Abstract
Contagious bovine pleuropneumonia (CBPP) is a severe respiratory disease that is widespread in sub-Saharan Africa. It is caused by Mycoplasma mycoides subsp. mycoides, a bacterium belonging to the Mycoplasma mycoides cluster. In the absence of an efficient CBPP vaccine, improved and easy-to-use diagnostic assays for recurrent testing combined with isolation and treatment of positive animals represent an option for CBPP control in Africa. Here we describe the comprehensive screening of 17 immunogenic Mycoplasma mycoides subsp. mycoides proteins using well-characterized bovine sera for the development of a novel cocktail enzyme-linked immunosorbent assay (ELISA) for laboratory use. Two recombinant Mycoplasma immunogens, MSC_0136 and MSC_0636, were used to set up a standardized cocktail ELISA protocol. According to the results from more than 100 serum samples tested, the sensitivity and specificity of the novel cocktail ELISA were 85.6% and 96.4%, respectively, with an overall diagnostic accuracy comparable to that of the Office International des Epizooties (OIE)-prescribed serological assays. In addition, we provide a proof of principle for a field-applicable, easy-to-use commercially produced prototype lateral-flow test for rapid (<30-min) diagnosis of CBPP.
INTRODUCTION
Contagious bovine pleuropneumonia (CBPP) is one of the most severe diseases affecting cattle in most countries of sub-Saharan Africa (1). CBPP is caused by Mycoplasma mycoides subsp. mycoides, a member of the Mycoplasma mycoides cluster, comprising four additional closely related mycoplasmas, i.e., M. mycoides subsp. capri, M. capricolum subsp. capricolum, M. capricolum subsp. capripneumoniae, and M. leachii (2), all causing diseases in ruminants. CBPP is associated with major financial losses and imposes trade restrictions on live animals, thus being a major constraint to the cattle production industry in Africa. Furthermore, the disease has serious implications on food security and the livelihood of smallholder farmers in affected countries. Cattle infected with Mycoplasma mycoides subsp. mycoides can develop acute, subacute, or chronic disease. Acute disease is characterized by fever, loss of appetite, and respiratory signs, e.g., rapid and painful respiration, cough, and nasal discharge. Unilateral lung lesions and large volumes of pleural fluid are frequently observed postmortem (3). CBPP was eradicated in many developed countries through testing and slaughter of infected animals, quarantine, restricted movement, and mass vaccination (4–6). However, poor infrastructure as well as the practice of nomadic pastoralism in many African countries renders the implementation and sustainability of these control measures difficult. Furthermore, the current live vaccines against CBPP, i.e., T1/44 and T1SR, confer only partial protection and a short-lived immunity (7). Thus, in the absence of better vaccines, improved diagnostic assays for recurrent testing combined with isolation and treatment of positive animals represent an option for CBPP control in Africa (8). Of particular importance as an integrated part of a CBPP outbreak response would be rapid and easy-to-use diagnostic tests that are applicable for field use, such as the recently described assay for diagnosis of contagious caprine pleuropneumonia (9). The different disease outcomes (acute, subacute, or chronic disease) associated with different or a total absence of clinical signs hamper the diagnosis of CBPP, and the use of more than one serological test is currently recommended (10–12). Thus, culture of the causative organism from specimens such as pleural fluid or infected lung tissues is still the gold standard for confirmation of CBPP. However, due to the fastidious nature of Mycoplasma, culturing is time-consuming, and the isolation is often hindered by sample contamination and reduced bacterial viability as a result of long times of transportation of specimens from the sampling site to the laboratory in Africa (9). Furthermore, culture findings still require molecular confirmation using nucleic acid-based hybridization or amplification techniques. The Office International des Epizooties (OIE)-prescribed serological tests for CBPP diagnosis, i.e., the complement fixation test (CFT) (13) and the competitive enzyme-linked immunosorbent assay (cELISA) (14) are, due to the risk of false-positive or false-negative results in individual animals, recommended for diagnosis of CBPP at the herd level only. Previous research efforts have resulted in the identification of immunogenic Mycoplasma mycoides subsp. mycoides proteins with diagnostic potential, and a cocktail ELISA comprising eight selected mycoplasma antigens has been developed (15–18). Although sensitive and specific, the rather large number of included recombinants may limit the production practicality of this assay in Africa. Here we present a systematic comparison of 17 recombinant Mycoplasma mycoides subsp. mycoides immunogens, performed in order to identify the best candidate proteins to be included in a novel cocktail ELISA. A standardized ELISA protocol was developed, and well-defined serum samples were used to compare individual recombinant proteins to the protein combinations with respect to diagnostic sensitivity and specificity. Moreover, as a proof of principle, two recombinant proteins, selected for the cocktail ELISA and displaying strong serological responses, were transferred onto a commercially produced, easy-to-use lateral-flow rapid-test platform that was further evaluated using a limited number of serum samples.
MATERIALS AND METHODS
Recombinant Mycoplasma mycoides subsp. mycoides proteins.
The 17 Mycoplasma mycoides subsp. mycoides proteins included for evaluation in this study are listed in Table 1. Nine of the proteins were previously identified as immunogenic with diagnostic potential in collaborative studies (16, 17). Eight additional proteins displaying strong serological responses and high disease specificity were also included (15, 18–20). Among those were lipoprotein Q (LppQ), a highly immunogenic surface protein, previously used for diagnosis of CBPP (15, 21). Amino acid sequences for the proteins were retrieved from Mycoplasma mycoides subsp. mycoides strain PG1 (GenBank accession no. NC_005364.2). These sequences were compared to the African field isolates Mycoplasma mycoides subsp. mycoides Afadé and B237 (22), and when discrepancies were detected, sequences originating from the African isolates were chosen for design of the full-length recombinants. N-terminal signal peptides from lipoproteins, i.e., cleavage sites for signal peptidase II, were identified using the LipoP v 1.0 server (23) and were removed prior to gene synthesis. When applicable, large transmembrane regions were removed, and sequences, codon optimized for expression in Escherichia coli, were synthesized, expressed, and purified in full length by GenScript Corp. (USA).
TABLE 1.
Recombinant Mycoplasma mycoides subsp. mycoides proteins included in this studya
| PG1 locus tagb | Protein | Protein size (aa) | rProtein coverage (aa) | Reference(s) |
|---|---|---|---|---|
| MSC_0108 | Glycosyltransferase | 449 | 37–312 | 17 |
| MSC_0136 | Hypothetical transmembrane protein | 322 | 25–294 | 18 |
| MSC_0139 | Fructose-bisphosphate aldolase class II | 297 | 1–297 | 16 |
| MSC_0160 | Translation elongation factor Tu | 395 | 1–395 | 16 |
| MSC_0240 | Immunodominant protein P72 | 554 | 25–554 | 18, 20 |
| MSC_0265 | Pyruvate dehydrogenase (lipoamide), alpha chain | 370 | 1–370 | 16 |
| MSC_0266 | Pyruvate dehydrogenase (lipoamide), beta chain | 329 | 1–329 | 16, 17 |
| MSC_0397 | Prolipoprotein | 237 | 24–206 | 18, 20 |
| MSC_0431 | Prolipoprotein | 356 | 27–356 | 18, 20 |
| MSC_0453 | FKBP-type peptidylprolyl isomerase | 428 | 1–428 | 16 |
| MSC_0610 | Molecular chaperone DnaK | 591 | 1–590 | 16 |
| MSC_0636 | Hypothetical protein | 176 | 2–176 | 17 |
| MSC_0653 | Prolipoprotein | 385 | 24–385 | 18 |
| MSC_0679 | Glyceraldehyde-3-phosphate dehydrogenase | 338 | 1–338 | 16 |
| MSC_0813 | Variable surface protein | 507 | 22–425 | 18, 20 |
| MSC_0816 | Variable surface protein | 405 | 24–405 | 18–20 |
| MSC_1046 | Prolipoprotein Q | 445 | 22–218 | 15, 21 |
rProtein, recombinant protein; aa, amino acid.
