Abstract
Synthetic humanized antibody libraries are frequently generated by random incorporation of changes at multiple positions in the antibody hypervariable regions. Although these libraries have very large theoretical diversities (>1020), the practical diversity that can be achieved by transformation of E. coli is limited to about 1010. To constrain the practical diversity to sequences that more closely mimic the diversity of natural human antibodies, we generated a scFv phage library using entirely pre-defined complementarity determining regions (CDR). We have used this library to select for novel antibodies against four human protein targets and demonstrate that identification of enriched sequences at each of the six CDRs in early selection rounds can be used to reconstruct a consensus antibody with selectivity for the target.
1. Introduction
Phage display is an attractive approach for the rapid identification of antibodies for therapeutic, diagnostic, and research applications. The phage display selections are done entirely in vitro. The derived antibodies can recognize targets that are toxic or non-immunogenic in animals. They can also discriminate antigen conformations or post-translational modifications. In addition, the gene encoding the antibody is easily accessible in phage display and can be cloned and genetically engineered in many ways.
Generally, phage display libraries are engineered to display the variable, antigen-binding domains of antibodies, either as single chains (scFv) (1) or as heavy and light chain antigen binding fragments (Fabs) (2). Several methods have been used to produce antibody diversity in vitro. Antibody variable sequences can be amplified from the natural immunoglobulin gene repertoire in human B-cells (3–6). Alternatively, recombinant antibody libraries can be built using synthetic diversity (7–9) in which antibody complementarity determining region (CDR) containing gene fragments are generated with mixed-nucleotide synthesis (10). A semi-synthetic approach can also be used to further diversify native heavy and light chain genes by synthetically randomizing the CDRs (11). Disadvantages to these library types include variable biophysical properties and expression levels of heterogeneous frameworks, and stop codons in mixed-nucleotide sequences in synthetic and semi-synthetic approaches. Libraries using triplet codon synthesis, in place of mixed single-base nucleotide synthesis have been developed (12), but are relatively expensive to produce and they do not generally allow one to vary the codon-frequency at more than a couple of sites in the gene. Sidhu et al. has developed an alternative approach by synthesizing libraries with reduced codon usage that eliminate the incorporation of stop codons (13). However, the total potential diversity with these libraries is still substantially higher (>1023) than the diversity that can be possibly sampled (typically 1010–1011 with phage libraries). Not all amino acids at a given CDR position will yield a functional antibody (14).
As an alternative to bulk synthesis of oligonucleotides with random, mixed bases, thousands of individual oligonucleotides with pre-defined CDR sequences could be synthesized on a microarray (15–18). LeProust et al. recently improved the quality of microarray-synthesized oligonucleotides by controlling depurination during the synthesis process (19), and several groups have used oligonucleotide library synthesis (OLS) pools in DNA capture technologies, promoter analysis, and DNA barcode development (16, 20–22). By pre-defining the diversity in an antibody display library, stop codons, immunogenic sequences, or sequences with the potential to interfere with production could be eliminated (15). Additionally, rapid identification of enriched sequences and clonal sequence reconstruction may be facilitated (15).
In this study, a single framework of human germline lineage was used to build a diverse synthetic scFv library with pre-defined CDRs. Each CDR was designed to exclude stop codons, cysteines, and restriction sites, and to include changes that mimic the mutation rate found in vivo. Single frameworks enable library amplification and sequencing using a single set of primers, facilitating analysis and providing similar biophysical properties to the resultant antibodies (e.g., expression, thermal stability, etc.) (23, 24). We used this library to select for novel antibodies against four human protein targets, EZH2 (25), ZMAT3 (26), ZNF622 (27), and TDP-43 (28) by phage display. We show that identification of enriched sequences at each of the six CDRs can be used to reconstruct a consensus antibody with selectivity for the target.
2. Materials and Methods
2.1 Bacterial strains and vectors
The E. coli strain CJ236 (Genotype: FΔ(HinDIII)::cat (Tra+, Pil+, CamR)/ ung-1, relA1, dut-1, thi-1, spoT1, mcrA) was purchased from New England BioLabs (NEB; Waverly, MA). This strain lacks functional dUTPase and uracil N-glycosylase, and yields uracilated, single-stranded DNA template when infected with M13 bacteriophage. The E. coli strain TG1 [F' (traD36, proAB+ lacIq, lacZΔM15), supE, thi-1, Δ(lac-proAB), Δ(mcrB-hsdSM)5, (rK−mK−)] was purchased from Lucigen Corporation (Middleton, WI). Electrocompetent cells were produced by Lucigen Corporation. The template plasmid for library construction (pAX1519) is a derivative of the phagemid, pAP-III6 (6) with a single-chain variable fragment (scFv) antibody fused to the coat protein III of bacteriophage M13. The scFv was based on the human frameworks VH5-3 and VL3. Complementarity determining regions (CDRs) L1, L2, H1 and H2 of the scFv in pAX1519 were modified to contain a BssHII restriction endonuclease recognition site. CDRs L3 and H3 of the scFv in pAX1519 were modified to contain a SacII (Eco29kI isoschizomer) restriction endonuclease recognition site and a TGA (opal) stop codon. In this template, non-recombinant clones are non-functional with respect to display of the scFv.
2.2 Oligonucleotide Synthesis
Approximately 4000, 60–100mer oligonucleotides, encoding pre-defined CDR sequences, were synthesized on each of two oligonucleotide microarrays (LC Biosciences, Houston, TX). The randomized CDR sequences were flanked on each end by at least 20 nucleotides complementary to the framework region flanking the CDR. The oligonucleotides were cleaved from the array and provided as a pool. The design of oligonucleotides for the library are discussed in detail below.
2.3 Oligonucleotide Amplification
Oligonucleotide primers were designed to the 5’ and 3’ flanking positions of each CDR, such that the resulting production from the polymerase chain reaction (PCR) was > 100 base pairs. The reverse primers were designed to contain a 5’ phosphate and four phosphorothioate linkages beginning at the 5’ end. For amplifying the oligonucleotide pool corresponding to each CDR, a separate PCR reaction was set up using 45 µl of Platinum Taq polymerase (Thermo Fisher Scientific, Waltham, MA), 1 µl of a 10 µM stock of forward PCR primer, 1 µl of 10 µM stock of reverse PCR primer, and 5 ng of the array-synthesized oligonucleotide pool. The resulting PCR product was cleaned up following the QIAquick standard protocol (Qiagen, Valencia, CA) and incubated with 25 µl of T7 exonuclease (NEB) at 25°C for 1 hr. Thesample was purified using the QIAquick PCR Purification Kit and eluted in 40 µl EB buffer (NEB). Approximately 2 micrograms of single-stranded product, corresponding to the amplified oligonucleotide pool for each CDR, was recovered.
