Abstract
Nutrient deficiencies restrict apple (Malus sp.) tree growth and productivity in Northwest China. The process of autophagy, a conserved degradation pathway in eukaryotic cells, has important roles in nutrient-recycling and helps improve plant performance during periods of nutrient-starvation. Little is known about the functioning of autophagy-related genes (ATGs) in apple. In this study, one of the ATG8 gene family members MdATG8i was isolated from Malus domestica. MdATG8i has conserved putative tubulin binding sites and ATG7 interaction domains. A 1865-bp promoter region cloned from apple genome DNA was predicated to have cis-regulatory elements responsive to light, environmental stresses, and hormones. MdATG8i transcriptions were induced in response to leaf senescence, nitrogen depletion, and oxidative stress. At cellular level, MdATG8i protein was expressed in the nucleus and cytoplasm of onion epidermal cells. Yeast two-hybrid tests showed that MdATG8i could interact with MdATG7a and MdATG7b. In Arabidopsis, its heterologous expression was associated with enhanced vegetative growth, leaf senescence, and tolerance to nitrogen- and carbon-starvation. MdATG8i-overexpressing “Orin” apple callus lines also displayed improved tolerance to nutrient-limited conditions. Our results demonstrate that MdATG8i protein could function in autophagy in a conserved way, as a positive regulator in the response to nutrient-starvation.
Keywords: apple, ATG8, Arabidopsis, autophagy, leaf senescence, nutrient deficiency
Introduction
Plants utilize sophisticated mechanisms for recycling intracellular constituents when nutrient supplies are limited (Doelling et al., 2002). In eukaryotes, two main pathways—the ubiquitin-26S proteasome system and autophagy—are involved in nutrient-recycling, turnover of organelles and aberrant/aggregated proteins, and the precise control of regulators necessary for growth and development (Marshall et al., 2015).
Autophagy is an intracellular degradation system conserved among eukaryotes, with generation of a double-membrane structure (i.e., “autophagosome”) that emerges and sequesters cytoplasmic components as a landmark event (Mizushima et al., 2011), followed by breaking down and recycling cellular materials during development or upon encountering stress conditions (Noda and Inagaki, 2015; Yang and Bassham, 2015). Studies with Saccharomyces cerevisiae have identified many autophagy-related proteins (AuTophaGy, or ATGs) that participate in this process (Tsukada and Ohsumi, 1993; Thumm et al., 1994; Klionsky et al., 2003). Among these proteins, several functional groups help form the autophagosome, including the ATG1 kinase complex, ATG2/9/18 transmembrane complex, phosphatidylinositol 3-kinase (PI3K) complex and ATG8/ATG12 conjugation systems (Mizushima et al., 2011; Li et al., 2015; Noda and Inagaki, 2015).
In higher plants, a suite of homologous ATG proteins directs the autophagy process in response to internal and external factors (Li and Vierstra, 2012; Liu and Bassham, 2012; Feng et al., 2014). The ATG5-ATG12 and ATG8-phosphatidylethanolamine (PE) conjugation systems are two essential components of the autophagic system (Li and Vierstra, 2012; Ohsumi, 2014), and in vitro reconstitution of them has indicated that the mechanism of autophagy is conserved from yeasts to plants (Fujioka et al., 2008). During conjugation, ATG8 and ATG12 are activated by a common ATP-dependent E1-activating enzyme, ATG7, which subsequently binds them to a conserved cysteine within ATG7 via a thioester linkage and then transfers them to their respective E2-conjugating enzymes, ATG3 and ATG10. The activated ATG12 is connected covalently to ATG5, then the dimeric ATG16 protein self-oligomerizes and interacts with ATG5 within ATG12-ATG5 conjugate, forming a tetrameric complex which serves as the E3-like ligase that conjugates ATG8 to PE (Chung et al., 2010). This ATG8-PE adduct coats expanding phagophores and serves as a docking platform for factors that promote vesicle closure and tonoplast fusion, as well as for receptors that capture specific cargo (Li and Vierstra, 2012; Rogov et al., 2014; Li et al., 2015; Noda and Inagaki, 2015). Through those receptors, autophagy can selectively remove unwanted large protein complexes such as ribosomes (ribophagy; Hillwig et al., 2011) and proteasomes (proteaphagy; Marshall et al., 2015); insoluble protein aggregates (aggrephagy; Zhou et al., 2013); damaged or inactive organelles including mitochondria (mitophagy; Li et al., 2014), chloroplasts (chlorophagy; Ishida et al., 2008; Wada et al., 2009), peroxisomes (perophagy; Farmer et al., 2013; Kim et al., 2013; Shibata et al., 2013), and endoplasmic reticulum (ER-phagy; Bernales et al., 2007); or invading pathogens (xenophagy; Gutierrez et al., 2004; Nakagawa et al., 2004). In addition to its decisive role in selective autophagy, ATG8 protein and its homologs in mammals and plants have been used as very reliable markers for monitoring autophagic activity, since they are localized to the isolated membranes and autophagosomes (Ichimura et al., 2000). In Arabidopsis, all nine ATG8 isoforms display high sequence similarity to yeast ATG8 and have a conserved glycine residue at their C-terminus (Doelling et al., 2002; Hanaoka et al., 2002; Yoshimoto et al., 2004). Among them, however, AtATG8h and AtATG8i differ from the others because they lack extra residues that follow this glycine residue (Avin-Wittenberg et al., 2011), as well as less studied than others.
Plant autophagy has several possible roles in seed development and germination, photomorphogenesis, pathogen resistance, senescence, protections against the negative effects of stress, and nutrient-recycling under starvation conditions (Thompson and Vierstra, 2005; Liu and Bassham, 2012; Hanamata et al., 2014). Autophagy is the major contributor to cellular housekeeping and the reuse of nutrients for alleviating nutrient stress (Li and Vierstra, 2012; Liu and Bassham, 2012; Ohsumi, 2014). In Arabidopsis, most ATG genes are transcriptionally up-regulated by nutrient-starvation and during leaf senescence (Doelling et al., 2002; Rose et al., 2006; Thomas, 2013). Loss-of-function autophagy mutants are hypersensitive to nitrogen (N)- and fixed-carbon (C)-limiting conditions, and plants show accelerated senescence even under nutrient-rich conditions (Hanaoka et al., 2002; Phillips et al., 2008). Autophagy is active in N-remobilization under either starvation conditions or normal growth (Guiboileau et al., 2012, 2013; Xia et al., 2012; Li et al., 2015). Therefore, all of these reports indicate that autophagy is associated with nitrogen metabolism and plant yields.
Apple (Malus domestica Borkh.) is one of the most economically important fruit trees grown worldwide. In China, the major region for apple fruit production is within the Northwest Loess Plateau. However, nutrient deficiencies are a challenge there because of reduced soil fertility and a lack of sufficient rainfall. Given that autophagy is critical for maintaining cell homeostasis, plant vitality, and yields under nutrient-starvation conditions, and ATG8s proteins play key roles in autophagy, we isolated one of the ATG8s gene, MdATG8i, from M. domestica. We further characterized MdATG8i gene through expression analysis, promoter isolation and analysis, yeast two-hybrid (Y2H), subcellular localization, and heterologous expression in Arabidopsis and “Orin” apple callus. The overexpression analysis demonstrated its functions in response to nutrient stress, showing its potential for breeding crops with improvement toward nutrient-starvation.