Locus tag in the Mycoplasma mycoides subsp. mycoides strain PG1 genome (GenBank accession no. NC_005364.2).
Protein expression and purification for the lateral-flow rapid test.
For the development of the field-applicable lateral-flow rapid test, two recombinant full-length proteins (MSC_0136 and MSC_0636, locus tags in the PG1 genome) were expressed and purified in-house. The codon usage of the gene sequences was Escherichia coli optimized and inserted into the pGS-21a expression vector by GenScript Corp. (USA). The expression vectors were transformed into the host strain, Escherichia coli BL21 Star (DE3) (Invitrogen, Germany). Expression of the recombinant proteins was induced in 250 ml exponentially growing bacterial cultures (optical density at 600 nm [OD600], 0.5) by addition of isopropyl β-d-thiogalactopyranoside (IPTG; final concentration, 1 mM) followed by overnight incubation at 28°C at 225 rpm. The hexahistidine-glutathione S-transferase labeled (6×His-GST) recombinant proteins were purified using glutathione column chromatography (Qiagen, Germany) and evaluated for size and purity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (24). Total protein concentrations were determined using the OD280/OD260 ratio in a photometer (Eppendorf, Germany) according to standard methods (24).
Serum samples.
In total, 227 well-characterized bovine sera were used in this study. These sera were collected from experimentally infected cattle (25, 26) or were OIE reference sera, sera from vaccinated animals (27), and archived field sera (CBPP positive and negative). The collection included sera from cattle infected with different Mycoplasma mycoides subsp. mycoides isolates from Africa and Europe as well as from CBPP-naive animals. The serum samples were divided into four panels, A, B, C, and D, which have been used for the four rounds of screening and evaluation: panel A, identification of the best-performing individual recombinant proteins for inclusion in a cocktail ELISA; panel B, identification of the best protein combinations with respect to diagnostic sensitivity and specificity; panel C, evaluation of three selected cocktail combinations; panel D, final screening of the best-performing cocktail.
Panel A comprised 86 sera from experimentally infected cattle in Kenya (n = 17) (3, 25) and from CBPP outbreaks in Namibia (n = 5) and Portugal (n = 21) as well as sera from naive German (n = 18) and Kenyan (n = 25) cattle. A subset of panel A, including 26 CBPP-positive and 21 CBPP-negative serum samples, constituted serum panel B. Serum panel C comprised 43 cattle sera from CBPP outbreaks in Kenya (n = 4) and Namibia (n = 39) and 63 sera collected at nine time points (0, 1, 2, 3, 6, 12, 18, 24, 34 weeks postinfection) during an experimental infection with Mycoplasma mycoides subsp. mycoides in Kenya (26). The experimental sera were grouped according to infection stage, i.e., preinfection stage (day of infection, n = 7), acute-disease stage (days 1 to 22 postinfection, n = 21), and post-acute-disease stage (days 23 to 238 postinfection, n = 35) (26). Serum panel D encompassed 29 serum samples collected from cattle vaccinated with the live CBPP vaccine, i.e., T1/44. Eight samples were collected 7 weeks after vaccination (27), while 21 samples were collected at three time points: 1 week prevaccination and 1 and 6 weeks after immunization. Throughout this study, the same CBPP positive (BD119 and Rd15)- and CBPP negative (Rd660)-control serum samples were used. BD119 was collected during an experimental infection with Mycoplasma mycoides subsp. mycoides (3), and Rd15 is an OIE reference sample from Portugal, while the negative control is from a healthy German calf. All sera were additionally tested using the OIE-prescribed tests for CBPP diagnosis, i.e., the complement fixation test (CFT) (13) and/or the competitive ELISA (cELISA) (14).
Selection of the best-performing individual recombinant proteins. (i) Optimization of the ELISA protocol.
The optimal protein coating concentration (0.1 μg/ml, 1 μg/ml, or 5 μg/ml) and dilution of sera (2-fold dilutions 1:100 to 1:12,800 of BD119 and Rd15 [CBPP positive-control sera] and Rd660 [CBPP negative-control serum]) were determined in triplicates for each of the 17 recombinant proteins. The signal-to-noise ratio (S/N), which is the ratio of the mean OD value of the positive-control sera to that of the negative-control serum, was calculated for each combination, i.e., protein concentration and dilution of sera, and the combinations that gave the highest S/N ratio were considered to be ideal for that particular protein. The optimal surface treatment of the 96-well plate, i.e., affinity to hydrophobic molecules (Nunc-Immunoplate PolySorp; Thermo Fisher Scientific, USA) or molecules with mixed hydrophilic/hydrophobic domains (Nunc-Immunoplate MaxiSorp; Thermo Fisher Scientific, USA), was evaluated in a similar way. In brief, S/N ratios were determined for the respective surface treatments using three recombinant proteins (MSC_0136, MSC_0397, and MSC_0636), three protein coating concentrations (1 μg/ml, 2.5 μg/ml, or 5 μg/ml), and 2-fold dilutions of the three control sera as described above.
The optimized ELISA procedure was performed as follows: 96-well polystyrene plates (Nunc-Immunoplate PolySorp; Thermo Fisher Scientific, USA) were coated with 100 μl protein (1 μg/ml to 5 μg/ml) in carbonate coating buffer (15 mM Na2CO3 and 35 mM NaHCO3, pH 10.6) overnight at 4°C. After coating, the wells were washed twice with 300 μl wash buffer (1× phosphate-buffered saline with 0.05% Tween 20 [PBST]) followed by blocking with 300 μl blocking buffer (PBST and 3% skimmed milk [Bärenmarke Vertriebsgesellschaft, Germany]) for 2 h at room temperature (RT). The wells were washed three times with 300 μl wash buffer prior to adding 100 μl serum samples (diluted 1:100 to 1:400 in PBST) followed by 1 h of incubation at RT and three additional washes as described above. In total, 100 μl peroxidase-conjugated goat-anti-bovine IgG antibody (Jackson Immuno Research, USA), diluted 1:5,000 in PBST, was added to each well prior to incubation for 1 h at RT. Following four washes as above, 100 μl tetramethylbenzidine substrate (TMB; Sigma-Aldrich, USA) was added. The reaction mixture was incubated at RT for 5 to 15 min before the reaction was stopped with 100 μl 1 M H2SO4. The absorbance (OD) was measured at 450 nm in a Tecan SpectraFluor Plus reader (Tecan, Germany).
(ii) Cutoff value, diagnostic sensitivity, and specificity of individual proteins.
Once the optimal ELISA conditions had been determined, the recombinant proteins were screened against serum panel A. The positive- and negative-control sera were included in quadruplicates in each plate, i.e., a duplicate in the first row and a duplicate in the last row. Mean OD values were expressed as percent positivity (PP), calculated as follows: PP = (ODsample − ODneg · control)/(ODpos · control − ODneg · control) × 100%. The normalized data were plotted in scatter plots, and individual cutoff values were set to distinguish CBPP-positive and CBPP-negative samples. Based on the cutoff value, a diagnostic sensitivity and specificity could be determined for each individual protein. Diagnostic sensitivity was defined as the number of true positive samples/(true positive samples + false-negative samples) × 100%, while specificity was calculated as the number of true negative samples/(true negative samples + false-positive samples) × 100%.
(iii) Analytical specificity.
The potential cross-reactivity of the recombinant proteins was tested against a panel of reference hyperimmune sera from rabbits experimentally immunized with type strains of 15 Mycoplasma spp. and three Acholeplasma spp. (28). Plates (Nunc-Immunoplate PolySorp; Thermo Fisher Scientific, USA) were coated with individual Mycoplasma mycoides subsp. mycoides proteins (1.0 μg/ml or 5 μg/ml) diluted in coating buffer and incubated at 4°C overnight. After washing three times with 300 μl/well PBST, the wells were blocked (PBST containing 3% skimmed milk) for 1 h at RT followed by three additional washes as described above. Rabbit sera (100 μl/well) were added at a dilution of 1:800 in PBST and were incubated for 2 h at 37°C. After washing three times as described above, peroxidase-conjugated goat-anti-rabbit IgG (H+L) antibody (Dianova, Germany) diluted 1:5,000 in PBST was added, and the plates were incubated for 1 h at 37°C. After three additional washes, the TMB substrate was added prior to incubation for 5 to 15 min at RT. The reaction was stopped with 100 μl/well of 1 M H2SO4. OD values were measured at 450 nm. An OD cutoff value of 0.5 was set for distinguishing positive and negative reactions.