2.4 Library Construction
Uracilated, circular, single-stranded template (dU-ssDNA) was produced following a published protocol (13). Seventy µl (~2 micrograms) each of amplified single strand L3 and H3 oligonucleotide was annealed to 12 µl (2 micrograms) of a ssDNA preparation of the library template, pAX1519 (2:1 molar ratio of oligonucleotide to dU-ssDNA, respectively) in 25 µl of 10× TM buffer (0.1 M MgCl2, 0.5 M Tris, pH 7.5) buffer and dH20 for a final reaction volume of 250 µl. The annealing reaction was carried out by heating the mixture at 90°C for 2 min (min), followed by a temperature decrease of 1 °C per min to 25°C in a thermal cycler. To the annealed product, 10 µl of 10 mM ATP, 10 µl of 100 mM dNTP mix, 15 µl of 100 mM DTT, 0.5 µl (30 U) T4 DNA ligase and 3 µl (30 U) T7 DNA polymerase were added. The mixture was distributed equally into five PCR tubes and incubated 16 hr (hr) at 20°C. The DNA was desalted and purified using a Qiagen QIAquick DNA purification kitusing 1 ml of buffer QC per sample and eluted with 70 ul of buffer EB.
2.5 Library transformation and virion production
Ten electroporations were conducted using 1 µl of heteroduplex product from above and 45 µl TG1 cells (Lucigen), following standard protocols. After a 1 hr recovery of the transformed cells in 1 ml recovery media (Lucigen), with shaking at 37 °C, the ten reactions were diluted into 50 ml of LB media containing ampicillin (100 µg/ml) and grown to an OD600 of 1.0. Cultures were then centrifuged at 1800 × g for 15 min, and DNA was isolated from the cell pellet using the Qiagen Midi prep kit following the standard protocol. Ten micrograms of resulting, pooled DNA was then digested with the SacII enzyme (NEB) at 37°C for 3 hr and cleaned up using the Qiaquick PCR purification kit (Qiagen). A second set of 10 electroporations was conducted using 0.5 µl (250 ng) of the SacII-digested DNA into 45 µl CJ236 cells (Lucigen), following the standard protocol. After 1 hr recovery of the transformed cells in 1 ml recovery media (Lucigen) shaking at 37°C, all 10 reactions (10ml total) were combined together with 20 ml of LB media, containing ampicillin (100 µg/ml), and grown at 37 °C, with shaking until the OD600 reached 0.4. Once the cells reached mid-log phase, 2 µl of M13K07 (1013 phage/ml) was added, and the culture was incubated at 37°C without shaking for 30 min. The culture was then centrifuged at 1800 × g for 15 min, and the pellet was resuspended in 30 ml of LB supplemented with ampicillin (100 µg/ml), kanamycin (50 µg/ml), and uridine (0.3 µg/ml) in a 250-ml baffled flask and grown overnight at 30°C with shaking. The next day, ssDNA was isolated using the method described in section 2.3 above. The resulting ssDNA was used in an annealing reaction with the amplified L1, L2, H1, and H2 oligonucleotides to generate the final library following the same procedure as described above. 100 electroporations of the final heteroduplex into TG1 cells were conducted as above and after 1 hr recovery, 25 ml of the transformed cells were diluted into each of four flasks containing 250 ml LB supplemented with ampicillin (100 µg/ml). The cells were grown to an OD600 of 0.4, and 25 ml were removed from each flask for phage production. Helper phage (M13K07; 1.25 × 1010) was added, and the cultures were incubated overnight at 30°C in LB containing ampicillin (100 µg/ml) and kanamycin (50 µg/ml), as described elsewhere (13). The remaining cells were pelleted and resuspended in LB supplemented with 15% glycerol. The cell stocks were frozen and stored at −80°C. The phage library from the overnight cultures was precipitated using PEG-NaCl, as described previously (13).
2.6 Antigen production, biotinylation, and purification
cDNA for all antigens was purchased from DNASU Plasmid Repository (Arizona State University, Tempe, AZ). Antigen fragments (see Table 1) were produced by PCR. EZH2, ZNF622, and ZMAT3 fragments were cloned into an expression vector with N-terminal Maltose Binding Protein (MBP) tag followed by Avi tag for in vivo biotinylation. TDP-43 fragment was cloned into an expression vector with N-terminal hexahistidine (6xHis), SUMO, and Avi tags. To enable the production of biotinylated protein these constructs were co-transfected into BL21 (DE3) cells (NEB) with an expression plasmid of E. coli BirA ligase (29).
Table 1.
Biotinylated antigens used in phage display screen
The amino acid sequences of each avi-tagged protein fragment is shown. The antigens were produced as either MBP (EZH2, ZNF622, ZMAT3) or SUMO fusions (TDP-43). Post-purification the antigens were captured on a neutravidin coated ELISA plate and screened with the PDC library for either 2 or 3 rounds.
