | ANKFY1 | 0.0091 | 0.0350 |
Abbreviations: ACVT, ‘A'-cancelation and vigilance test; FDR, false discovery rate; FSIQ, full scale intelligence quotient; PIQ, performance intelligence quotient; ROCFT, Rey-Osterrieth complex figure test; SNP, single nucleotide polymorphism; WISC, Wechsler Intelligence Scale for Children.
Figure 2.
Manhattan plot illustrating the independently analyzed endophenotypes with age and sex as covariates for chromosomal regions 4q, 5q, 11p, 17p and 20q. The vertical axis represents –log10(P-value); –lg(P)>1.30 was defined as a significant association. ROCFT copy was highly associated with LPHN3 variants harbored in the same region associated to ADHD. ACVTCR, <<A>> Cancellation and Vigilance Test copy response; ACVTO, <<A>> Cancellation and Vigilance Test Omissions; ROCFT_copy, Rey-Osterrieth Complex Figure Test Copy subtest; WISC_FSIQ, Wechsler Intelligence Scale for Children Full Scale Intelligence Quotient; WISC_PIQ, Wechsler Intelligence Scale for Children Performance Intelligence Quotient; WISC_block, Wechsler Intelligence Scale for Children Block Design.
Discussion
One of the long-term goals of finding associations between endophenotypes and specific loci and/or genes is the prediction of disease risk before clinical symptoms manifest fully. As a preliminary approach, this exploratory study sought possible associations between neuropsychological impairments frequently observed in patients with ADHD and novel genes. By using neuropsychological and genetic data from multigenerational families from the Paisa genetic isolate, we performed genetic linkage and association studies for several neuropsychological endophenotypes of ADHD. Linkage analysis was used to identify potential chromosomal regions. Then, association analysis was applied to specific loci identified as linked to ADHD in these pedigrees with the aim of identifying potential new candidate genes or to evaluate loci previously defined within the LPHN3 gene.
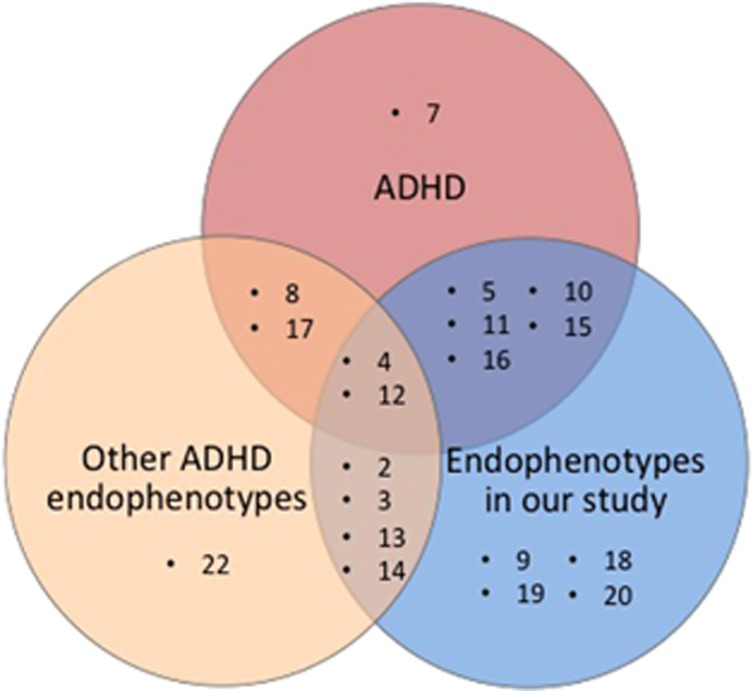
Our linkage analyses yielded nominal linkage at multiple chromosomal regions for WISC Block Design, WISC PIQ, WISC FSIQ, ACVT Correct Responses and ACVT Omissions. The six endophenotypes tested for a diverse range of performance, from fluid intelligence to sustained attention and visual-motor skills. We found genomic regions previously implicated in ADHD linked to our endophenotypes in chromosomes 4q13.2, 5q33.3, 11p15.5, 12p11.23 and 17p11. The overlapping of our linked regions to endophenotypes, that is, 2p24.2, 13q31.1, 15q23 and 12p11.23 to those reported by other studies is very difficult to demonstrate without applying a formal meta-analysis. However, our linkage results suggest that there is convergence with other endophenotype linkage studies in chromosomes 2, 3, 4, 12, 13 and 14 (ref. 15, 16) (Figure 3).