Office International des Epizooties-prescribed tests for CBPP diagnosis.
The CFT (13) and cELISA (14) were purchased from Cirad (France) and Idexx Livestock—Poultry and Dairy (France), respectively. The assays and the interpretation of results were performed according to the manufacturers' instructions and the OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (29).
Selection of the best-performing recombinant protein combinations. (i) ELISA procedure for the combined recombinant proteins.
The ELISA for the combined antigens was performed as described above but with the following modifications: two or three proteins were mixed in five different combinations using their individual optimal coating concentrations in a total volume of 100 μl/well, and the bovine serum was added in a 1:400 dilution. For the initial screening, 26 CBPP-positive and 21 CBPP-negative sera (serum panel B) were tested in duplicates with each plate being repeated three times. The normalized data (PP values) were plotted in a frequency distribution graph (30) to set a fixed cutoff value to be used for all combinations. Diagnostic sensitivity and specificity for the combined proteins were determined as described above. Based on the results from this screening, a more narrow selection of combinations was made, and these combinations were screened in duplicate against serum panel C as described above. The best-performing cocktail was additionally screened against serum panel D.
(ii) Diagnostic accuracy of the selected cocktail ELISA.
The diagnostic accuracy of the best-performing cocktail was compared to that of CFT and the cELISA, using the combined results from the screenings of serum panels B and C. Diagnostic sensitivity and specificity, positive and negative predictive values, and overall diagnostic accuracy were calculated and compared for all three tests. Furthermore, the percent agreement and Cohen's kappa index value (31) were determined to assess the diagnostic agreement between the tests. A 95% confidence interval was used.
(iii) Repeatability of the selected cocktail ELISA.
The repeatability of the best-performing cocktail was validated in a separate experiment. One strong positive serum sample (SPRN) and one weak positive serum sample (H000840) were run in duplicate wells in 10 separate runs, i.e., two plates per day for 5 consecutive days. Mean OD values were used to assess the intraplate variance as well as intra- and interassay variance. The coefficients of variability (CVs) were calculated using Statgraphics Plus (Manugistics, Inc., USA).
A field-applicable, easy-to-use lateral-flow rapid test for diagnosis of CBPP.
The proof of principle for a field-applicable lateral-flow rapid test was elucidated. Two recombinant proteins, MSC_0136 and MSC_0636, were expressed, purified (as described above), and transferred to an existing lateral-flow rapid-test platform by Senova immunoassay systems (Senova, Gesellschaft für Biowissenschaft und Technik GmbH, Germany). Forty-four prototype dipsticks containing the two recombinant proteins (test region, 0.25 μg/test) and immobilized anti-mouse IgG (control region, 0.25 μg/test) on nitrocellulose membranes were initially manufactured for the purpose of the work presented here. The strips were evaluated using 32 positive sera and 12 negative sera (randomly selected from serum panel A and including the positive and negative controls), according to manufacturer instructions. In brief, serum samples were diluted 1:100 in sample dilution buffer in a 96-well plate (working station). The test strips were subsequently incubated standing for 5 min in 30 μl of diluted serum sample followed by a single wash step in 50 μl sample dilution buffer for 2 min. After the wash step, the test strips were incubated in 30 μl detection antibody conjugate (gold particles coated with mouse anti-bovine IgG1 antibodies, 0.25 μg/test) for 10 min. The results, i.e., the appearance of either two colored lines (one in the control region and another one in the test region, indicating a positive test result) or one colored line (one line in the control region only, indicating a negative test result), were read within 15 min after removing the rapid test from the final incubation in detection antibody.
RESULTS
Performance of individual recombinant proteins. (i) Expression and purification of the recombinant Mycoplasma mycoides subsp. mycoides proteins.
Expression and purification (85% purity), performed by GenScript, USA, were successful for 16 of the 17 recombinant Mycoplasma mycoides subsp. mycoides proteins. Protein MSC_1046 (locus tag in the PG1 genome) was expressed and purified, but due to product degradation the purity of the resulting protein was low (>20%). Four proteins, MSC_0108, MSC_0136, MSC_0397, and MSC_0813, had large transmembrane regions removed prior to gene synthesis to increase expression efficiency. Total protein concentrations of 2.1 mg/ml and 4.44 mg/ml were obtained from the in-house-expressed proteins MSC_0136 and MSC_0636, respectively.
(ii) Diagnostic sensitivity and specificity.
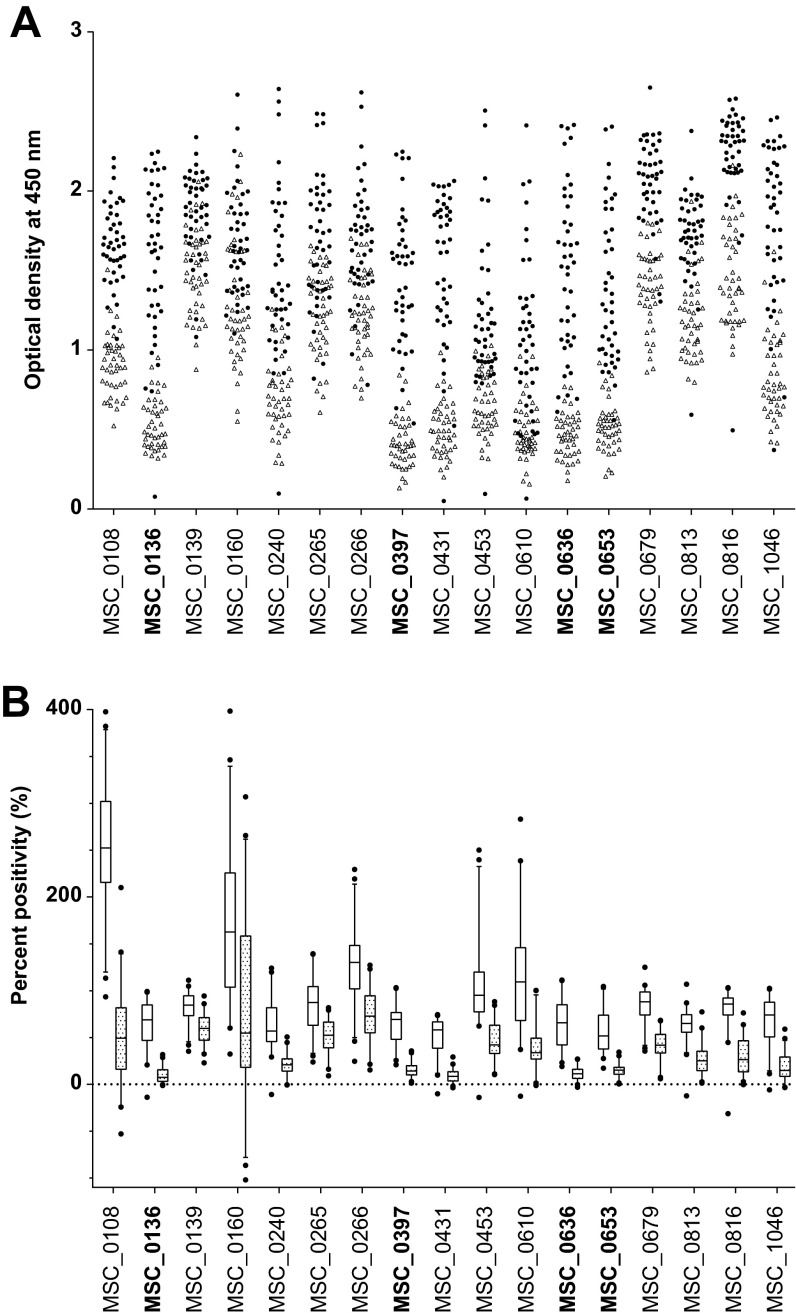
The optimized ELISA conditions, i.e., protein coating concentrations (μg/ml) and serum dilutions, are presented in Data Set S1 in the supplemental material. The highest signal-to-noise (S/N) ratio between CBPP-positive and CBPP-negative samples was obtained using a 96-well plate with affinity to hydrophobic molecules (Nunc-Immunoplate PolySorp; Thermo Fisher Scientific, USA) (data not shown). The mean optical density (OD) values from duplicate samples repeated five times and percent positivity (PP) for all 17 proteins screened against 86 sera (panel A) are presented in Fig. 1A and B, respectively. Diagnostic sensitivities between 67% and 95% were obtained, while the measured specificities ranged between 60% and 100% (see Data Set S2 in the supplemental material). Four proteins, MSC_0136, MSC_0397, MSC_0636, and MSC_0653, with the highest ranked sensitivities (>90%) and specificities (>95%), were selected for further testing in combinations.