| Antigen | Fragment size (aa) |
Sequence |
|---|---|---|
| ZMAT3 | 65 | MILLQHAVLPPPKQPSPSPPMSVATRSTGTLQLPPQKPFGQEASLPLAGEEELSKGGEQDCALEE |
| EZH2 | 100 | GGRRRGRLPNNSSRPSTPTINVLESKDTDSDREAGTETGGENNDKEEEEKKDETSSSSEANSRCQTPIKMKPNIEPPENV EWSGAEASMFRVLIGTYYDN |
| TDP-43 | 100 | MSEYIRVTEDENDEPIEIPSEDDGTVLLSTVTAQFPGACGLRYRNPVSQCMRGVRLVEGI LHAPDAGWGNLVYVVNYPKDNKRKMDETDASSAVKVKRAV |
| ZNF622 | 103 | ASMAPVTAEGFQERVRAQRAVAEEESKGSATYCTVCSKKFASFNAYENHLKSRRHVELEKKAVQAVNRKVEMMNEKN LEKGLGVDSVDKDAMNAAIQQAIKAQ |
For MBP fusion protein fragments: a single colony was picked off a LB + 100ug/mL Ampicillin + 34ug/mL of chloramphenicol plate and grown in 5mL of LB/Amp/CAM overnight at 37°C with shaking. The culture was diluted 1:100 in 400mL of HDA media (10g/L tryptone, 5g/L yeast extract) enriched with G (0.5% glycerol, 0.05% glucose, 0.2% alpha-D-lactose monohydrate) and M (25mM Na2HPO4, 25mM KH2PO4, 50mM NH4Cl, 5mM Na2SO4) + 100µg/mL Amp + 50uM biotin and grown at 30°C for 7 hrs and then at 16°C for 18–20 hrs with shaking. Cultures were centrifuged at 3500 × g for 20 min and the cell pellets were frozen at −80°C for at least 15 min. The pellets were thawed at room temperature and lysed in 4mL of B-Per bacterial protein extraction reagent (Pierce) supplemented with 250mM NaCl, 1mM PMSF, 1mM DTT, and protease inhibitor cocktails (5 µg/ml of each aprotinin, leupeptic, pestatin and chymostatin). Lysates were incubated at room temperature for 30 min while shaking and clarified by centrifugation at 15,000 × g for 15 min at 4°C. The clarified lysate was then purified on 2mL amylose resin bed volume (NEB). Columns were equilibrated with 10 column volumes of dH20 followed with PBS + 1mM PMSF. After loading the lysate onto the column it was washed with 12 column volumes of PBS + 1mM PMSF and then eluted with 3 column volumes of PBS + 0.01% Igepal + 1mM DTT + 10mM Maltose. Elutions were analyzed by SDS-PAGE and Coomassie Brilliant Blue staining.
For SUMO fusion of TDP-43 protein fragment: a single colony was picked off a LB + 100ug/mL Ampicillin + 34ug/mL of chloramphenicol plate and grown in 5mL of TYH (20g/L tryptone, 10g/L yeast extract, 11g/L Hepes, 5g/L NaCl, 1g/L MgSO4) + 100µg/mL Amp + 34 µg/mL CAM overnight at 37°C. The overnight culture was diluted by adding 0.65mL into 50mL of TYH/Amp and grown to OD600nm of 0.7. 1.5mM IPTG and 50µM Biotin was added to the culture and further incubated at 30°C overnight shaking at 225rpm. The cultures were centrifuged at 4000 × g for 20 min and the cell pellets were frozen at −80°C for at least 15 min. The pellets were thawed at room temperature and resuspended in 3mL of B-Per bacterial protein extraction reagent (Pierce) supplemented with 100mM PMSF and protease inhibitor cocktails (5 µg/ml of each aprotinin, leupeptic, pestatin and chymostatin) using a vortex. Lysates were incubated at room temperature for 30 min while shaking. Imidazole was added to the lysate for a final concentration of 7.5mM. The lysate was centrifuged at 15,000 × g for 20 min at 4°C. The clarified lysate was then purified on 0.5mL bed volume of HisPur Cobalt Resin (Thermo Fisher Scientific). Columns were equilibrated with 10 column volumes of dH20 followed with TBS (25mM Tris pH 7.4, 250mM NaCl) + 10% glycerol + 100mM PMSF+ 7.5mM Imidazole. After loading the lysate onto the column it was washed with 20 column volumes of TBS + 10% glycerol + 100mM PMSF+ 7.5mM Imidazole and eluted with 3 column volumes of TBS + 10% glycerol + 100mM PMSF + 4M Imidazole. Elutions were analyzed as above.
2.7 Phage display screen
Immunoplates (Nunc Maxisorp) were coated with NeutrAvidin (100 µl/well at 10 µg/ml; ThermoFisher Scientific, Rockford, IL) overnight at 4°C. After washing with PBS, plates were blocked with 250 µl/well of 2% nonfat dry milk in PBS (MPBS). To capture the biotinylated antigens, EZH2, ZMAT3, ZNF622, or TDP-43 on the NeutrAvidin-coated plates, the plates were washed 3 times with PBS, and 8 wells per target were coated with the corresponding biotinylated target at a concentration of 10 µg/ml in PBS (100 µl/well) at room temperature for 1 hr. The plates were again washed 3 times with PBS and blocked with MPBS. Phage library (around 1 × 1013 transducing units per ml in MPBS) was added to the plates (100 µl/well) and incubated for 2 hr at room temperature. After rigorous washing with PBS containing 0.1% (v/v) Tween 20, bound phage was eluted by addition of 2M glycine, pH 2.0 (100 µl/well). The eluate was neutralized by addition of 2M Tris, pH 10. The eluate was used to infect exponentially growing E. coli TG1 (200 µl/well) for 30 min at 37°C without shaking. The transduced cells were pelleted and resuspended in 600 µl/well LB media containing ampicillin (100 µg/ml) for overnight growth at 30°C. The following day, 5 µl/well of the overnight cultures were diluted into fresh LB media containing ampicillin (200 µl/well) and grown at 37°C until absorbance at 600 nm reached 0.4. M13K07 helper phage was added (1 × 109 transducing units per well) and the plates were incubated at 37 °C without shaking for 30 min to allow phage infection and then pelleted. The cells were resuspended in 600 µl/well LB containing ampicillin and kanamycin (50 µg/ml) and grown overnight at 30 °C with shaking. The next day the blocks were centrifuged at 1800 × g for 10 min and 100 µl/well of the phage supernatant was added directly to antigen-coated wells for a second round of selection following the protocol as above. Three total selection rounds were conducted. After the third round, transduced cells were plated onto LB-agar plates, containing ampicillin (100 µg/ml), for single colony isolation. The clones were amplified by colony PCR and sequenced at Quintara Biosciences (Albany, CA). Re-synthesis of scFvs with the enriched consensus CDRs was carried out by Gene Art service (Thermo Fisher Scientific).