Materials and methods
Apple plant materials and treatments for gene cloning and expression analysis
Two-year-old plants of apple (M. domestica Borkh. “Golden Delicious”), grafted onto rootstock M. hupehensis, were grown in pots in the greenhouse at the Horticultural Experimental Station of Northwest A&F University, Yangling, China. Standard horticultural and management practices were followed for disease and pest control.
To analyze the tissue-specific expression of MdATG8i, buds and flowers were collected on 20 March and 15 April 2014, while stems, shoots, and roots were sampled on 6 June 2014. The fruits were collected 150 days after blooming (15 September 2014). Mark the leaves after they emerged. Young, mature, and senescent leaves were collected on 1 May, 1 August, and 30 October 2014, respectively.
To examine the expression of MdATG8i under N-starvation, we applied N-starvation to hydroponically grown seedlings of M. hupehensis Rehd. (Bai et al., 2008; Li et al., 2012). For this treatment, the Ca(NO3)2 and KNO3 in our Hoagland's nutrient solution were replaced by CaCl2 and KCl, respectively, while the control plants continue to receive the standard solution. Leaves and white roots were sampled on Days 0, 2, 4, 6, and 8 after treatments. To induce oxidative stress, we supplemented the nutrient solution with 50 mM methyl viologen (MV) and collected the white roots at 0, 3, 6, 9, 12, 24, and 36 h post-treatments.
RNA extraction, cloning, and quantitative real-time PCR
Total RNA was extracted according to a CTAB method (Chang et al., 1993). Residual DNA was removed by treating with RNase-free DNase I (Invitrogen, Carlsbad, CA, USA). First-strand cDNA was generated by using a RevertAidTM First Strand cDNA synthesis kit (Fermentas, Thermo Scientific, Waltham, MA, USA). The coding sequence (CDS) of MdATG8i was amplified from cDNA with gene-specific primers (Table 1) using high fidelity Platinum® Taq DNA Polymerase (Invitrogen).
Table 1.
Primers used in this study.
| Primer | Purpose/Vector | Sequence (5′–3′) |
|---|---|---|
| MdATG8i–F | Clone/pMD19T–simple | F: ATGGGGAAGATCCAATCTTTCAAG |
| MdATG8i–R | R: TTAGCCAAAGGTTTTCTCGCTGCT | |
| BK–MdATG8i–EcoR I –F | Yeast two–hybrid/pGBKT7 | F: CCGGAATTCATGGGGAAGATCCAATC |
| BK–MdATG8i–BamH I –R | R: CGCGGATCCTTAGCCAAAGGTTTTC | |
| AD–MdATG7a–EcoR I –F | Yeast two–hybrid/pGADT7 | F: CCGGAATTCATGGAGGGAGGC |
| AD–MdATG7a–Cla I –R | R: CCATCGATTCAAACTTCAACCAAATCATC | |
| AD–MdATG7b–EcoR I –F | F: CCGGAATTCATGGAGCGAGGC | |
| AD–MdATG7b–Cla I –R | R: CCATCGATTCAAACCTCAACCAAATCATC | |
| pro–MdATG8i–F | Promoter clone/pMD19T–simple | F: GAACAACAAACAATGATTATTTGACA |
| pro–MdATG8i–R | R: GATTGGATCTTCCCCATAAAGC | |
| OE–MdATG8i–F | Plant overexpression and Subcellular localization /pGWB406–GFP | F: AAAAAAGCAGGCTTCATGGGGAAGATCCAATC |
| OE–MdATG8i–R | R: CAAGAAAGCTGGGTTTTAGCCAAAGGTTTTCTC | |
| q MdATG8i–F | qRT–PCR | F: GCAGCAGGCTTCACTTGACTCC |
| q MdATG8i–R | R: GGAATCCATGCGACTGGCTGTT | |
| AtUBQ10–F | F: TTCACTTGGTCCTGCGTCTTC | |
| AtUBQ10–R | R: CATCAGGGATTATACAAGGCCCC | |
| attB1 | Adapter primer for gateway vector | F: GGGGACAAGTTTGTACAAAAAAGCAGGCT |
| attB2 | R: GGGGACCACTTTGTACAAGAAAGCTGGGT |
Restriction sites and adapters are underlined.
The same amount of mRNA (2 μg) from each sample was used to synthesize first-strand cDNA. qRT-PCR was performed on an iQ 5.0 instrument (Bio-Rad, Hercules, CA, USA), using a SYBR Premix Ex Taq kit (TaKaRa, Kyoto, Japan) according to the manufacturer's instructions. Transcripts of the Malus elongation factor 1 alpha gene (EF-1α; DQ341381) were used to standardize the cDNA samples for different genes. The specific primer sequences for expression analysis are shown in Table 1. The experiments were performed using three biological samples with three technical PCR repeats for each RNA extract.
Genomic DNA extraction and MdATG8i promoter isolation
Apple genomic DNA was extracted from mature leaves of “Golden Delicious” using an improved CTAB method (Modgil et al., 2005). The DNA with good purity and quality was used to amplify the MdATG8i promoter. PCR was performed with high fidelity Platinum® Taq DNA Polymerase (Invitrogen) with specific primers (Table 1).
Analyses of sequences and phylogenetics
Putative conserved domains were predicted from the National Center for Biotechnology Information (NCBI), using the BLASTp program (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Homologous sequences from other species were aligned with the ClustalW program (Thompson et al., 1994). Phylogenetic trees were constructed with MEGA 5.0 software (Tamura et al., 2011), using the Neighbor-Joining method and bootstrap tests with 1000 replications. Putative cis-regulatory elements in MdATG8i promoter region were examined using the PlantCARE web tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Yeast two-hybrid assays
To generate constructs for Y2H assays, we cloned the CDS of MdATG8i into the pGBKT7 vector (Clontech Laboratories, Inc., http://www.clontech.com/) to produce a translational fusion with a GAL4 DNA binding domain. Likewise, we separately introduced the CDS of MdATG7a (KF438034) and MdATG7b (KF438035) into the pGADT7 vector (Clontech) to produce a translational fusion with a GAL4 activation domain. Primers with restriction sites used for the constructs are listed in Table 1. The constructs were co-transformed into yeast strain AH109 according to the Clontech protocol. Protein–protein interactions were examined by checking the growth of the transformed colonies on SD-Leu-Trp and SD-Leu-Trp-His-Ade+X-α-gal plates.
Plant-overexpressing vector construction for MdATG8i
The Gateway binary vector pGWB406 with a GFP fusion at the N-terminus was used for constructing the MdATG8i-overexpressing vector. We followed the Gateway Technology protocol (Invitrogen) and inserted MdATG8i into pGWB406 destination vectors by BP and LR reactions, using pDONR222 as the donor vector. These vectors were driven by the CaMV 35S promoter and carried the kanamycin (Kan) selectable marker in plants.
Subcellular localization analysis of MdATG8i
Plasmids of GFP-pGWB406/MdATG8i and control were transiently transformed, via the gene gun method (Bio-Rad), into onion epidermal cells. After bombardment, the cells were cultured under darkness for 16–20 h in an MS medium at 22°C before being observed with a confocal microscope (Zeiss LSM).