FIG 1.
Scatter plot showing the optical density (OD) (A) and box plots representing percentage positivity (%) (B) for the individual recombinant Mycoplasma mycoides subsp. mycoides proteins when screened against serum panel A. Filled circles, CBPP-positive sera; open triangles, CBPP-negative sera; open boxes, CBPP-positive sera; patterned boxes, CBPP-negative sera. The recombinant antigens that were selected for testing in a cocktail ELISA are highlighted in bold.
(iii) Cross-reactivity.
Cross-reactivity with the recombinant immunogens was seen with antibodies against nine mycoplasmas and three acholeplasmas (see Table S1 in the supplemental material). Most recombinant proteins showed cross-reactions with antisera from rabbits immunized with M. alkalescens (n = 12) and antisera against members of the Mycoplasma mycoides cluster, i.e., M. capricolum subsp. capricolum (n = 11) and M. mycoides subsp. capri (n = 8). The four proteins selected for further evaluation in a cocktail ELISA showed cross-reactivity to three or four Mycoplasma isolates (see Table S1 in the supplemental material).
Performance of the cocktail ELISAs. (i) Diagnostic sensitivity and specificity of the cocktails.
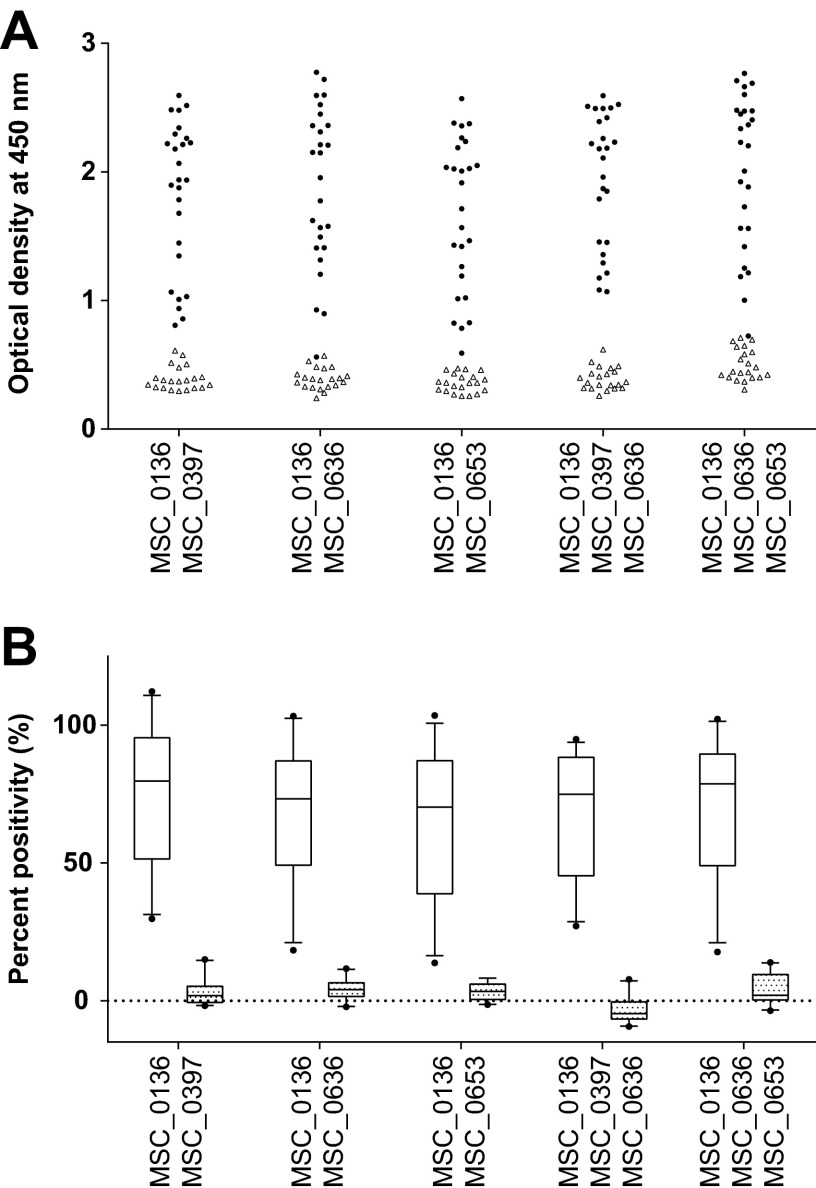
In total, five different ELISA cocktails were run in duplicates using serum panel B. A 25-PP cutoff was set to distinguish positive from negative samples (see Fig. S1 in the supplemental material) (30). The mean OD values as well as PP for all five protein cocktails screened against 47 sera are presented in Fig. 2A and B, respectively. All cocktails correctly identified all negative CBPP samples, yielding a diagnostic specificity of 100%. Compared to the performance of the individual proteins, all but one of the cocktails resulted in equal or higher numbers of correctly identified CBPP-positive samples (see Data Set S2 in the supplemental material). Increasing the number of antigens from two to three did not improve the sensitivity; therefore, the two combinations containing two antigens with the highest ranking sensitivities, i.e., MSC_0136/MSC_0397 and MSC_0136/MSC_0636, were selected for further testing.
FIG 2.
Scatter plot representing the optical density (OD) (A) and box plots showing percent positivity (%) (B) for combinations of recombinant Mycoplasma mycoides subsp. mycoides proteins when screened against serum panel B. Filled circles, CBPP-positive sera; open triangles, CBPP-negative sera; open boxes, CBPP-positive sera; patterned boxes, CBPP-negative sera.
(ii) Performance of the cocktail ELISAs in a long-term experimental infection with Mycoplasma mycoides subsp. mycoides.
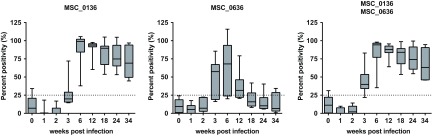
Among the outbreak serum samples (n = 43, panel C), both cocktails identified >93% of the samples as CBPP positive (Table 2). In the group of samples collected during an experimental infection trial, one sample was false positive using cocktail MSC_0136/MSC_0636, while two day 0 samples were positive using cocktail MSC_0136/MSC_0397. During the acute stage of disease (days 1 to 22 postinfection), 6 of the 21 samples were positive using cocktail MSC_0136/MSC_0636. All samples (n = 35) collected during the postacute stage (days 23 to 238 postinfection) were positive using the two cocktails. When comparing the cocktails to their respective single proteins at the individual time points (0, 1, 2, 3, 6, 12, 18, 24, 34 weeks postinfection), earlier antibody detection was achieved using cocktail MSC_0136/MSC_0636 (Fig. 3). Taking the performance of the cocktails during the evaluation and the long-term trial into consideration, as well as the sizes of the recombinant proteins, expression efficacy, and purity, cocktail MSC_0136/MSC_0636 was considered the best-performing cocktail. It was furthermore used for screening 29 serum samples (panel D) from T1/44-vaccinated animals. Only one sample, taken 6 weeks postvaccination was found to be positive (see Data Set S2 in the supplemental material).