2.8 Phage ELISA
Single colonies were picked from the plates into 200 µl of LB containing ampicillin (100 µg/ml) in 96 well deep-well blocks. Bacteria were grown overnight at 37°C and 2 µl samples transferred to 200 µl of medium in fresh blocks. Once the OD at 600 nm reached 0.4, 25 µl aliquots of M13K07 helper phage, each containing around 1 × 109 viruses, were added to each well to initiate superinfection, and the plates were incubated at 37°C without shaking for 30 min. The plates were then centrifuged at 1800 × g for 10 min and the cell pellets were resuspended in 200 µl of LB containing ampicillin and kanamycin (50 µg/ml) and the deep-well blocks incubated at 30°C with shaking overnight to allow viral replication. The plates were then centrifuged at 1800 × g for 10 min and supernatants were transferred directly to an ELISA plate that had been coated with the antigen. For coating of the ELISA plates, NeutrAvidin (10 µg/ml in PBS; 100 µl/well) was added and plates were incubated overnight at 4°C. The wells were washed three times with PBS (250 µl/well) and blocked for 1 hour with 2% nonfat dry milk in PBS (MPBS). The wells were again washed three times with PBS and biotinylated target was added at 10 µg/ml in PBS. After an hour incubation with biotinylated protein fragments, the wells were again blocked for an hour with MPBS and washed three times with PBS. Phage-containing supernatants were then incubated with the antigen-coated wells for 1 hour at room temperature. The wells were washed three times with PBS containing 0.1% (w/v) Tween 20 (250 µg/well) and binding of the phage was detected using a monoclonal anti-M13 Ab, conjugated to horseradish peroxidase (HRP; GE Healthcare, Piscataway, NJ).
2.9 Production and purification of soluble scFv antibodies
Antibodies fused to the gpIII coat protein of the phage were converted to soluble scFv proteins by infection into E. coli Mach I cells (Thermo Fisher Scientific). Single colonies were picked into 50 ml C.R.A.P. media [0.3 M ammonium sulfate, 0.002 M sodium citrate, 0.014 M potassium chloride, 0.5% yeast extract, 0.5% Hy-Case SF casein hydrolysate (Sigma #C9386), pH 7.3] supplemented with 7mM MgSO4, 14mM glucose, and 100µg/ml ampicillin and grown overnight in 250-ml baffled flasks with shaking at 30°C to induce expression from the phoA promoter. The following day, the cells were pelleted at 3000 × g for 20 min and the pellets were lysed in 2.5 ml B-Per (Thermo Fisher Scientific) containing benzonase (EMD Chemicals, Gibbstown, NJ), protease inhibitors, and 1mM PMSF by gentle rocking at room temperature for 30 min. Lysates were clarified by centrifugation at 15,000 × g in a Sorvall SS34 rotor for 30 min at 4°C. The scFvs were purified from the clarified lysates by cobalt chelation chromatography on 0.2 ml columns of Cobalt His-Pur resin (Thermo Scientific). After binding the scFv proteins, the columns were washed with 25mM Tris buffer pH7.4 containing 0.25M sodium chloride, 10% glycerol, and 1mM PMSF. The scFvs were eluted with the same buffer containing 250mM imidazole. Eluted protein was collected and analyzed by SDS-PAGE. For ELISA, binding of the scFvs to the protein fragment antigen on NeutrAvidin-coated plates was detected using an anti-FLAG-HRP conjugate (Abcam, Cambridge, UK).
3. Results
3.1 Library design
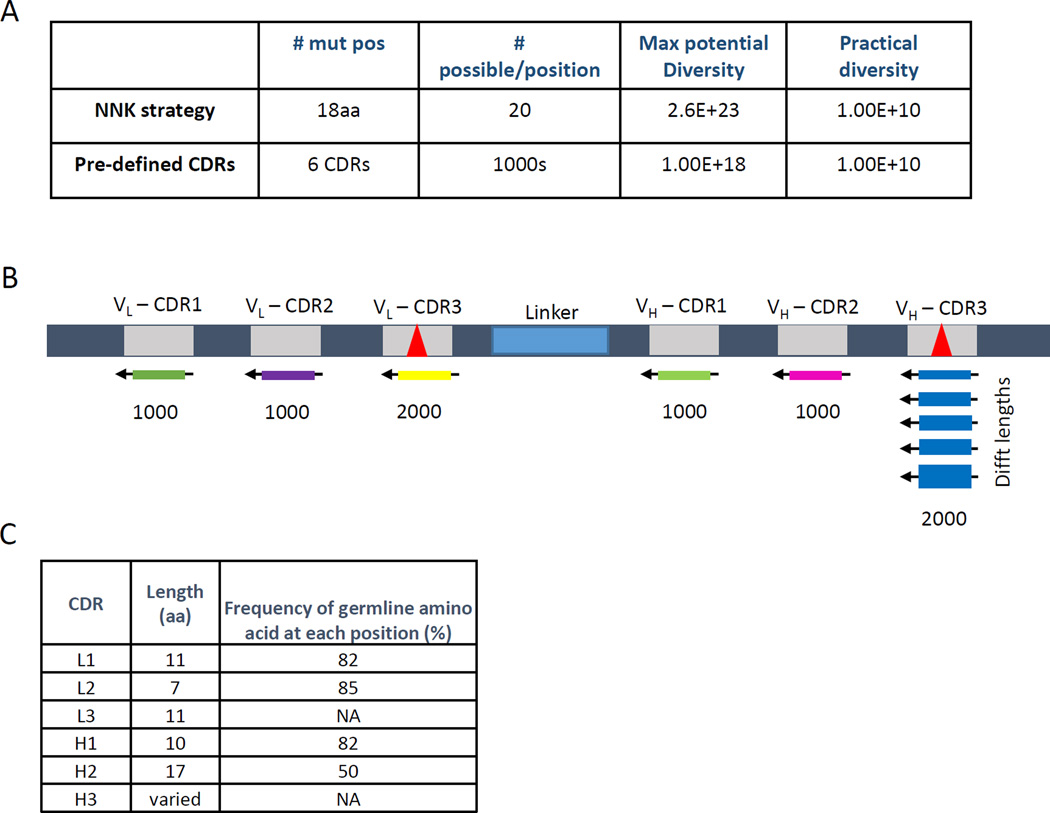
Recombinant antibody libraries based on random incorporation of changes at each of 18 positions in the antibody CDRs have theoretical diversities of greater than 1020, but the practical diversity that can be achieved by transformation of E. coli is only about 1010 (Fig. 1a). In order to constrain the practical diversity to sequences that more closely mimic the diversity of natural human antibodies, we generated a scFv phage library using entirely pre-defined diversity.
Fig. 1. Library Strategy.