Figure 3.
Venn diagram comparing chromosomes implicated in ADHD linkage analyses, our endophenotype linkage analyses and ADHD endophenotype analyses by other investigators.
The implicated chromosomes described here and in other endophenotype studies could lead to the identification of novel candidate genes for ADHD, for example, promising linkage results from chromosome 13 led to the identification of TUBA3 as a potential ADHD candidate gene.15 These results support the concept that endophenotype measures could be a good indicator of ADHD status, but it is important to emphasize that by definition an endophenotype is not identical to a diagnosis, for example, an endophenotype is presumed to be more directly related to genetic factors, but many individuals will exhibit the endophenotype without the diagnosis, and vice versa.4
Association analyses carried out on selected chromosomal regions (4q, 5q, 11p, 17p and 20q) revealed significant associations for all tested endophenotypes, particularly close to the genes LPHN3, FGF1, POLR2A, SPNS3, SPNS2, KCNQ2, CHRNA4 and ANKFY1. Interestingly, ROCFT was the endophenotype with the greatest significance in association studies even though it did not yield any nominal linkage signals. From the ROCFT association studies, two genes of interest were identified, LPHN3 and FGF1. LPHN3 encodes a member of the latrophilin subfamily of G protein-coupled receptors and has already been implicated in ADHD.3 Latrophilins are thought to function in cell adhesion and signal transduction. LPHN3 is highly expressed in the brain, particularly in the amygdala, caudate nucleus, pontine nucleus and cerebellum.10 A loss of LPHN3 function caused a reduction in the number and misplacement of dopamine-positive neurons in the ventral diencephalon of zebrafish which displayed a hyperactive/impulsive motor phenotype.17 Hyperactivity/impulsivity or a lack of attention and motor control, prominent symptoms of ADHD, should impair the ability to perform the ROCFT copy test. Thus, the significant association between LPHN3 and the ROCFT suggests that this endophenotype may be useful in dissecting the complex pathophysiology of ADHD.
Our ROCFT analysis also showed significant associations with FGF1. FGF1 encodes for a protein in the fibroblast growth factor (FGF) family. The FGF family is involved in a broad range of mitogenic and cell survival activities ranging from embryonic development, cell growth, morphogenesis, tissue regeneration, tumor growth and invasion.18 The FGF1 protein has a role in neuronal survival in Alzheimer's disease and is thought to be involved in its pathophysiology.19 This gene could represent a novel candidate gene for ADHD by virtue of its involvement in neuronal survival, which may underlie the consistently decreased brain volume observed in ADHD.20 FGF1 was found to be highly expressed in brain regions related to major depression that might also be relevant for ADHD such as the dorsolateral prefrontal cortex and the anterior cingulate cortex.21
The association analyses also showed that CHRNA4 was significantly associated with WISC FSIQ and WISC PIQ endophenotypes. This gene encodes a subunit of the neuronal nicotinic acetylcholine receptor, which is widely distributed in the brain and is involved in attention, memory and perception.22 This gene has previously been associated with ADHD and is strongly implicated in tobacco addiction.22, 23, 24 On the basis of the comorbidity of substance use disorders (particularly nicotine addiction) with ADHD,3, 8, 25 and the distribution of receptors that would be affected by CHRNA4 mutations, this gene and the nicotinic pathway should be examined more closely to better understand the pathophysiology of ADHD.