TABLE 2.
Performance of individual and combined recombinant Mycoplasma mycoides subsp. mycoides proteinsa
| Panel ID and serum category | No. of sera | No. (%) of sera testing positive or negative |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MSC_0136 |
MSC_0397 |
MSC_0636 |
MSC_0136/MSC_0397 |
MSC_0136/MSC_0636 |
|||||||
| Pos. | Neg. | Pos. | Neg. | Pos. | Neg. | Pos. | Neg. | Pos. | Neg. | ||
| Serum panel B (CBPP positive)b | 26 | 23 (88.5) | 3 (11.5) | 23 (88.5) | 3 (11.5) | 25 (96.2) | 1 (3.8) | 26 (100.0) | 0 (0.0) | 25 (96.2) | 1 (3.8) |
| Serum panel C | |||||||||||
| Outbreak | 43 | 40 (93.0) | 3 (7.0) | 13 (30.2) | 23 (69.8) | 39 (90.7) | 4 (9.3) | 40 (93.0) | 3 (7.0) | 41 (95.3) | 2 (4.6) |
| Preinfection | 7 | 1 (14.3) | 6 (85.7) | 0 (0.0) | 7 (100.0) | 0 (0.0) | 7 (100.0) | 2 (28.6) | 5 (71.4) | 1 (14.3) | 6 (85.7) |
| Acute stage | 21 | 1 (4.8) | 20 (95.2) | 2 (9.5) | 19 (90.5) | 5 (23.8) | 16 (76.2) | 2 (9.5) | 19 (90.5) | 6 (28.6) | 15 (71.4) |
| (1–22 dpi) | |||||||||||
| Postacute stage (23–238 dpi) | 35 | 35 (100.0) | 0 (0.0) | 20 (57.1) | 15 (42.9) | 12 (34.3) | 23 (65.7) | 35 (100.0) | 0 (0.0) | 35 (100.0) | 0 (0.0) |
This table represents results obtained through the use of panel B and C serum samples only. Pos., positive; Neg., negative.
All CBPP-negative samples (n = 21) from panel B were correctly identified as negative.
FIG 3.
Box plot showing percent positivity (%) for the single and combined recombinant Mycoplasma mycoides subsp. mycoides proteins when screened against sera from a long-term Mycoplasma mycoides subsp. mycoides infection trial.
(iii) Diagnostic performance of the selected cocktail ELISA in comparison to CFT and cELISA.
Sensitivity and specificity values of 85.6% and 96.4%, respectively, were obtained for the best-performing cocktail, MSC_0136/MSC_0636, when combining data from serum panels B and C (Table 3). The corresponding sensitivity and specificity percentages were 88.5% and 100% for CFT and 82.4% and 100% for the cELISA. The overall percentage agreements between the tests were 94.1% for cocktail MSC_0136/MSC_0636 versus cELISA and 88.7% for cocktail MSC_0136/MSC_0636 versus CFT, as supported by Cohen's kappa index values of 0.86 (very good) and 0.73 (good), respectively.
TABLE 3.
Diagnostic accuracy of the best-performing cocktail ELISA compared to the OIE-prescribed serological tests for CBPP diagnosisf
| ELISA components or test | No. diagnosed positive/total no. positive | No. diagnosed negative/total no. negative | Sensitivitya (95% CI) | Specificityb (95% CI) | PPVc (95% CI) | NPVd (95% CI) | Overall diagnostic accuracye (95% CI) | % agreement (95% CI) |
|---|---|---|---|---|---|---|---|---|
| MSC_0136/MSC_0636 | 107/125 | 27/28 | 85.6 (78.0–91.0) | 96.4 (79.8–99.8) | 99.1 (94.2–99.9) | 60.0 (44.4–73.9) | 87.6 (82.4–92.8) | NA |
| CFT | 101/114 | 28/28 | 88.5 (80.9–93.5) | 100 (85.0–100) | 100 (95.4–100) | 68.3 (51.8–81.4) | 90.8 (86.1–95.6) | NA |
| cELISA | 103/125 | 27/27 | 82.4 (74.3–88.4) | 100 (84.5–100) | 100 (95.5–100) | 55.1 (40.3–69.1) | 85.5 (79.9–91.1) | NA |
| Comparison of MSC_0136/0636 with CFT | 91 | 35 | NA | NA | NA | NA | NA | 88.7 (83.5–93.9), κ = 0.73 |
| Comparison of MSC_0136/0636 with cELISA | 101 | 42 | NA | NA | NA | NA | NA | 94.1 (90.3–97.8), κ = 0.86 |
Diagnostic sensitivity: TP/(TP + FN) × 100%, where TP is the number of true-positive samples and FN is the number of false-negative samples.
Diagnostic specificity: TN/(TN + FP) × 100%, where TN is the number of true-negative samples and FP is the number of false-positive samples.
PPV, positive predictive value: TP/(TP + FP) × 100%.
NPV, negative predictive value: TN/(TN + FN) × 100%.
Overall diagnostic accuracy: (TP + TN)/(TP + TN + FP + FN) × 100%.
This table represents results obtained through the use of panel B and C serum samples only. κ, Cohen's kappa index value, classified as very good (1 to 0.76), good (0.75 to 0.61), acceptable (0.6 to 0.4), and poor (<0.4) (31); NA, not applicable; CI, confidence interval.
(iv) Repeatability of the selected cocktail ELISA.
An intraplate coefficient of variability (CV) of 6.80% was obtained, while the intra-assay CV and interassay CV were 7.72% and 9.14%, respectively.
Proof of principle for a field-applicable lateral-flow rapid test for diagnosis of CBPP.
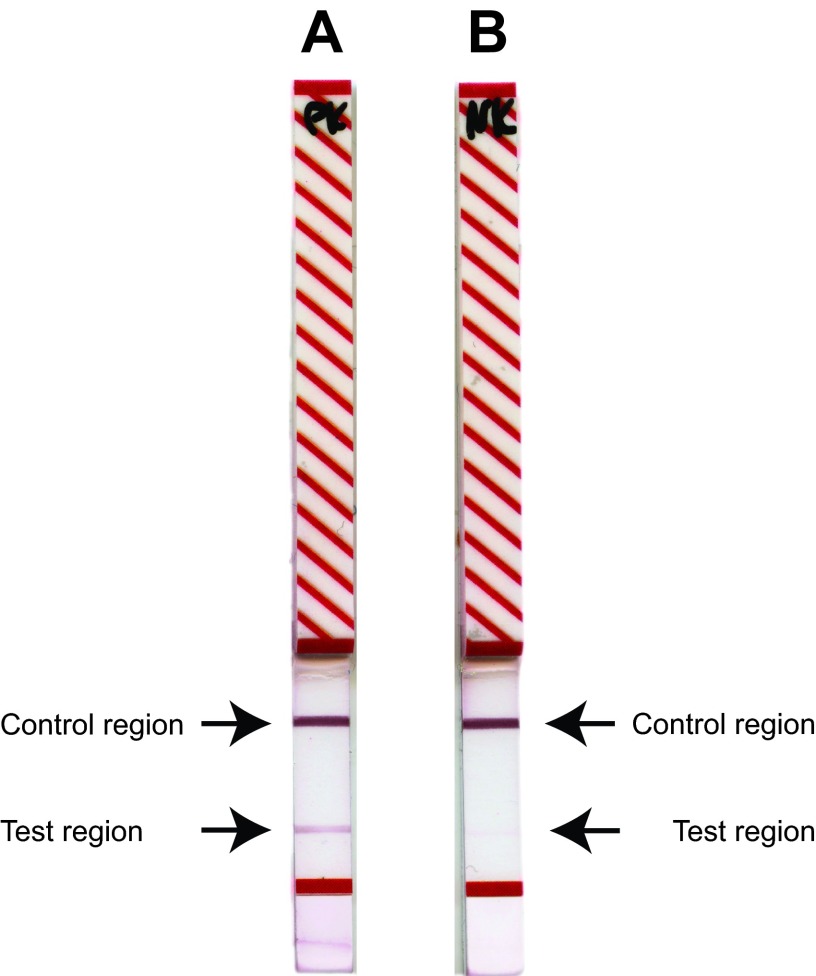
The in-house-expressed recombinant proteins MSC_0136 and MSC_0636 were successfully transferred to the lateral-flow rapid-test platform (Senova, Gesellschaft für Biowissenschaft und Technik GmbH, Germany). All test strips incubated in sera from CBPP-positive animals (n = 32) developed two colored lines within approximately 10 min after the final incubation in detection antibody, while only one line developed in the control region on the strips incubated in sera from CBPP-negative cattle (n = 12) (see Data Set S2 in the supplemental material). Thus, a definite diagnosis of CBPP was obtained within a total time of 30 min. Examples of the test strips are shown in Fig. 4.