(A) (top) A typical NNK-type synthetic library with 18 changes to the amino acid sequence of the 6 CDRs will have a maximum diversity of 2018 (=2.6 × 1023) and nearly 1 stop codon per clone. Suppression of these stop codons is used to allow expression of full length in E. coli, although at greatly reduced levels. (Bottom) By replacing the NNK strategy with pre-defined CDRs we can achieve a similar maximum potential diversity and the same practical diversity (ie. the actual size of the library; dependent on the transformation efficiency of the host bacterium and the number of transformations used to create the library). (B) A schematic of the pre-defined CDR library built using 1000 oligos each for CDRs L1, L2, H1, and H2 and 2000 oligos each for CDRs L3 and H3. (C) For the pre-defined CDR approach all nucleotides within CDRs L1, L2, H1, and H2 were changed at a frequency based upon the somatic hyper-mutation rate found in vivo for that specific CDR. Any oligos containing stop codons, cysteines, or restriction sites were eliminated. The mutation rate multiplied by the length of the CDR results in the theoretical frequency of finding the wildtype germline amino acid sequence. CDRs L3 and H3 have a much higher mutation rate in vivo and thus were synthesized as described in the text.
We synthesized a scFv, based on the human frameworks VH5-3 and VL3-10, which is expressed well on virions, and in E. coli, yeast, and mammalian expression systems (data not shown). CDRs 1 and 2 of the heavy and light chains are encoded in the germline and are constrained in their diversity. Natural diversity in these CDRs is introduced via somatic hypermutation, whereby single base mutations are accumulated. We designed 1000 different oligonucleotides for each of CDRs L1, L2, H1, and H2 with conservative changes that mimic somatic hypermutation (Fig. 1b). The frequency of changes was based on the natural diversity within the CDR in the Kabat database of sequenced antibodies (30). For CDR H2, for example, the original nucleotide was incorporated at each position 70% of the time, with 10% each of the other three possible nucleotides, resulting in the original amino acid at each position approximately 50% of the time (Fig. 1c). Cysteines, stop codons, and certain restriction enzyme sites were eliminated from the designed CDRs.
For CDRs L3 and H3, which are produced by random splicing of gene segments (V and J for L3, and V, D, and J for H3, respectively (31)), our designed oligonucleotides were more diverse. The frequencies of amino acids at each position within the CDR were based on the frequencies in the Kabat database of sequenced human antibodies, and a total of 2000 different oligonucleotides were designed. In addition to containing substantial sequence diversity, CDR H3 also varies in length between 3 and 20 amino acids. We used an algorithm based on the natural distribution of CDR H3 lengths in the Kabat database to vary the lengths of this CDR in our oligonucleotide design (Fig. 1b).
3.2 Library construction
We synthesized approximately 8000, 60–100mer oligonucleotide encoding the pre-defined CDR sequences on an oligonucleotide microarray (LC Biosciences). These oligonucleotides were chemically cleaved from the microarray, but the oligonucleotide yield was not sufficient for library construction without an amplification step. The oligonucleotides were amplified and the library was generated using a modified version of our published AXM cloning method (32) (Fig. 2). In this method, one of the primers in the PCR reaction contains four phosphorothioate linkages at its 5’ end, and treatment of the resulting dsDNA PCR product with T7 exonuclease is used to preferentially remove the strand synthesized with the non-modified primer. The resulting ssDNA fragment serves as a megaprimer to prime DNA synthesis on a uracilated, circular, single-stranded DNA template.
Fig. 2. Library construction approach.
Phase I – incorporation of diversity in L3 and H3. 2000 oligos each of L3 and H3 were synthesized and subsequently cleaved from a chip. Each pool of cleaved oligos is amplified using a reverse primer containing phosphorothioate linkages on its 5’ end. The resulting double-stranded DNA is treated with T7 exonuclease to selectively degrade the unmodified strand of the dsDNA molecule. The resulting single-stranded DNA, or ‘megaprimer’, is then annealed to the uracilated, circular, single-stranded phagemid DNA and used to prime in vitro synthesis by DNA polymerase. The ligated, heteroduplex product is then digested by SacII where the uracilated strand is cleaved by uracil N-glycosylase, favoring survival of the newly synthesized, recombinant strand containing the megaprimer and transformed into E. coli cells. Upon completion of a library that contains L3 and H3, the process is repeated in phase II with the incorporation of 1000 oligos each of L1, L2, H1, and H2 CDRs.
To facilitate library construction, we introduced SacII (Eco29kI isoschizomer; 5’-CCGCGG-3’) restriction sites in CDRs L3 and H3 of the library template (Fig. 2). The restriction sites were incorporated to allow in vitro or in vivo cleavage of parental clones that have not incorporated diversified oligonucleotides. The library was constructed in two steps, with CDRs L3 (2000 oligonucleotides) and H3 (2000 oligonucleotides) incorporated first to ensure all potential diversity (4 × 106) in these two critical CDRs was represented (Fig. 2). In the second phase, predefined oligonucleotides at CDRs L1, L2, H1, and H2 were incorporated within the library of L3 and H3 variants. The theoretical diversity of the library is 1018 (Fig. 1b) while the actual total library diversity was estimated to be greater than 108 based on the transformation efficiency and efficiency of oligonucleotide incorporation.
3.3 Phage display library screening and sequence analysis
The phage library was screened against protein fragments (50–100 amino acids) of human transcription factor or cell signaling proteins. A list of the antigens and their amino acid sequences is shown in Table 1. These antigens were chosen due to the high hit rate we’ve observed in previous screens with multiple proprietary phage display libraries. These protein fragments also express and purify in high quantities. Three successive rounds of affinity selection were performed, and 88 clones for each target were evaluated by phage ELISA. Phage binders were identified from the selections for each of four targets, EZH2, ZMAT3, ZNF622, and TDP-43 (Table 2).
Table 2.
Analysis of phage display hits
After 3 rounds of biopanning against each of four targets, ZMAT-3, EZH2, TDP-43, and ZNF622, eighty-eight individual clones from the selections were evaluated by phage ELISA. Binding was detected using an HRP-conjugated anti-M13 antibody. The number of ELISA positive hits (Fold over background (FOB >10)) are shown for each target along with the number of unique hits as determined by sequence analysis.