Association analyses also found significant associations for WISC Block Design and markers close to SPNS3/SPNS2 and POLR2A. SPNS3 is a gene that belongs to the solute carrier family 22 (SLC22) expressed in rat frontal cortex.26 However, the possible functions of SPNS3 within the brain remain unknown. With regard to SPNS2, there is no evidence of a specific role in brain, but it appears to be essential in cell trafficking from the bone marrow to blood.27 The POLR2A gene encodes the primary subunit of the RNA polymerase II, which synthesizes messenger RNA in eukaryotes.28 As there is scarce information about this gene, its possible involvement in ADHD remains to be studied.
A gene marker close to KCNQ2, a potassium channel encoding gene, was associated with WISC FSIQ. KCNQ2 mutations are involved in forms of benign familial neonatal epilepsy, a condition that can co-occur with ADHD.29, 30
Finally, the biological relevance of the association between the endophenotypes ACVT Omissions and ACVT Correct Responses with ANKFY1 remains unknown. ANKFY1 encodes an endosomal protein that participates in cell trafficking and is expressed in several tissues including the brain, but this gene has not been previously related to any neuropathological conditions.
Given that these pedigrees were ascertained because of ADHD clustering, our analyses controlled for ADHD as a predictor (covariate of interest) to determine genetic effects after adjusting for ADHD. By adding ADHD status to the conditional mean model and also using such status for the offset computation, we also increased the power of the family-based association test statistic substantially.31, 32
This study has several limitations. Chromosome 2 had the highest linkage score for WISC Block Design (LOD=2.51) in our analyses and was shown to be a locus with significant linkage when Motor Timing was studied as a potential ADHD endophenotype.15 However, we only performed association analyses on our six endophenotypes from chromosome 4 onwards. This was because of the limited availability of SNP genotyped data for chromosome 2 in our sample. As a result, we were unable to observe potential associations to candidate genes on chromosome 2.
Even though the number of individuals in this study was of sufficient size to capture endophenotype associations, the sample derived from a genetic isolate, the Paisas. This genetic isolate has been useful in identifying a number of genes associated to endophenotypes and potentially to ADHD,5 but our association findings need to be replicated in independent populations and meta-analyzed to determine whether our results will apply to diverse populations. We pursued such a strategy for LPHN3, in which the initial finding was made in the Paisa genetic isolate and replicated in German, Spanish, American and Norwegian samples.10 Thus, future replications in non-Paisa populations are planned. Further studies should also be carried out on the genes of interest that were significantly associated to the endophenotypes in this project, particularly FGF1 and CHRNA4, to determine whether their variants confer susceptibility or protection for ADHD.
In conclusion, we found that neuropsychological endophenotypes were useful in discovering potential candidate genes related to ADHD that can afford greater insight into the pathophysiology of the disorder. Our analyses support the concept that the six endophenotypes are potentially linked and/or associated to ADHD. The ROCFT and WISC endophenotypes are specially promising, relative to the use of clinical categorical criteria alone, as they are able to identify individuals along a continuum. These are also widely available well-standardized measures. Furthermore, using different endophenotypes may also allow narrowing the specific neurobiological issue faced by individual subjects, because different pathways were implicated. For example, lower scores on the WISC could implicate the nicotinic pathway (because CHRNA4 was associated with WISC endophenotypes) while lower scores on the ROCFT might implicate frontal-parietal circuits given FGF1 association with the endophenotype. Thus, from linkage and association analyses, endophenotypes provide a powerful, objective and independent framework to current syndromic psychiatric classification in assessing the potential genetic causes underpinning ADHD susceptibility.