FIG 4.
Readout of the lateral-flow rapid test. (A) Serum from a CBPP-positive cow (BD119) shows a clear band in the test and control regions. (B) Serum from a CBPP-negative cow (Rd 660) shows a band only in the control region.
DISCUSSION
Serological tests for diagnosis of animal diseases are widespread in African laboratories, mainly due to the availability of equipment, ease of sampling, transport, and storage of serum specimens, and relatively low costs per test. Moreover, no preculturing or extensive sample preparation is needed. In addition, many sensitive and specific nucleic acid-based detection methods are available for CBPP (32–34), yet the rather elaborate requirements for the laboratory facilities, sample costs, and processing associated with these techniques have limited their routine use in Africa. Recently, a sensitive and easy-to-use loop-mediated isothermal amplification (LAMP)-based assay for CBPP was published, which overcomes many of the above-mentioned constraints (35). However, the time window of Mycoplasma shedding varies with disease status (acute or chronic), and this impacts the diagnostic capacity of all nucleic acid-based detection methods. For CBPP diagnosis, the current OIE-prescribed serological tests are the CFT (13) and the cELISA (14). As both have limited sensitivity, in particular during chronic disease, they are recommended for diagnosis at the herd level only. Moreover, both methods require a fully equipped laboratory, and the CFT in particular is considered hard to perform and standardize. Thus, the development of improved and easy-to-use diagnostic assays comprising antigens able to identify animals in all stages of disease is considered a research priority. Several studies employing different experimental approaches, e.g., in silico analysis, proteomics, cloning, etc., have described the identification of immunogenic Mycoplasma mycoides subsp. mycoides proteins (15–18, 20), yet as expected, each methodology had its limitation. This study is based on a thorough comparison of recombinant immunogens, using a standardized expression system employing synthetic, codon-optimized genes. The Mycoplasma protein sequences were furthermore based on African outbreak strains and not only on the widely used laboratory type strain PG1, which has been shown to be phylogenetically different from African field strains (36). The removal of the N-terminal signal peptides in lipoproteins and large transmembrane regions led to improved expression and solubility of the recombinant proteins, thus making them more suitable for a future large-scale production. All immunogens, except the N-terminal part of the strong immunogen LppQ, were expressed and purified properly. Thus, to ensure standardization and comparability of results, we excluded LppQ after the initial screening. The development of the cocktail ELISA was a two-stage process starting with the selection of the four best-performing recombinant immunogens, which subsequently were tested in cocktails to improve performance. We used well-characterized sera from CBPP outbreaks as well as from naive cattle. The sera were derived from different geographical regions, cattle breeds, and isolation dates in order to account for variability of strains and humoral immune responses. To foster the identification of chronically infected animals, we screened immunogens against sera from a long-term experimental infection with Mycoplasma mycoides subsp. mycoides (26). Indeed, several recombinant proteins displaying high diagnostic sensitivity (95%) and specificity (100%) (see Data Set S2 in the supplemental material) were identified, thus confirming their suitability for inclusion in a diagnostic assay. The four best individual immunogens, i.e., a hypothetical transmembrane protein (MSC_0136), a hypothetical protein (MSC_0636), and two prolipoproteins (MSC_0397 and MSC_0653), were selected. All but one of these recombinants (MSC_0636) have been shown to elicit strong antibody responses in vaccinated cattle (37). Furthermore, the surface localization of two of the proteins, i.e., MSC_0397 and MSC_0653, has been confirmed (38). Notably, the former protein is also a constituent of the Mycoplasma mycoides subsp. mycoides in vitro surface core proteome and hence likely to be involved in host-pathogen interactions (38). Due to the limited genetic diversity within the Mycoplasma mycoides cluster (36, 39–41), cross-reactivity with antibodies induced by other Mycoplasma species is likely to be detected and has been reported previously (18). The four selected immunogens reacted with sera from rabbits immunized with type strains of Mycoplasma alkalescens, Mycoplasma leachii, Mycoplasma capricolum subsp. capricolum, and Mycoplasma mycoides subsp. capri. Mycoplasma alkalescens is a natural inhabitant of the respiratory tract in cattle but can, like Mycoplasma leachii, cause clinical disease. Such bovine infections have to the best of our knowledge not been reported in Africa; hence, the cross-reactivity described here is not likely to affect the diagnostic potential of the included recombinant proteins, albeit this needs further investigation. In addition, the experimental serological response of rabbits is likely to be different from the humoral response of naturally exposed ruminants.
In an attempt to increase the diagnostic sensitivity, different protein cocktails were tested. We did, however, limit the number of recombinants in a cocktail to a maximum of three to ensure a smooth future large-scale production with limited quality control efforts required, in comparison to cocktail ELISAs that contain additional antigens (18). Indeed, increasing the number of immunogens from one to two resulted in the identification of more CBPP-positive animals (Table 2). However, increasing the number of antigens from two to three did not improve the sensitivity. Therefore, two cocktails comprising two immunogens (MSC_0136/MSC_0397 and MSC_0136/MSC_0636) were investigated further. A prerequisite for a diagnostic assay for CBPP is the ability to identify animals in all stages of disease; therefore, the selected cocktails were screened against sera from an experimental infection with Mycoplasma mycoides subsp. mycoides in which samples were collected for a time period of >230 days (26). Cocktail MSC_0136/MSC_0636 identified more animals in the acute stage of disease than the other cocktails (Table 2) and allowed for earlier detection than single antigens (Fig. 3). All cocktails presented here are combinations with protein MSC_0136. Based on the data regarding the performance of the different cocktails and to harmonize and simplify the standardization of the ELISA protocol, we consider MSC_0636 the optimal partner protein. Furthermore, this cocktail had a performance comparable to those of the OIE-prescribed tests, i.e., the CFT and the cELISA, with 88.7% and 94.1% agreement, respectively (Table 3). However, a fraction of the serum panel, i.e., the outbreak sera used for these calculations, were selected and included depending on previous positivity in either cELISA or CFT, thus excluding the possibility of a superiority of the immunogens tested over cELISA and CFT. The cocktail ELISA was also more straightforward and faster to perform and standardize than the CFT and cELISA. Although the assay is not able to distinguish T1/44-vaccinated animals from infected animals, it is likely to be a very useful tool for screening animals with suspected CBPP and for epidemiological studies investigating the prevalence and impact of Mycoplasma mycoides subsp. mycoides on animal welfare and productivity. Albeit applicable for CBPP diagnosis, the cocktail ELISA described here requires a laboratory. Thus, as proof of principle for a rapid and field-applicable diagnostic assay for CBPP, we transferred the two highly immunogenic proteins (MSC_0136 and MSC_0636) to a prototype lateral-flow rapid-test format. This is to our knowledge the first prototype CBPP lateral-flow test produced by a company. The test was evaluated with a limited number of sera representing naïve and experimentally and naturally infected animals. All CBPP-infected animals (n = 32) tested positive within 10 min of postincubation, after a total handling and readout time of less than 30 min. Though a field-applicable diagnostic test is likely to be more expensive than a laboratory-based ELISA, it overcomes transport costs, provides timely test results, and excludes the risk associated with unsatisfactory laboratory quality control concomitant with financial constraints in many sub-Saharan laboratories. Albeit a successful proof-of-principle experiment, additional work is required to optimize the assay conditions. It is crucial to embed the diagnostic assays into effective disease control strategies that motivate livestock owners and veterinary services to use the latter. Future studies should thus include ring testing of the novel cocktail ELISA and the lateral-flow assay, performed by African national laboratories in cooperation with international bodies such as OIE, African Union Inter-African Bureau for Animal Resources (AU-IBAR), or FAO to pave the way for rollout of the assays.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge receiving four field sera from Rose Matua from the Kenyan Department of Veterinary Services. We thank Anne-Marie Gad and Hans Herrmann Söffing from Senova GmbH, Weimar, Germany, for their help in developing the lateral-flow rapid test. We thank Elise Schieck for her help on the figures and Beate Burkert for her help on the data analysis. We also thank Victoria Andres, Kerstin Nitzsche, Susann Bahrmann, Simone Bettermann, Christine Grajetzki, and Sabine Scharf for their technical assistance. In addition we thank Phil Toye and Henry Kiara for critiquing the manuscript.