| Target | # of hits (FOB >10) | # unique hits |
|---|---|---|
| ZMAT3 | 64 | 3 |
| EZH2 | 86 | 19 |
| TDP-43 | 80 | 5 |
| ZNF622 | 81 | 1 |
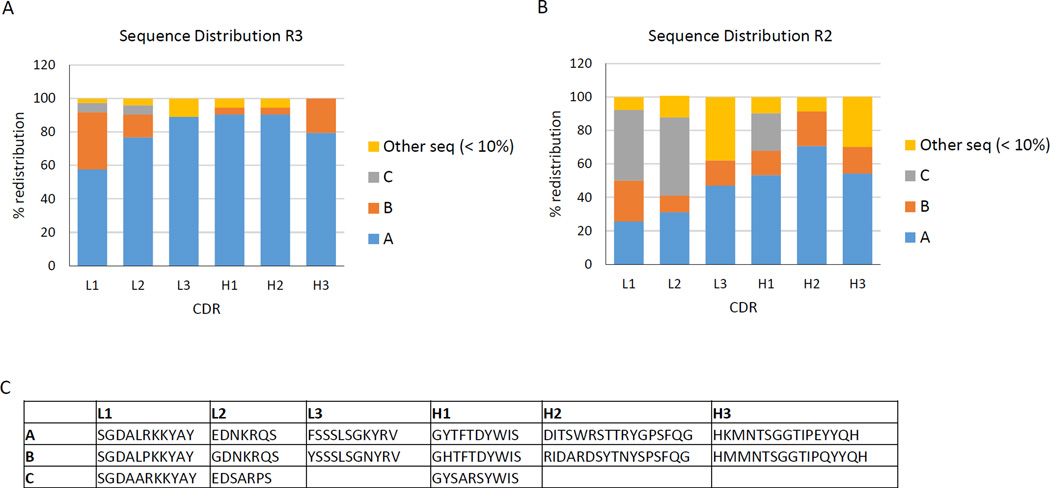
A hypothesis of the pre-defined CDR approach is that rapid identification of enriched CDR sequences could replace the need for many rounds of phage selection and extensive ELISA screening of recovered clones. Ninety-six clones from each round of selection were analyzed by standard Sanger sequencing. Strong enrichment of consensus sequences at each of the six CDRs could be found after three rounds of selection for all four targets. Representative data is shown for the EZH2 target (Fig. 3a). As shown in Figure 3b, those same enriched CDR sequences could be identified in two rounds of selection against the target, EZH2 (Fig. 3b). The consensus sequences A, B, and C enriched after two and three rounds of panning are shown in figure 3c. None of the enriched CDR sequences were found in ninety-six clones sequenced from the un-enriched library (data not shown), and different CDR sequences were found in the libraries selected against the different targets. This data suggests that fewer rounds of biopanning may be sufficient to find the most enriched clones.
Fig. 3. Sequence analysis of selected clones.
Specific CDR sequences were enriched during selection. The sequences identified with the highest frequency are labeled sequence A, B, and C. All other sequences were found at a frequency of less than 10%. (A) Oligonucleotide distribution of CDRs found against the EZH2 target after three rounds of biopanning shows a strong consensus in each CDR (represented as sequence A). (B) Oligonucleotide distribution after two rounds of biopanning against the EZH2 target. The same sequences A, B, and C found in round 3 were also identified in round 2. (C) The enriched sequences A, B, and C found after two and three rounds of panning against EZH2 protein fragment.
3.4 Characterization of consensus CDRs from early-round screening pools
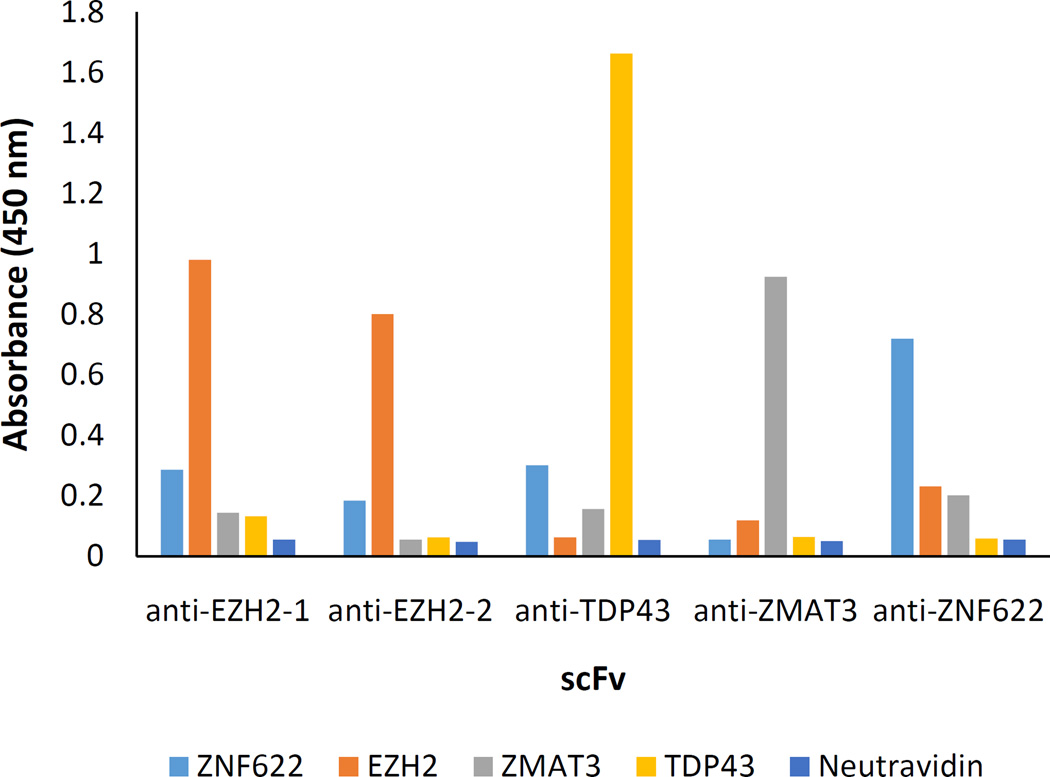
To confirm that scFvs with specificity for the target could be obtained from the consensus CDR sequences, we re-synthesized scFv antibodies containing the most frequent CDR sequences in the enriched pools against each of the four targets. Since several enriched CDR sequences were found in the pool of selected scFvs against EZH2 (Fig. 3), and two different pairings of L1 and H3 were noted, two different engineered anti-EZH2 antibodies were generated with the same L2, L3, H1, and H2 sequences, but differing in sequence at L1 and H3. Five scFv proteins, two against EZH2 and one scFv protein against each target, EZH2, ZMAT3, and TDP-43, were expressed in E. coli and purified. The soluble scFv proteins were tested for binding to each of the four targets in an ELISA. All of the synthetic antibodies showed selectivity for their respective targets in the ELISA (Fig. 4). From this data, we conclude that identification of enriched sequences at each of the six CDRs can be used to reconstruct a consensus antibody with selectivity for the target.
Fig. 4. Analysis of scFv specificity.