Acknowledgments
Claudio Mastronardi, Eva Pillai and Mauricio Arcos-Burgos are supported by John Curtin School of Medical Research, ANU College of Medicine, Biology and Environment, The Australian National University, Canberra, ACT, Australia, Project R42100-12. Maximilian Muenke, Ariel F Martinez, Maria T Acosta and Mauricio Arcos-Burgos are supported by the Division of Intramural Research, National Human Genome Research Institutes, National Institutes of Health, Bethesda, MD, USA. Part of this research was funded by the NHGRI-NIH and by COLCIENCIAS which funded the Project: ‘Genética del Trastorno de Atención-Hiperactividad: los fenotipos complejos, los endofenotipos y la asociación con genes mayores y de susceptibilidad' Code: 1115-04-18083, Contract: 459-2005, to David Pineda, University of Antioquia and University of San Buenaventura.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- Polanczyk G, De Lima MS, Horta BL, Biederman J, Rohde LA. The worldwide prevalence of ADHD: A systematic review and metaregression analysis. Am J Psychiatry 2007; 164: 942–948. [DOI] [PubMed] [Google Scholar]
- Faraone SV, Mick E. Molecular genetics of attention deficit hyperactivity disorder. Psychiatr Clin North Am 2010; 33: 159–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcos-Burgos M, Vélez J, Solomon B, Muenke M. A common genetic network underlies substance use disorders and disruptive or externalizing disorders. Hum Genet 2012; 131: 917–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: Etymology and strategic intentions. Am J Psychiatry 2003; 160: 636–645. [DOI] [PubMed] [Google Scholar]
- Pineda DA, Lopera F, Puerta IC, Trujillo-Orrego N, Aguirre-Acevedo DC, Hincapie-Henao L et al. Potential cognitive endophenotypes in multigenerational families: segregating ADHD from a genetic isolate. Atten Defic Hyperact Disord 2011; 3: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcos-Burgos M, Castellanos FX, Lopera F, Pineda D, Palacio JD, Garcia M et al. Attention-deficit/hyperactivity disorder (ADHD): feasibility of linkage analysis in a genetic isolate using extended and multigenerational pedigrees. Clin Genet 2002; 61: 335–343. [DOI] [PubMed] [Google Scholar]
- Arcos-Burgos M, Castellanos FX, Pineda D, Lopera F, Palacio JD, Palacio LG et al. Attention-deficit/hyperactivity disorder in a population isolate: linkage to loci at 4q13.2, 5q33.3, 11q22, and 17p11. Am J Hum Genet 2004; 75: 998–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacio JD, Castellanos FX, Pineda DA, Lopera F, Arcos-Burgos M, Quiroz YT et al. Attention-deficit/hyperactivity disorder and comorbidities in 18 Paisa Colombian multigenerational families. J Am Acad Child Adolesc Psychiatry 2004; 43: 1506–1515. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 2002; 30: 97–101. [DOI] [PubMed] [Google Scholar]
- Arcos-Burgos M, Jain M, Acosta MT, Shively S, Stanescu H, Wallis D et al. A common variant of the latrophilin 3 gene, LPHN3, confers susceptibility to ADHD and predicts effectiveness of stimulant medication. Mol Psychiatry 2010; 15: 1053–1066. [DOI] [PubMed] [Google Scholar]
- Arcos-Burgos M, Castellanos FX, Konecki D, Lopera F, Pineda D, Palacio JD et al. Pedigree disequilibrium test (PDT) replicates association and linkage between DRD4 and ADHD in multigenerational and extended pedigrees from a genetic isolate. Mol Psychiatry 2004; 9: 252–259. [DOI] [PubMed] [Google Scholar]
- Wong ML, Dong C, Flores DL, Ehrhart-Bornstein M, Bornstein S, Arcos-Burgos et al. Clinical outcomes and genome-wide association for a brain methylation site in an antidepressant pharmacogenetics study in Mexican Americans. Am J Psychiatry 2014; 171: 1297–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Filho G, Boguszewski MC, Mastronardi CA, Patel HR, Johar AS, Chuah A et al. Whole exome sequencing of extreme morbid obesity patients: translational implications for obesity and related disorders. Genes (Basel) 2014; 5: 709–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johar AS, Mastronardi C, Rojas-Villarraga A, Patel HR, Chuah A, Peng K et al. Novel and rare functional genomic variants in multiple autoimmune syndrome and Sjögren's syndrome. J