Generation and characterization of transgenic Arabidopsis overexpressing MdATG8i
Plant-overexpressing vectors GFP-pGWB406/MdATG8i was introduced into Agrobacterium tumefaciens strain EHA105 by electroporation. Arabidopsis thaliana ecotype “Columbia” (Col-0) was transformed via the floral dip method (Zhang et al., 2006). Growing conditions for these Arabidopsis plants included 22°C, 100 μmol photons m−2 s−1, 70% relative humidity, and a long-day (LD, 16-h) photoperiod. Transgenic seeds (T1) were harvested, surface-sterilized in 50% bleach, and selected on an MS medium supplemented with 50 mg L−1 Kan. Kan-resistant plants were PCR-confirmed and further selected for T2 homozygous lines. The collected T3 seeds from two independent T2 lines and the wild-type (WT) were used for the following treatments.
To monitor differences in performance between WT and transgenic Arabidopsis plants, we sowed T3 seeds in soil and exposed them to a short-day (SD, 10-h) photoperiod. The maximum radius of each basal rosette was measured on alternate days with an electronic digital caliper. Other T3 seeds were germinated and grown under LD conditions. Photographs were taken to depict the status of natural flowering and leaf senescence during the experiments.
To analyze bolting time, 5-day-old plate-grown Arabidopsis seedlings were transferred to either standard MS or sugar-depleted media, where they were grown under LD conditions. Bolting was scored as the time at which the main inflorescence shoot had elongated to 5 mm long (Hanaoka et al., 2002).
To conduct dark-induced leaf senescence, we detached the first and seconds true leaves from 2-week-old seedlings grown on MS plates under LD conditions, then placed them adaxial side-up (Oh et al., 1996) on papers wetted with 3 mM MES (pH 5.7), and held them at 22°C under darkness (Doelling et al., 2002). Chlorophyll was extracted with 80% acetone and contents were determined spectrophotometrically according to the method of Lichtenthaler and Wellburn (1983).
To study the response of WT and transgenic Arabidopsis to nitrogen-starvation, we used 5-day-old seedlings grown under LD and transferred them to MS control, N-depleted, or N/sugar-depleted agar media (Doelling et al., 2002). The N-depleted medium was prepared by replacing KNO3 and Ca(NO3)2 with KCl and CaCl2, respectively. After 10 days, the plants were photographed under a light microscope (DM2000; Leica, Germany).
The response of WT and transgenic Arabidopsis to C-starvation was evaluated with 7-day-old seedlings that had grown under LD conditions. They were transferred to either MS-only or sugar-depleted agar media. After the plates were wrapped with aluminum foil and incubated in the dark for 10 days, they were returned to LD conditions for 10 days. Following this recovery period, the plants were photographed and their root hairs were observed under a light microscope (DM2000; Leica, Germany). As a second test, T3 seeds of the WT and transgenic lines were germinated and held for 14 days on MS solid media with or without 1% sucrose. Afterward, the plates were wrapped with foil and incubated in the dark for another 14 days. And then the plants were placed under LD conditions for recovery and were photographed daily (Chung et al., 2010).
Generation of MdATG8i-transgenic “Orin” apple callus and treatments
Callus of “Orin” apple was used for gene transformation (Li et al., 2002) and growth assays. They were placed in the dark at 25°C on an MS medium containing 1.0 mg L−1 2,4-D, 1.0 mg L−1 6-BA, and 8 g L−1 agar, and were sub-cultured at 15-d intervals. For transformation, 7-day-old callus grown in a liquid medium were co-cultivated (10 min, gentle rotation, 25°C) with A. tumefaciens EHA105 that carried the GFP-pGWB406/MdATG8i plasmid. After 2 days co-cultivation, the callus was washed three times with sterile water containing 400 mg L−1 cefotaxime (Cef), then transferred to a subculture medium supplemented with 200 mg L−1 Cef and 30 mg L−1 Kan for transgene selection (Xie et al., 2012).
To investigated the response of transgenic “Orin” callus and the WT to nutrient deficiencies, we shifted 0.02-g samples of 10-day-old tissue to the following treatment media: (1) MS control (subculture); (2) Low-nitrogen, with the N concentration decreased from 60 to 5 mM; or (3) Low-carbon, with the sucrose concentration reduced from 30 to 3 g L−1. All plates were placed in the dark at 25°C, and the callus was photographed and weighed on Day 20.
Statistical analysis
The treatments were repeated three times with consistent results. Data from one representative experiment are shown here, expressed as means and standard deviation (SD). Statistical differences were compared by Student's t-tests at significance levels of *P < 0.05, **P < 0.01, or ***P < 0.001.
Results
Molecular cloning and sequence analysis of MdATG8i
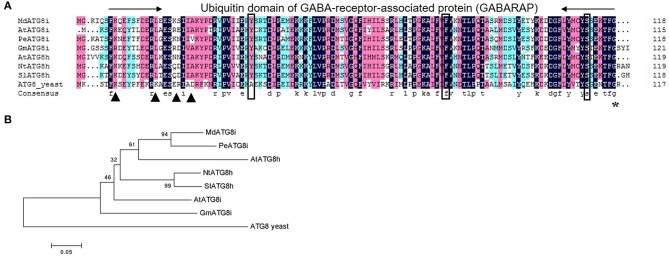
We identified a homologous sequence of AtATG8i from genomic database of M. domestica “Golden Delicious” (GenBank Accession No. KF438037), named as MdATG8i. The gene contains a 357-bp open reading frame (ORF) and encodes a deduced protein with 118 amino acids. Predictions of the conserved domains showed that it has putative tubulin binding sites and ATG7 binding sites, and a conserved ubiquitin domain of GABA-receptor-associated protein (GABARAP) that belongs to the UBQ superfamily (Figure 1A). Protein alignment with other ATG8h or ATG8i from A. thaliana, Populus euphratica, Glycine max, Nicotiana tabacum, and Solanum lycopersicum (Table 2) showed a 79.33% similarity. Like AtATG8h and AtATG8i, MdATG8i protein lacks extra residues at the C-terminus that follows the conserved glycine residue (Figure 1A). The phylogenetic tree analyses indicated that MdATG8i protein forms a close cluster with PeATG8i and AtATG8h (Figure 1B).
Figure 1.
Analysis of amino acid sequence of MdATG8i. (A) Alignment of deduced amino acid sequence from MdATG8i with other ATG8h and ATG8i members. Black triangle, putative tubulin binding sites; Box, ATG7 binding sites; *, Conserved glycine residue; (B) Phylogenetic analysis of ATG8 proteins from Malus domestica (Md), Arabidopsis thaliana (At), Populus euphratica (Pe), Glycine max (Gm), Nicotiana tabacum (Nt), Solanum lycopersicum (Sl), and Saccharomyces cerevisiae (yeast). GenBank Accession Numbers are listed in Table 2.
Table 2.