The consensus sequence scFvs found after biopanning against four targets, ZMAT-3, EZH2, TDP-43, and ZNF622, were tested in an ELISA against the specific target antigen and the other three, non-relevant targets. For EZH2, two different scFvs with different L1 and H3 sequences were tested. All target proteins were biotinylated and attached to a neutravidin coated ELISA plate. Binding was detected using an HRP-conjugated anti-FLAG antibody that recognizes the tag on the scFv. All of the scFvs bound specifically to their cognate antigens.
4. Discussion
We have demonstrated an approach to generate scFv phage libraries using chip-synthesized oligonucleotides encoding pre-defined CDR sequences. The library was successfully used to select novel antibodies against several human target proteins. For this study, a library of 108 diversity was generated, but larger libraries can be made by increasing the number of E. coli transformations. In addition, secondary affinity maturation libraries can be produced using degenerate oligonucleotides that incorporate the original nucleotide at 99%, 97.5%, or 95% frequency at each position and equal amounts of the other three nucleotides at the remaining frequencies. Since all of the CDRs are pre-designed, oligonucleotides containing the desired degeneracy can be made a priori, facilitating the affinity maturation process.
Although it is relatively inexpensive to synthesize thousands of oligonucleotides on a microarray, several challenges still remain. First, we found that the amount of DNA (nanograms) recovered from the chip was insufficient to generate a large library without a subsequent amplification step. Additional unsolicited diversity may be generated through crossover between oligonucleotides during the amplification step, and approaches like emulsion PCR may be needed to minimize the introduction of this diversity (33, 34). In addition, the error rate in oligonucleotide synthesis on the chip can result in a significant frequency of sequences that differ by one or two nucleotides from the design. A 50% mutation frequency resulting in 39% non-functional clones was recently reported using microarray-synthesized oligonucleotides (15). We expect microarray synthesis to be more accurate in the future as the technique matures.
Our algorithms for generating pre-defined CDR diversity in L1, L2, H1, and H2 were based on incorporating single base changes from the germline sequence and thereby mimicking the diversity generated in natural human antibodies. To improve the utility of the library, additional algorithms can be designed to incorporate structural data about sequence biases that influence antigen-antibody interactions and to eliminate potential immunogenic sequences (15). To facilitate expression of the soluble antibody and downstream processing, we were able to eliminate cysteine residues and stop codons in the CDRs as well as restriction sites that were needed for cloning and re-engineering the scFv. These restrictions cannot be easily applied using traditional library generation approaches.
An advantage of the library based on pre-designed diversity is that rapid identification of enriched CDR sequences could replace the need for many rounds of phage selection and extensive ELISA screening of recovered clones. In this study, we demonstrated that identification of enriched sequences at each of the six CDRs after two rounds of affinity selection can be used to reconstruct a consensus antibody with selectivity for the target. One way to rapidly and cost-effectively identify the enriched sequences would be to use microarrays containing complements of the 8000 pre-defined CDRs. The limitation of this approach is that substantial oligonucleotide design optimization may be needed to prevent cross-hybridization, and the linkages of particular CDRs to each other will be lost.
Alternatively, there have been significant advances in next-generation sequencing technology, and massively parallel DNA sequencing is being used more frequently to assist antibody selection by comprehensively monitoring libraries during selection (35). Native heavy- and light-chain framework libraries are difficult to analyze with short-read sequencing and require many steps for clonal isolation. In contrast, the use of a rationally designed, fully-defined library using a constant scFv framework allows for analysis with short-read deep sequencing (15). Since the sequences are defined before the library is constructed, we should be able to decode 1000 CDRs by sequencing as little as 18 bases at each paired end. In CDR H3, where the sequence diversity is so high, it is unlikely that any of the 2000 pre-defined CDRs will contain 18 bases in common. However, at some of the less variable CDRs, it may be possible to incorporate unique barcodes to the pre-defined oligonucleotides by introducing different bases in the wobble position of a codon where they would be translationally silent. Incorporating each possible nucleotide at the wobble position in each of 5 codons would allow for 45 or 1024 possible barcodes. Advances in DNA sequencing depth and read length will improve the ability to quantify clonal abundances and could eventually eliminate the need for mate-pair reconstruction of full-length sequences. We believe that with these approaches, it will be possible to identify consensus sequences and re-construct functional antibodies after a single round of selection using pre-defined CDR libraries.
Highlights.
A scFv phage library using entirely pre-defined complementarity determining regions (CDR).
Constrain the practical diversity of the library to sequences that more closely mimic the diversity of natural human antibodies.
Identification of enriched sequences at each of the six CDRs in early selection rounds can be used to reconstruct a consensus antibody with selectivity for the target.