Transl Med 2015; 13: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommelse NN, Arias-Vasquez A, Altink ME, Buschgens CJ, Fliers E, Asherson P et al. Neuropsychological endophenotype approach to genome-wide linkage analysis identifies susceptibility loci for ADHD on 2q21.1 and 13q12.11. Am J Hum Genet 2008; 83: 99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle AE, Ferreira MA, Sklar PB, Lasky-Su J, Petty C, Fusillo SJ et al. Multivariate genomewide linkage scan of neurocognitive traits and ADHD symptoms: suggestive linkage to 3q13. Am J Med Genet B Neuropsychiatr Genet 2008; 147B: 1399–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange M, Norton W, Coolen M, Chaminade M, Merker S, Proft F et al. The ADHD-susceptibility gene lphn3.1 modulates dopaminergic neuron formation and locomotor activity during zebrafish development. Mol Psychiatry 2012; 17: 946–954. [DOI] [PubMed] [Google Scholar]
- Yun Y-R, Won JE, Jeon E, Lee S, Kang W, Jo H et al. Fibroblast growth factors: biology, function, and application for tissue regeneration. J Tissue Eng 2010; 1: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashayekhi F, Hadavi M, Vaziri HR, Naji M. Increased acidic fibroblast growth factor concentrations in the serum and cerebrospinal fluid of patients with Alzheimer's disease. J Clin Neurosci 2010; 17: 357–359. [DOI] [PubMed] [Google Scholar]
- Krain AL, Castellanos FX. Brain development and ADHD. Clin Psychol Rev 2006; 26: 433–444. [DOI] [PubMed] [Google Scholar]
- Evans SJ, Choudary PV, Neal CR, Li JZ, Vawter MP, Tomita H et al. Dysregulation of the fibroblast growth factor system in major depression. Proc Natl Acad Sci USA 2004; 101: 15506–15511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itier V, Bertrand D. Neuronal nicotinic receptors: from protein structure to function. FEBS Lett 2001; 504: 118–125. [DOI] [PubMed] [Google Scholar]
- Wallis D, Arcos-Burgos M, Jain M, Castellanos FX, Palacio JD, Pineda D et al. Polymorphisms in the neural nicotinic acetylcholine receptor alpha4 subunit (CHRNA4) are associated with ADHD in a genetic isolate. Atten Defic Hyperact Disord 2009; 1: 19–24. [DOI] [PubMed] [Google Scholar]
- Banaschewski T, Becker K, Scherag S, Franke B, Coghill D. Molecular genetics of attention-deficit/hyperactivity disorder: an overview. Eur Child Adolesc Psychiatry 2010; 19: 237–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta MT, Arcos-Burgos M, Muenke M. Attention deficit/hyperactivity disorder (ADHD): complex phenotype, simple genotype? Genet Med 2004; 6: 1–15. [DOI] [PubMed] [Google Scholar]
- Jacobsson JA, Haitina T, Lindblom J, Fredriksson R. Identification of six putative human transporters with structural similarity to the drug transporter SLC22 family. Genomics 2007; 90: 595–609. [DOI] [PubMed] [Google Scholar]
- Fukuhara S, Simmons S, Kawamura S, Inoue A, Orba Y, Tokudome T et al. The sphingosine-1-phosphate transporter Spns2 expressed on endothelial cells regulates lymphocyte trafficking in mice. J Clin Invest 2012; 122: 1416–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Asbroek AL, Fluiter K, Van Groenigen M, Nooij M, Baas F. Polymorphisms in the large subunit of human RNA polymerase II as target for allele-specific inhibition. Nucleic Acids Res 2000; 28: 1133–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JE, Watson R, Sheth R, Caplan R, Koehn M, Seidenberg M et al. Psychiatric comorbidity in children with new onset epilepsy. Dev Med Child Neurol 2007; 49: 493–497. [DOI] [PubMed] [Google Scholar]
- Kattimani S, Mahadevan S. Treating children with attention-deficit/hyperactivity disorder and comorbid epilepsy. Ann Indian Acad Neurol 2011; 14: 9–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C, Laird NM. On a general class of conditional tests for family-based association studies in genetics: the asymptotic distribution, the conditional power, and optimality considerations. Genet Epidemiol 2002; 23: 165–180. [DOI] [PubMed] [Google Scholar]
- Lange C, Laird NM. Power calculations for a general class of family-based association tests: dichotomous traits. Am J Hum Genet 2002; 71: 575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.