GenBank Accession Numbers for proteins used to construct phylogenetic trees.
| Protein | Species | GenBank Accession Number |
|---|---|---|
| MdATG8i | Malus domestica | AID50965.1 |
| AtATG8h | Arabidopsis thaliana | NP_566283.1 |
| AtATG8i | Arabidopsis thaliana | NP_566518.1 |
| PeATG8i | Populus euphratica | XP_011011529.1 |
| GmATG8i | Glycine max | NP_001235960.1 |
| NtATG8h | Nicotiana tabacum | AIS71902.1 |
| SlATG8h | Solanum lycopersicum | NP_001234639.1 |
| Yeast ATG8 | Saccharomyces cerevisiae | NP_009475.1 |
Expression analysis of MdATG8i
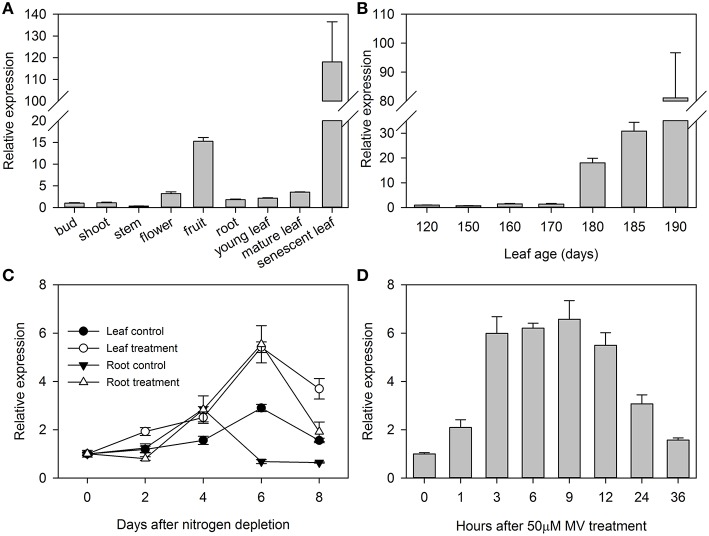
qRT-PCR analysis showed that MdATG8i transcripts were detected in the buds, shoots, stems, flowers, roots, young and mature leaves, and were especially highly accumulated in the fruits and senescent leaves (Figure 2A). Expression in the leaves was greatly enhanced upon aging (Figures 2A,B). To monitor patterns of expression for MdATG8i during the senescence process, we analyzed samples of fully mature leaves over time. MdATG8i transcript levels were relatively low from Days 120 to 170, but were significantly up-regulated when leaves began to turn yellow. In 190-day-old leaves, MdATG8i expression was up-regulated by 80-fold (Figure 2B).
Figure 2.
Expression profiles for MdATG8i in different tissues (A), during leaf senescence (B), under nitrogen-starvation (C), and under MV-induced oxidative stress (D). Total mRNA was extracted from samples and qRT-PCR was performed with gene-specific primers. Expression levels were calculated relative to expression of Malus EF-1α mRNA. Data are means ± SD of three replicate samples.
Here, we used hydroponics culturing system with N-depleted Hoagland's nutrient solution to examine the responses of MdATG8i to nitrogen-starvation. It showed that MdATG8i expression was differentially induced in the leaves and roots, with respective upregulation by 87% and seven-fold at Day 6 (Figure 2C). Using the same hydroponics system, we found that treatment in the roots with an oxidative stress-inducer MV, stimulated gradual upregulation of MdATG8i in the first 3–12 h, with a 5.6-fold increment at 9 h (Figure 2D). Therefore, the results demonstrated that MdATG8i was responsive to leaf senescence, nitrogen-depletion, and oxidative stress.
MdATG8i promoter sequence analysis
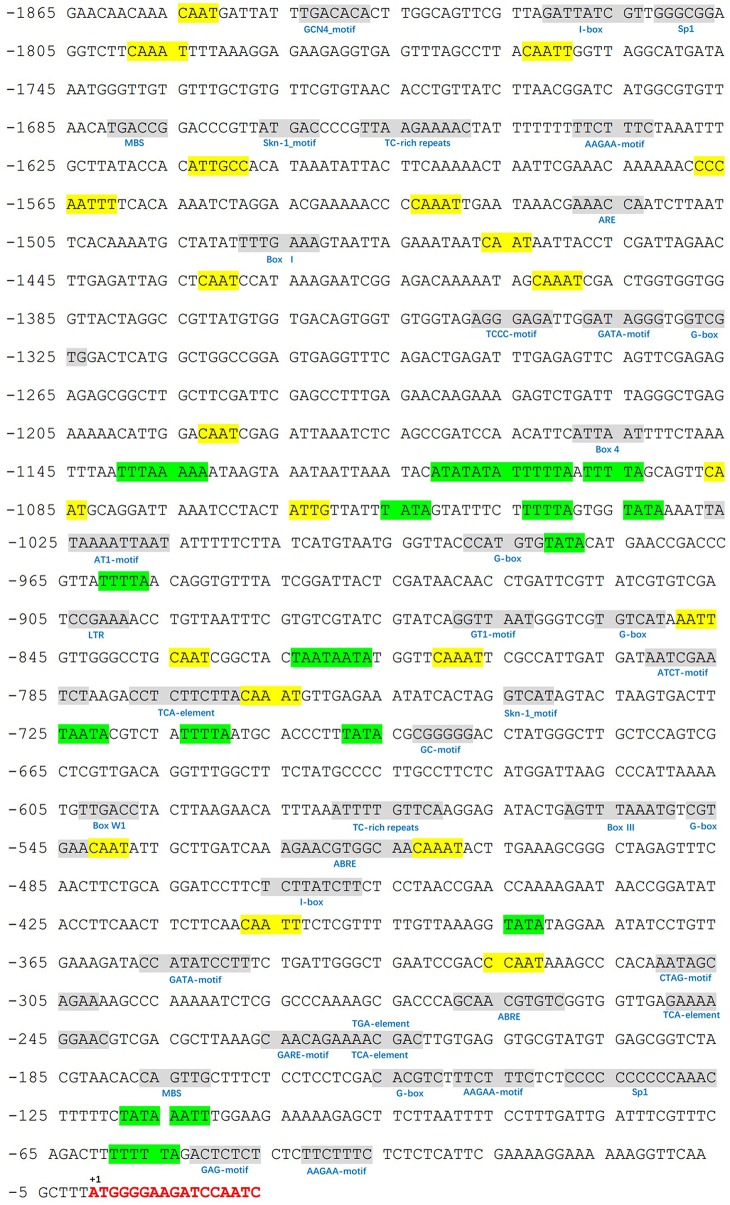
We identified a 1865-bp fragment (GenBank Accession No. KU379689) from the upstream of MdATG8i CDS region. Analyses of potential regulatory cis-acting elements by PlantCARE program and revealed that MdATG8i promoter contains typical CAAT and TATA boxes, as well as many light-responsive elements, including Box I, G-box, ATCT-motif, Box 4, GT1-motif, Sp1, GATA-motif, I-box, TCCC-motif, GAG-motif, and AT1-motif (Figure 3; Table 3). There were also MBS, TC-rich repeats, Box-W1, ARE, GC-motif, and LTR that involved in responses to environmental stresses (Figure 3; Table 3). Beyond that, some important cis-regulatory elements in responsive to hormones were also identified, such as ABRE involved in the abscisic acid responsiveness, gibberellin-responsive GARE-motif, salicylic acid-responsive TCA-element, and auxin-responsive TGA-element (Figure 3; Table 3). In this promoter region, there were also a protein binding site Box III, and two motifs (GCN4 and Skn-1) required for endosperm expression (Figure 3; Table 3). These analyses suggested that MdATG8i might be regulated by various factors, such as light, environmental stresses and hormone signaling.
Figure 3.