M.H., J.J., and A.L. conceived and designed the experiments; M.H., N.G., C.M., J.J., and A.L. performed the experiments; M.H., N.G., M.Y., J.J., and A.L. analyzed the data; G.T.-Z., A.B., M.Y., and J.N. contributed reagents/materials/analysis tools; J.J. and A.L. drafted the manuscript.
Funding Statement
This work was funded by the German Ministry of Economic Cooperation and Development (project no. 09.7860.1-001.00, contract no. 81121408, and project no. 09.7860.1-001.00, contract no. 81170269). Anne Liljander was supported by the Centre for International Migration and Development (CIM). Additional support was received from the CGIAR research program on Livestock and Fish.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. We declare that we have no conflicts of interest in the research.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03259-15.
REFERENCES
- 1.Jores J, Mariner JC, Naessens J. 2013. Development of an improved vaccine for contagious bovine pleuropneumonia: an African perspective on challenges and proposed actions. Vet Res 44:122. doi: 10.1186/1297-9716-44-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manso-Silvan L, Vilei EM, Sachse K, Djordjevic SP, Thiaucourt F, Frey J. 2009. Mycoplasma leachii sp. nov. as a new species designation for Mycoplasma sp. bovine group 7 of Leach, and reclassification of Mycoplasma mycoides subsp. mycoides LC as a serovar of Mycoplasma mycoides subsp. capri. Int J Syst Evol Microbiol 59:1353–1358. doi: 10.1099/ijs.0.005546-0. [DOI] [PubMed] [Google Scholar]
- 3.Sacchini F, Naessens J, Awino E, Heller M, Hlinak A, Haider W, Sterner-Kock A, Jores J. 2011. A minor role of CD4+ T lymphocytes in the control of a primary infection of cattle with Mycoplasma mycoides subsp. mycoides. Vet Res 42:77. doi: 10.1186/1297-9716-42-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fisher J. 2003. To kill or not to kill: the eradication of contagious bovine pleuro-pneumonia in western Europe. Med Hist 47:314–331. [PMC free article] [PubMed] [Google Scholar]
- 5.Kusiluka LJ, Sudi FF. 2003. Review of successes and failures of contagious bovine pleuropneumonia control strategies in Tanzania. Prev Vet Med 59:113–123. doi: 10.1016/S0167-5877(03)00087-4. [DOI] [PubMed] [Google Scholar]
- 6.Windsor RS, Wood A. 1998. Contagious bovine pleuropneumonia. The costs of control in central/southern Africa. Ann N Y Acad Sci 849:299–306. [DOI] [PubMed] [Google Scholar]
- 7.Thiaucourt F, Dedieu L, Maillard JC, Bonnet P, Lesnoff M, Laval G, Provost A. 2003. Contagious bovine pleuropneumonia vaccines, historic highlights, present situation and hopes. Dev Biol (Basel) 114:147–160. [PubMed] [Google Scholar]
- 8.Ssematimba A, Jores J, Mariner JC. 2015. Mathematical modelling of the transmission dynamics of contagious bovine pleuropneumonia reveals minimal target profiles for improved vaccines and diagnostic assays. PLoS One 10:e0116730. doi: 10.1371/journal.pone.0116730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liljander A, Yu M, O'Brien E, Heller M, Nepper JF, Weibel DB, Gluecks I, Younan M, Frey J, Falquet L, Jores J. 2015. A field-applicable recombinase polymerase amplification assay for rapid detection of Mycoplasma capricolum subsp. capripneumoniae. J Clin Microbiol 53:2810–2815. doi: 10.1128/JCM.00623-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amanfu W, Sediadie S, Masupu KV, Raborokgwe MV, Benkirane A, Geiger R, Thiaucourt F. 2000. Comparison between c-ELISA and CFT in detecting antibodies to Mycoplasma mycoides mycoides biotype SC in cattle affected by CBPP in Botswana. Ann N Y Acad Sci 916:364–369. [DOI] [PubMed] [Google Scholar]
- 11.Muuka G, Hang'ombe BM, Nalubamba KS, Kabilika S, Mwambazi L, Muma JB. 2011. Comparison of complement fixation test, competitive ELISA and LppQ ELISA with post-mortem findings in the diagnosis of contagious bovine pleuropneumonia (CBPP). Trop Anim Health Prod 43:1057–1062. doi: 10.1007/s11250-011-9805-5. [DOI] [PubMed] [Google Scholar]
- 12.Schubert E, Sachse K, Jores J, Heller M. 2011. Serological testing of cattle experimentally infected with Mycoplasma mycoides subsp. mycoides Small Colony using four different tests reveals a variety of seroconversion patterns. BMC Vet Res 7:72. doi: 10.1186/1746-6148-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Etheridge JR, Buttery SH. 1976. Improving the specificity and yield of the contagious bovine pleuropneumonia complement fixation test antigen. Res Vet Sci 20:201–206. [PubMed] [Google Scholar]
- 14.Le Goff C, Thiaucourt F. 1998. A competitive ELISA for the specific diagnosis of contagious bovine pleuropneumonia (CBPP). Vet Microbiol 60:179–191. doi: 10.1016/S0378-1135(98)00156-4. [DOI] [PubMed] [Google Scholar]
- 15.Bruderer U, Regalla J, Abdo el-M, Huebschle OJ, Frey J. 2002. Serodiagnosis and monitoring of contagious bovine pleuropneumonia (CBPP) with an indirect ELISA based on the specific lipoprotein LppQ of Mycoplasma mycoides subsp. mycoides SC. Vet Microbiol 84:195–205. doi: 10.1016/S0378-1135(01)00466-7. [DOI] [PubMed] [Google Scholar]
- 16.Jores J, Meens J, Buettner FF, Linz B, Naessens J, Gerlach GF. 2009. Analysis of the immunoproteome of Mycoplasma mycoides subsp. mycoides small colony type reveals immunogenic homologues to other known virulence traits in related Mycoplasma species. Vet Immunol Immunopathol 131:238–245. doi: 10.1016/j.vetimm.2009.04.016. [DOI] [PubMed] [Google Scholar]
- 17.Naseem S, Meens J, Jores J, Heller M, Dubel S, Hust M, Gerlach GF. 2010. Phage display-based identification and potential diagnostic application of novel antigens from Mycoplasma mycoides subsp. mycoides small colony type. Vet Microbiol 142:285–292. doi: 10.1016/j.vetmic.2009.09.071. [DOI] [PubMed] [Google Scholar]
- 18.Neiman M, Hamsten C, Schwenk JM, Bolske G, Persson A. 2009. Multiplex screening of surface proteins from Mycoplasma mycoides subsp. mycoides small colony for an antigen cocktail enzyme-linked immunosorbent assay. Clin Vaccine Immunol 16:1665–1674. doi: 10.1128/CVI.00223-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamsten C, Westberg J, Bolske G, Ayling R, Uhlen M, Persson A. 2008. Expression and immunogenicity of six putative variable surface proteins in Mycoplasma mycoides subsp. mycoides SC. Microbiology 154:539–549. doi: 10.1099/mic.0.2007/010694-0. [DOI] [PubMed] [Google Scholar]