Acknowledgements
This work was supported by Small Business Innovative Research (SBIR) grants from the National Institutes of Health [1R43GM112385-01 and 1R43GM105080-01].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, et al. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 2.Hoogenboom HR, Griffiths AD, Johnson KS, Chiswell DJ, Hudson P, et al. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 1991;19:4133–4137. doi: 10.1093/nar/19.15.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheets MD, Amersdorfer P, Finnern R, Sargent P, Lindquist E, et al. Efficient construction of a large nonimmune phage antibody library: the production of high-affinity human single-chain antibodies to protein antigens. Proc Natl Acad Sci U S A. 1998;95:6157–6162. doi: 10.1073/pnas.95.11.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaughan TJ, Williams AJ, Pritchard K, Osbourn JK, Pope AR, et al. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat Biotechnol. 1996;14:309–314. doi: 10.1038/nbt0396-309. [DOI] [PubMed] [Google Scholar]
- 5.de Haard HJ, van Neer N, Reurs A, Hufton SE, Roovers RC, et al. A large non-immunized human Fab fragment phage library that permits rapid isolation and kinetic analysis of high affinity antibodies. J Biol Chem. 1999;274:18218–18230. doi: 10.1074/jbc.274.26.18218. [DOI] [PubMed] [Google Scholar]
- 6.Haidaris CG, Malone J, Sherrill LA, Bliss JM, Gaspari AA, et al. Recombinant human antibody single chain variable fragments reactive with Candida albicans surface antigens. J Immunol Methods. 2001;257:185–202. doi: 10.1016/s0022-1759(01)00463-x. [DOI] [PubMed] [Google Scholar]
- 7.Knappik A, Ge L, Honegger A, Pack P, Fischer M, et al. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J Mol Biol. 2000;296:57–86. doi: 10.1006/jmbi.1999.3444. [DOI] [PubMed] [Google Scholar]
- 8.Sidhu SS, Li B, Chen Y, Fellouse FA, Eigenbrot C, et al. Phage-displayed antibody libraries of synthetic heavy chain complementarity determining regions. J Mol Biol. 2004;338:299–310. doi: 10.1016/j.jmb.2004.02.050. [DOI] [PubMed] [Google Scholar]
- 9.Rauchenberger R, Borges E, Thomassen-Wolf E, Rom E, Adar R, et al. Human combinatorial Fab library yielding specific and functional antibodies against the human fibroblast growth factor receptor 3. J Biol Chem. 2003;278:38194–38205. doi: 10.1074/jbc.M303164200. [DOI] [PubMed] [Google Scholar]
- 10.Nelson B, Sidhu SS. Synthetic antibody libraries. Methods Mol Biol. 2012;899:27–41. doi: 10.1007/978-1-61779-921-1_2. [DOI] [PubMed] [Google Scholar]
- 11.Strachan G, McElhiney J, Drever MR, McIntosh F, Lawton LA, et al. Rapid selection of anti-hapten antibodies isolated from synthetic and semi-synthetic antibody phage display libraries expressed in Escherichia coli. FEMS Microbiol Lett. 2002;210:257–261. doi: 10.1111/j.1574-6968.2002.tb11190.x. [DOI] [PubMed] [Google Scholar]
- 12.Yin CC, Ren LL, Zhu LL, Wang XB, Zhang Z, et al. Construction of a fully synthetic human scFv antibody library with CDR3 regions randomized by a split-mix-split method and its application. J Biochem. 2008;144:591–598. doi: 10.1093/jb/mvn103. [DOI] [PubMed] [Google Scholar]
- 13.Fellouse FA, Esaki K, Birtalan S, Raptis D, Cancasci VJ, et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J Mol Biol. 2007;373:924–940. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Haidar JN, Zhu W, Lypowy J, Pierce BG, Bari A, et al. Backbone flexibility of CDR3 and immune recognition of antigens. J Mol Biol. 2014;426:1583–1599. doi: 10.1016/j.jmb.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 15.Larman HB, Xu GJ, Pavlova NN, Elledge SJ. Construction of a rationally designed antibody platform for sequencing-assisted selection. Proc Natl Acad Sci U S A. 2012;109:18523–18528. doi: 10.1073/pnas.1215549109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li JB, Levanon EY, Yoon JK, Aach J, Xie B, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 17.Kosuri S, Eroshenko N, Leproust EM, Super M, Way J, et al. Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips. Nat Biotechnol. 2010;28:1295–1299. doi: 10.1038/nbt.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rothberg J, Weiner M. Methods of generating antibody diversity in vitro. 20060160178. [Filed Nov. 17, 2004];U.S. Patent Application.
- 19.LeProust EM, Peck BJ, Spirin K, McCuen HB, Moore B, et al. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process. Nucleic Acids Res. 2010;38:2522–2540. doi: 10.1093/nar/gkq163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan W, Li L, Xu Q, Wang Z, Yao Y, et al. Error removal in microchip-synthesized DNA using immobilized MutS. Nucleic Acids Res. 2014;42:e102. doi: 10.1093/nar/gku405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlabach MR, Hu JK, Li M, Elledge SJ. Synthetic design of strong promoters. Proc Natl Acad Sci U S A. 2010;107:2538–2543. doi: 10.1073/pnas.0914803107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patwardhan RP, Hiatt JB, Witten DM, Kim MJ, Smith RP, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steinhauer C, Wingren C, Hager AC, Borrebaeck CA. Single framework recombinant antibody fragments designed for protein chip applications. Biotechniques. 2002;(Suppl):38–45. [PubMed] [Google Scholar]
- 24.Worn A, Pluckthun A. Different equilibrium stability behavior of ScFv fragments: identification, classification, and improvement by protein engineering. Biochemistry. 1999;38:8739–8750. doi: 10.1021/bi9902079. [DOI] [PubMed] [Google Scholar]
- 25.Chen H, Rossier C, Antonarakis SE. Cloning of a human homolog of the Drosophila enhancer of zeste gene (EZH2) that maps to chromosome 21q22.2. Genomics. 1996;38:30–37. doi: 10.1006/geno.1996.0588. [DOI] [PubMed] [Google Scholar]
- 26.Levine AJ, Hu W, Feng Z. The P53 pathway: what questions remain to be explored? Cell Death Differ. 2006;13:1027–1036. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- 27.Seong HA, Gil M, Kim KT, Kim SJ, Ha H. Phosphorylation of a novel zinc-fingerlike protein, ZPR9, by murine protein serine/threonine kinase 38 (MPK38) Biochem J. 2002;361:597–604. doi: 10.1042/0264-6021:3610597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ou SH, Wu F, Harrich D, Garcia-Martinez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashraf SS, Benson RE, Payne ES, Halbleib CM, Gron H. A novel multi-affinity tag system to produce high levels of soluble and biotinylated proteins in Escherichia coli. Protein Expr Purif. 2004;33:238–245. doi: 10.1016/j.pep.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 30.Kabat EA, Wu TT. Identical V region amino acid sequences and segments of sequences in antibodies of different specificities. Relative contributions of VH and VL genes, minigenes, and complementarity-determining regions to binding of antibodycombining sites. J Immunol. 1991;147:1709–1719. [PubMed] [Google Scholar]
- 31.Potter M. Structural correlates of immunoglobulin diversity. Surv Immunol Res. 1983;2:27–42. doi: 10.1007/BF02918394. [DOI] [PubMed] [Google Scholar]
- 32.Holland EG, Buhr DL, Acca FE, Alderman D, Bovat K, et al. AXM mutagenesis: an efficient means for the production of libraries for directed evolution of proteins. J Immunol Methods. 2013;394:55–61. doi: 10.1016/j.jim.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 33.DeKosky BJ, Ippolito GC, Deschner RP, Lavinder JJ, Wine Y, et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat Biotechnol. 2013;31:166–169. doi: 10.1038/nbt.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, et al. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat Med. 2015;21:86–91. doi: 10.1038/nm.3743. [DOI] [PubMed] [Google Scholar]
- 35.D'Angelo S, Kumar S, Naranjo L, Ferrara F, Kiss C, et al. From deep sequencing to actual clones. Protein Eng Des Sel. 2014;27:301–307. doi: 10.1093/protein/gzu032. [DOI] [PMC free article] [PubMed] [Google Scholar]