Putative cis-regulatory elements (shaded in gray) in promoter region of MdATG8i, as predicted from PlantCARE database. A 1865-bp fragment was cloned from apple genome DNA, with a GenBank Accession Number KU379689. Key sequence elements—TATA box and CAAT box—are shaded in green and yellow, respectively. Letters in red indicates the coding sequence of MdATG8i.
Table 3.
Predicted cis-regulatory elements with putative functions identified in promoter of MdATG8i using PLANTCARE database.
| Cis-element | Positions | Sequences | Putative function |
|---|---|---|---|
| MBS | 185 (−) | CGGTCA | MYB binding site involved in drought-inducibility |
| 1689 (−) | CAACTG | ||
| TC-rich repeats | 208 (−) | GTTTTCTTAC | Element involved in defense and stress responsiveness |
| 1286 (+) | ATTTTCTTCA | ||
| ABRE | 1341 (+) | AGTACGTGGC | Element involved in the abscisic acid responsiveness |
| 1344 (+) | ACGTGGC | ||
| 1597 (+) | GCAACGTGTC | ||
| GARE-motif | 1640 (−) | TCTGTTG | Gibberellin-responsive element |
| TCA-element | 1088 (−) | TCAGAAGAGG | Involved in salicylic acid responsiveness |
| 1614 (+), 1643 (+) | CAGAAAAGGA | ||
| TGA-element | 1648 (+) | AACGAC | Auxin-responsive element |
| Box III | 1307 (−) | CATTTACACT | Protein binding site |
| Box-W1 | 1263 (+) | TTGACC | Fungal elicitor responsive element |
| ARE | 227 (−) | TGGTTT | Regulatory element essential for the anaerobic induction |
| GC-motif | 1173 (−) | CCCCCG | Enhancer-like element involved in anoxic specific inducibility |
| LTR | 962 (+) | CCGAAA | Involved in low-temperature responsiveness |
| GCN4_motif | 22 (−) | TGTGTCA | Involved in endosperm expression |
| Skn-1_motif | 184 (−), 199 (−), 1016 (+), 1121 (+) | GTCAT | Regulatory element required for endosperm expression |
| Box I | 377 (−) | TTTCAAA | Light responsive |
| G-box | 537 (−), 1316 (−), 1006 (−) | CACGAC | Light responsive |
| 1343 (−), 1599 (−) | CACGTT | ||
| 1710 (+) | CACGTC | ||
| 877 (−) | CACATGG | ||
| ATCT-motif | 1074 (+) | AATCTAATCT | Light responsiveness |
| Box 4 | 707 (+), 845 (+) | ATTAAT | Light responsiveness |
| GT1-motif | 997 (+) | GGTTAAT | Light responsive element |
| Sp1 | 54 (+) | GGGCGG | Light responsive element |
| 532 (−), 1727 (+) | CC(G/A)CCC | ||
| GATA-motif | 528 (+) | GATAGGG | Part of a light responsive element |
| 1509 (−) | AAGGATAAGG | ||
| I-box | 44 (−) | GATAATC | Part of a light responsive element |
| 527 (+) | GATAAGGTG | ||
| 524 (−) | CATATCCAAT | ||
| 1400 (−) | AGATAAGA | ||
| 1509 (−) | GATATGG | ||
| TCCC-motif | 518 (−) | TCTCCCT | Part of a light responsive element |
| GAG-motif | 1814 (−) | AGAGAGT | Part of a light responsive element |
| AT1-motif | 839 (−) | ATTAATTTTACA | Part of a light responsive module |
| CAAT-box | In yellow shade | CAAAT/CCAAT/CAAT | Common cis-acting element in promoter and enhancer regions |
| TATA-box | In green shade | TATA | Core promoter element around −30 of transcription start |
Interaction of MdATG8i with MdATG7s in yeast
In yeast, protein ATG8-ATG7 interaction complex was detected in vitro and residues involved in this interaction were also structurally examined (Hanada et al., 2009; Taherbhoy et al., 2011). We found that MdATG8i protein has putative ATG7 binding sites (Figure 1A) as well as the active site Cys-575 within MdATG7s. To test whether MdATG8i interacts with MdATG7s, we conducted Y2H assays using MdATG7a (KF438034) and MdATG7b (KF438035) as prey inserts and MdATG8i as the bait insert. Our data indicated that MdATG8i could interact with both in yeast (Figure 4), thereby implying that MdATG8i protein might participate in the ATG8-PE conjugation system in a conserved way in Malus.
Figure 4.
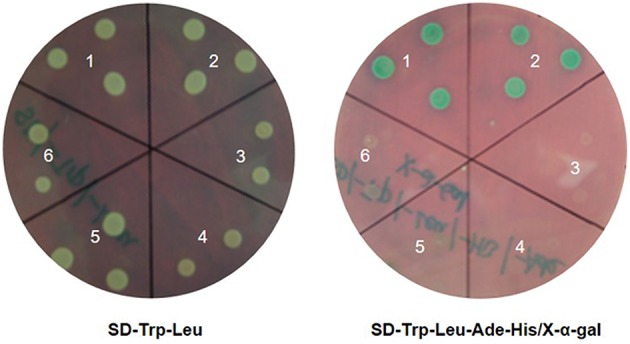
Yeast two-hybrid assays of full-length MdATG8i and MdATG7s proteins. For testing protein–protein interactions, transformed yeast cells were grown on selective SD medium lacking Trp and Leu (Left) or SD medium lacking Trp, Leu, Ade, and His nutrients (Right). Empty vectors pGBKT7 and pGADT7 and their combinations with MdATG8i and MdATG7s were used as negative controls. Combinations: 1, MdATG8i BK + MdATG7a AD; 2, MdATG8i BK + MdATG7b AD; 3, MdATG8i BK + AD; 4, BK + MdATG7a AD; 5, BK + MdATG7b AD; and 6, BK + AD.
Subcellular localization of MdATG8i protein
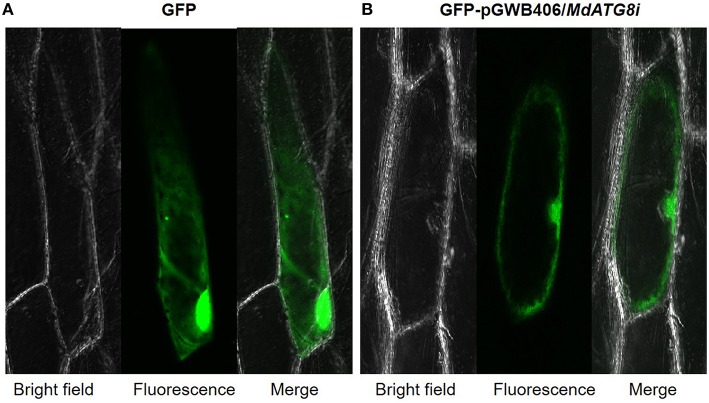
Driven by the 35S promoter, MdATG8i protein with N-terminus GFP fusion was constitutively expressed in the nucleus and cytoplasm of onion epidermal cells after gene gun bombardment (Figure 5).
Figure 5.
Subcellular localization analysis of MdATG8i in onion epidermal cells. (A) GPF control; (B) MdATG8i-GFP fusion protein.