- 20.Hamsten C, Neiman M, Schwenk JM, Hamsten M, March JB, Persson A. 2009. Recombinant surface proteomics as a tool to analyze humoral immune responses in bovines infected by Mycoplasma mycoides subsp. mycoides small colony type. Mol Cell Proteomics 8:2544–2554. doi: 10.1074/mcp.M900009-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdo EM, Nicolet J, Frey J. 2000. Antigenic and genetic characterization of lipoprotein LppQ from Mycoplasma mycoides subsp. mycoides SC. Clin Diagn Lab Immunol 7:588–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer A, Santana-Cruz I, Hegerman J, Gourle H, Schieck E, Lambert M, Nadendla S, Wesonga H, Miller RA, Vashee S, Weber J, Meens J, Frey J, Jores J. 2015. High quality draft genomes of the Mycoplasma mycoides subsp. mycoides challenge strains Afadé and B237. Stand Genomic Sci 10:89. doi: 10.1186/s40793-015-0067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahman O, Cummings SP, Harrington DJ, Sutcliffe IC. 2008. Methods for the bioinformatic identification of bacterial lipoproteins encoded in the genomes of Gram-positive bacteria. World J Microbiol Biotechnol 24:2377–2382. doi: 10.1007/s11274-008-9795-2. [DOI] [Google Scholar]
- 24.Green MR, Sambrook J. 2012. Molecular cloning: a laboratory manual, 4th ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 25.Jores J, Nkando I, Sterner-Kock A, Haider W, Poole J, Unger H, Muriuki C, Wesonga H, Taracha EL. 2008. Assessment of in vitro interferon-gamma responses from peripheral blood mononuclear cells of cattle infected with Mycoplasma mycoides ssp. mycoides small colony type. Vet Immunol Immunopathol 124:192–197. doi: 10.1016/j.vetimm.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 26.Schieck E, Liljander A, Hamsten C, Gicheru N, Scacchia M, Sacchini F, Heller M, Schnee C, Sterner-Kock A, Hlinak A, Naessens J, Poole J, Persson A, Jores J. 2014. High antibody titres against predicted Mycoplasma surface proteins do not prevent sequestration in infected lung tissue in the course of experimental contagious bovine pleuropneumonia. Vet Microbiol 172:285–293. doi: 10.1016/j.vetmic.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 27.Mwirigi M, Nkando I, Aye R, Soi R, Ochanda H, Berberov E, Potter A, Gerdts V, Perez-Casal J, Naessens J, Wesonga H. 2016. Experimental evaluation of inactivated and live attenuated vaccines against Mycoplasma mycoides subsp. mycoides. Vet Immunol Immunopathol 169:63–67. doi: 10.1016/j.vetimm.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Rosengarten R, Behrens A, Stetefeld A, Heller M, Ahrens M, Sachse K, Yogev D, Kirchhoff H. 1994. Antigen heterogeneity among isolates of Mycoplasma bovis is generated by high-frequency variation of diverse membrane surface proteins. Infect Immun 62:5066–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.OIE. 2014. Chapter 2.4.9, p 6–9. In Manual of diagnostic tests and vaccines for terrestrial animals. OIE, Paris, France. [Google Scholar]
- 30.Wright PF, Nilsson E, Van Rooij EM, Lelenta M, Jeggo MH. 1993. Standardisation and validation of enzyme-linked immunosorbent assay techniques for the detection of antibody in infectious disease diagnosis. Rev Sci Tech 12:435–450. [DOI] [PubMed] [Google Scholar]
- 31.Fleiss JL, Cohen J. 1973. The equivalence of weighted Kappa and the intraclass correlation coefficient as measures of reliability. Educ Psychol Measurement 33:613–619. doi: 10.1177/001316447303300309. [DOI] [Google Scholar]
- 32.Gorton TS, Barnett MM, Gull T, French RA, Lu Z, Kutish GF, Adams LG, Geary SJ. 2005. Development of real-time diagnostic assays specific for Mycoplasma mycoides subspecies mycoides Small Colony. Vet Microbiol 111:51–58. doi: 10.1016/j.vetmic.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 33.Schnee C, Heller M, Jores J, Tomaso H, Neubauer H. 2011. Assessment of a novel multiplex real-time PCR assay for the detection of the CBPP agent Mycoplasma mycoides subsp. mycoides SC through experimental infection in cattle. BMC Vet Res 7:47. doi: 10.1186/1746-6148-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorenzon S, Manso-Silvan L, Thiaucourt F. 2008. Specific real-time PCR assays for the detection and quantification of Mycoplasma mycoides subsp. mycoides SC and Mycoplasma capricolum subsp. capripneumoniae. Mol Cell Probes 22:324–328. doi: 10.1016/j.mcp.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Mair G, Vilei EM, Wade A, Frey J, Unger H. 2013. Isothermal loop-mediated amplification (LAMP) for diagnosis of contagious bovine pleuro-pneumonia. BMC Vet Res 9:108. doi: 10.1186/1746-6148-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dupuy V, Manso-Silvan L, Barbe V, Thebault P, Dordet-Frisoni E, Citti C, Poumarat F, Blanchard A, Breton M, Sirand-Pugnet P, Thiaucourt F. 2012. Evolutionary history of contagious bovine pleuropneumonia using next generation sequencing of Mycoplasma mycoides Subsp. mycoides “Small Colony.” PLoS One 7:e46821. doi: 10.1371/journal.pone.0046821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez-Casal J, Prysliak T, Maina T, Wang Y, Townsend H, Berverov E, Nkando I, Wesonga H, Liljander A, Jores J, Naessens J, Gerdts V, Potter A. 2015. Analysis of immune responses to recombinant proteins from strains of Mycoplasma mycoides subsp. mycoides, the causative agent of contagious bovine pleuropneumonia. Vet Immunol Immunopathol 168:103–110. doi: 10.1016/j.vetimm.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 38.Krasteva I, Liljander A, Fischer A, Smith DG, Inglis NF, Scacchia M, Pini A, Jores J, Sacchini F. 2014. Characterization of the in vitro core surface proteome of Mycoplasma mycoides subsp. mycoides, the causative agent of contagious bovine pleuropneumonia. Vet Microbiol 168:116–123. doi: 10.1016/j.vetmic.2013.10.025. [DOI] [PubMed] [Google Scholar]
- 39.Fischer A, Shapiro B, Muriuki C, Heller M, Schnee C, Bongcam-Rudloff E, Vilei EM, Frey J, Jores J. 2012. The origin of the ‘Mycoplasma mycoides cluster’ coincides with domestication of ruminants. PLoS One 7:e36150. doi: 10.1371/journal.pone.0036150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cottew GS, Breard A, DaMassa AJ, Erno H, Leach RH, Lefevre PC, Rodwell AW, Smith GR. 1987. Taxonomy of the Mycoplasma mycoides cluster. Isr J Med Sci 23:632–635. [PubMed] [Google Scholar]
- 41.Dupuy V, Verdier A, Thiaucourt F, Manso-Silvan L. 2015. A large-scale genomic approach affords unprecedented resolution for the molecular epidemiology and evolutionary history of contagious caprine pleuropneumonia. Vet Res 46:74. doi: 10.1186/s13567-015-0208-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.