Constitutive expression of MdATG8i promoted vegetative growth, bolting, and leaf senescence in transgenic Arabidopsis
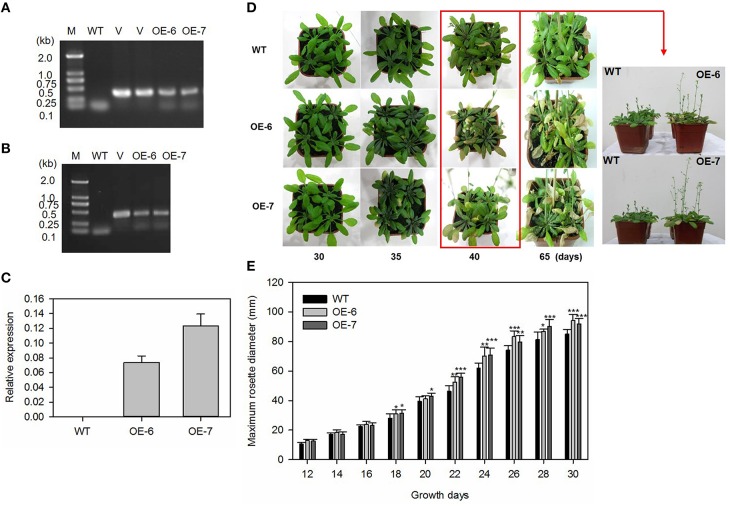
The construct GFP-pGWB406/MdATG8i was introduced into Arabidopsis by the floral-dip method (Zhang et al., 2006). Two representative T2 homozygous overexpression lines (OE, #6 and #7) with relatively higher expression of MdATG8i were selected for further analyses (Figures 6A–C).
Figure 6.
Heterologous expression of MdATG8i in Arabidopsis and growth phenotype. (A) PCR with gDNA; (B) PCR with cDNA; Lanes M, molecular marker DL2000; V, positive vector containing pGWB406-MdATG8i plasmid; WT, non-transformed wild-type; OE-6 and -7, MdATG8i-transgenic lines; (C) qRT-PCR analysis of MdATG8i transcripts in Arabidopsis lines OE-6 and OE-7; (D) Growth phenotype comparison of WT and transgenic lines under LD photoperiod; (E) Growth for rosettes from WT or transgenic lines under SD photoperiod; Data are means ± SD of 20 replicates. *, ** or *** indicates the statistically significant differences determined by Student's t-tests at P < 0.05, P < 0.01, or P < 0.001, respectively.
Under LD conditions, all of the transgenic plants bolted earlier and had higher main inflorescence height than the WT (Figure 6D). By Day 35, some leaves from transgenic plants were yellowing and beginning to senesce (Figure 6D). As a growth parameter, we measured the maximum rosette radius of MdATG8i transgenic Arabidopsis plants and the WT under SD conditions. During the first 12–16 days, the radius values did not differ significantly among genotypes. However, MdATG8i transgenic plants began to grow faster than the WT after Day 18 (Figure 6E).
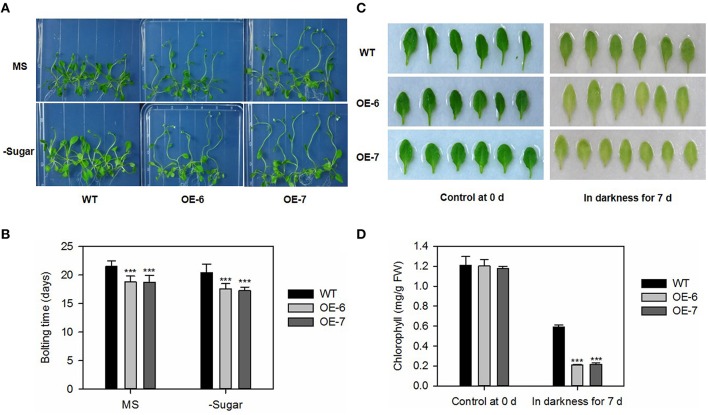
To evaluate the difference in bolting days, we transferred 7-day-old Arabidopsis seedlings to plates containing either a standard MS medium (Murashige and Skoog, 1962) or one that was sugar-depleted. In both media types, plants of the overexpressing lines were taller, and had bolted significantly earlier than the WT (Figures 7A,B). We also tested the senescence progress of detached leaves under darkness, and found that, after 7 days, the transgenic lines showed earlier yellowing (Figure 7C) and lower chlorophyll contents (Figure 7D) when compared with the WT.
Figure 7.
Bolting and dark-induced leaf senescence is promoted by heterologous expression of MdATG8i in Arabidopsis. (A) Bolting comparison between WT and OE lines on Day 25; (B) Statistical analysis of bolting time. Data are means ± SD of 18 replicates; (C) Representative samples of control and dark-treated tissue from WT and OE lines; (D) Chlorophyll contents. Data are means ± SD of five replicates. *, ** or *** indicates the statistically significant differences determined by Student's t-tests at P < 0.05, P < 0.01, or P < 0.001, respectively.
MdATG8i enhanced tolerance to N- and C starvation in transgenic Arabidopsis
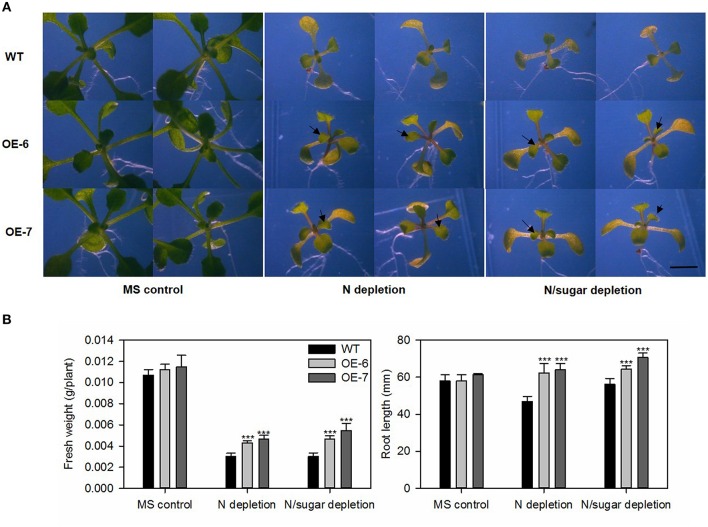
We transferred 5-day-old Arabidopsis seedlings of the transgenic lines and WT into media that were either nitrogen-depleted or both nitrogen- and sugar-depleted. After exposure to LD conditions for 10 days, the cotyledons of all plants turned yellow. However, we noted distinct differences in the size and growth patterns of the true leaves. As shown in Figure 8A, all transgenic lines produced more and larger true leaves than the WT under both N-depleted and N/sugar-depleted conditions. All OE lines had significantly higher fresh weights and longer roots than the WT after 10 days of growth on N-depleted and N/sugar-depleted media (Figure 8B). This phenotype suggested that, under a nitrogen deficiency, overexpression of MdATG8i enables Arabidopsis seedlings to perform better than the WT.
Figure 8.
Tolerance to nitrogen-starvation is enhanced by heterologous expression of MdATG8i. Five-day-old Arabidopsis seedlings were transferred to MS control, N-depleted, or N/sugar-depleted agar media. After 10 days under LD photoperiod, they were photographed (A) before recording fresh weights and root lengths (B). Black arrowheads indicate newly emerged true leaves. Scale bar = 5 mm. Data are means ± SD of 15 replicates. *, ** or *** indicates the statistically significant differences determined by Student's t-tests at P < 0.05, P < 0.01, or P < 0.001, respectively.
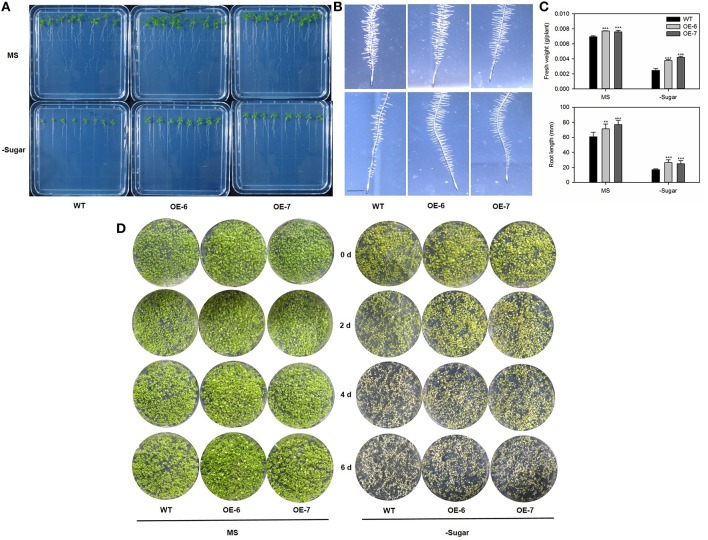
Generally, Arabidopsis atg mutants are more sensitive to environments in which the supply of carbon is limited (Hanaoka et al., 2002; Thompson, 2005; Phillips et al., 2008). To address whether MdATG8i-overexpressing plants have enhanced tolerance to C-starvation, we transferred 7-day-old Arabidopsis seedlings to a medium without supplemental sugar and exposed them to continuous darkness for 10 days. Afterward, the plates were returned to the light. Following 10 days of recovery, the transgenic lines had higher fresh weights, more root hair and longer roots, especially when growing on the sugar-depleted medium (Figures 9A–C). It was interesting to find differences in the distribution and density of root hairs. When sugar was omitted from the media, the WT root hairs were sparse and short, while two OE lines showed no obvious variations from those grown on the standard MS medium (Figure 9B). This improved adaptation to carbon-starvation was also confirmed by placing seeds of the Arabidopsis WT and transgenic lines directly on either MS or sugar-depleted media for 14 days, then transferring the plates to darkness for 14 days, before allowing them to recover under LD conditions. On the standard media, plants of all genotypes were greener in the light and had produced new leaves, and performance by the transgenic plants was slightly better than that of the WT (Figure 9D). More obvious differences were observed on the sugar-depleted media, where WT plants wilted after 2 days before they quickly turned white and then died. Although some leaves from the transgenic plants also appeared bleached on Day 2, they did not wilt, and some plants survived after 6 days of recovery (Figure 9D). These results demonstrated that overexpression of MdATG8i contributes to greater endurance against N- and C-starvation.
Figure 9.
Results of analysis using plants to test tolerance to carbon-starvation, which is enhanced by heterologous expression of MdATG8i. Seven-day-old Arabidopsis seedlings were transferred to standard MS or sugar-depleted agar media. After incubation in dark for 10 days, plates were returned to light for recovery under LD photoperiod. (A) Seedling phenotypes for WT and OE lines; (B) Root hair phenotype; scale bar = 5 mm; (C) Fresh weights and root lengths. Data are means ± SD of 15 replicates; (D) Alternative analysis using seeds to test tolerance to carbon-starvation. Arabidopsis seeds were germinated and grown on MS solid medium with or without 1% sucrose for 14 days. Afterward, plates were incubated in dark for another 14 days. Plants from WT and OE lines were photographed on alternate days (0, 2, 4, 6) to monitor recovery phenotype under LD photoperiod. *, ** or *** indicates the statistically significant differences determined by Student's t-tests at P < 0.05, P < 0.01, or P < 0.001, respectively.
Constitutive expression of MdATG8i enhanced tolerance of “Orin” apple callus to limited nutrients
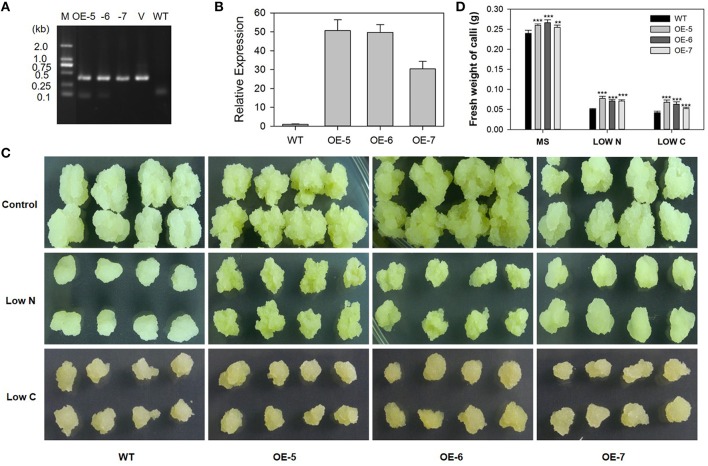
We obtained three transgenic “Orin” lines that over-express MdATG8i, as confirmed by PCR with gDNA (Figure 10A) and by qRT-PCR with cDNA (Figure 10B). The mRNA transcripts were increased by 49-, 48-, and 29-fold in OE-5, OE-6, and OE-7, respectively (Figure 10B). After 14 days of growth, transgenic and WT callus of similar size and weight were transferred to MS control, low-N, or low-C media. By Day 20, the WT callus on the low-N medium had turned white and stopped growing while the three transgenic lines were slightly yellow but showed better growth (Figure 10C). Although all of the callus grown on the low-C medium turned brown (Figure 10C), transgenic callus was significantly heavier than the WT under all treatments (Figure 10D). These results showed that overexpression of MdATG8i confers tolerance to nitrogen and sucrose deficiencies at the cellular level.
Figure 10.
Tolerance to limited supplies of nitrogen and carbon is enhanced by constitutive expression of MdATG8i in transgenic “Orin” apple callus. (A) PCR verification of constitutive expression. Lanes M, molecular marker DL2000; V, positive vector containing pGWB406-MdATG8i plasmid; WT, non-transformed wild-type; and OE-5 through -7, MdATG8i-transgenic callus lines; (B) qRT-PCR analysis of MdATG8i expression in WT and transgenic lines; (C) Morphology of callus after 20 days; (D) Fresh weights of callus after 20 days. Data are means ± SD of three biological replicates. *, ** or *** indicates the statistically significant differences determined by Student's t-tests at P < 0.05, P < 0.01, or P < 0.001, respectively.
Discussion
In response to low energy resources, eukaryotic cells initiate autophagy (Mizushima and Levine, 2010). During that degradative process, autophagosomes are formed. They then engulf a portion of the cytosol or organelles and fuse with a lysosome or vacuole before the cellular cytosolic contents are ultimately recycled (Nakatogawa et al., 2009). MdATG8i protein has the typical conserved glycine residue at the C-terminus. Moreover, this protein has putative tubulin binding sites, thereby suggesting that it is also involved in linking the autophagy pathway to the microtubule network, similar to what happens with AtATG8a and AtATG8d from Arabidopsis (Ketelaar et al., 2004). The MdATG8i protein has ATG7 binding sites as well. During autophagy, a thioester-bonded ATG7-ATG8 intermediate has been generated and could be detected in vitro (Hanada et al., 2009; Noda et al., 2011). We used Y2H assays to show that MdATG8i could interact with both MdATG7a and MdATG7b in yeast. This was evidence that MdATG8i protein can also participate normally in the ATG8-PE conjugation system, and the essential residues of MdATG8i were well conserved in M. domestica.
As the final stage in leaf development, senescence is characterized by chlorophyll degradation and the transition of nutrients from assimilation to remobilization (Masclaux et al., 2000). Autophagy might play important roles in degradation/remobilization during natural or stress-induced leaf senescence (Avila-Ospina et al., 2014; Li et al., 2015). Many ATGs in Arabidopsis are strongly up-regulated during natural-, dark-induced, and detachment-induced leaf senescence, thereby indicating that autophagy is critical for the efficient remobilization and recycling of nutrients (van der Graaff et al., 2006). The mRNA for ATG8 are preferentially accumulated as leaves senesce, suggesting that the autophagy conjugation pathways are up-regulated during that plant stage (Doelling et al., 2002). We found that transcriptional level for MdATG8i is much higher in senescent leaves than in any other tissues examined here. Moreover, it is sharply up-regulated in the late stages of natural senescence, implying that, MdATG8i participates in this process (Wang et al., 2014). Because chloroplast degradation involves autophagy (Ishida et al., 2008; Wada et al., 2009), most atg mutants from Arabidopsis exhibit a phenotype of accelerated and early senescence when compared with the WT (Doelling et al., 2002; Hanaoka et al., 2002; Xiong et al., 2005), a phenomenon thought to be due to excess production of salicylic acid (Yoshimoto et al., 2009). In our study, however, heterologous expression of MdATG8i in Arabidopsis led to a slight promotion of leaf senescence and an early transition into the reproductive growth stage. Although these phenotypes seem to be inconsistency with those reported for the atg mutants, they are similar to those observed when soybean GmATG8c is expressed in Arabidopsis. There, vegetative growth is stimulated, as evidenced by the development of larger rosettes and earlier bolting (Xia et al., 2012). In addition, the atg5 and atg7 mutants from Arabidopsis display a functional stay-green phenotype rather than the yellowing that occurs in the WT when exposed to mild abiotic stress (Sakuraba et al., 2014). Our results and these previous reports suggest that autophagy has roles in both plant survival as well as mortality depending upon environmental conditions. And this might be a positive strategy to improve nutrient use efficiency by facilitating the remobilization of chloroplast proteins during leaf senescence by MdATG8i.
Arabidopsis atg mutants like atg7-1 and atg9-1 are hypersensitive to N- and C-starvation (Doelling et al., 2002; Hanaoka et al., 2002). In Arabidopsis, most autophagy genes are transcriptionally up-regulated when nutrients are less available (Doelling et al., 2002; Xiong et al., 2005; Rose et al., 2006; Liu and Bassham, 2012). Furthermore, in maize, ATG transcripts and ATG8-PE are accumulated when N and C supplies are reduced (Chung et al., 2009). Consistent with this, MdATG8i respond to N-starvation through transcriptional upregulation. Furthermore, overexpression of MdATG8i in Arabidopsis leads to similar phenotypes, i.e., enhanced tolerance to nitrogen and carbon starvations. The transgenic lines not only grow better than the WT under such conditions, but they recover and have increased performance when returned to normal growth practices. In particular, transgenic plants produce more leaves, longer roots, and more root hairs, regardless of whether they are starved or are moved again to optimum conditions. This suggests that heterologous expression of MdATG8i contributes to the fitness and survival of nutrient-stressed Arabidopsis.
Previous studies have shown that transgenic Arabidopsis plants over-expressing GmATG8c produce more leaves under extended periods of N- and C-starvation (Xia et al., 2012). Likewise, expression of GFP-AtATG8f -HA under the control of the 35S promoter is associated with improved Arabidopsis growth when N and fixed-C are limited (Slavikova et al., 2008). Finally, Arabidopsis mutants defective in either the ATG8 or ATG12 conjugation system display the same sensitivity to N or fixed-C starvation (Chung et al., 2010). Therefore, the involvement of MdATG8i in autophagy conjugation systems has a positive role in the maintenance of plant fitness and adaptations to nutrient limitations.
For further evaluation of the function of MdATG8i in coping with nutrient-starvation, we constitutively over-expressed MdATG8i under the control of the 35S promoter in “Orin” callus. Transgenic callus had much better growth and were significantly heavier than the WT under low-N or -C conditions. Our finding is consistent with that of Xia et al. (2012), and confirms that MdATG8i functions against nutrient deficiencies at the cellular level. Because of the challenges associated with apple transformation (Seong and Song, 2008), it is still difficult to produce a large number of transgenic apple plants. Based on our current findings, MdATG8i has a physiological function to protect tissues from the negative effects of nutrient-deprivation, at both the cellular and whole-plant levels. Furthermore, our results demonstrate that this gene has potential as a target gene in research programs that focus on improving yields and making plants more adaptable to nutrient-deficient growing conditions.
In summary, we characterized an apple autophagy gene—MdATG8i, analyzed its sequence, expression and promoter, and also examined its function in response to nutrient deficiencies in both Arabidopsis and apple callus. The improved tolerance to N- and C-starvation by MdATG8i provided evidence that it owns conserved function in apple autophagy process, which might be linked to elevated fitness in apple plants that grow in challenging environment such as nutrient deficiencies.
Author contributions
PW and FM conceived and designed the experiments; PW and XS performed the experiments; NW, XJ, and XG collected data; PW wrote the manuscript; FM critically revised the article.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the earmarked fund for the China Agriculture Research System (CARS-28) and by the National High Technology Research and Development Program of China (2011AA100204). The authors have no conflict of interest to declare.
Glossary
Abbreviations
- ATG
AuTophaGy-related protein
- C
carbon
- LD
long-day
- MV
methyl viologen
- N
nitrogen
- OE lines
overexpressing lines
- SD
short-day
- WT
wild-type.
References
- Avila-Ospina L., Moison M., Yoshimoto K., Masclaux-Daubresse C. (2014). Autophagy, plant senescence, and nutrient recycling. J. Exp. Bot. 65, 3799–3811. 10.1093/jxb/eru039 [DOI] [PubMed] [Google Scholar]
- Avin-Wittenberg T., Honig A., Galili G. (2011). Variations on a theme: plant autophagy in comparison to yeast and mammals. Protoplasma 249, 285–299. 10.1007/s00709-011-0296-z [DOI] [PubMed] [Google Scholar]
- Bai T., Li C., Ma F., Shu H., Han M., Wang K. (2008). Physiological responses and analysis of tolerance of apple rootstocks to root-zone hypoxia stress. Sci. Agric. Sin. 41, 4140–4148. 10.3864/j.issn.0578-1752.2008.12.026 [DOI] [Google Scholar]
- Bernales S., Schuck S., Walter P. (2007). ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy 3, 285–287. 10.4161/auto.3930 [DOI] [PubMed] [Google Scholar]
- Chang S., Puryear J., Cairney J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11, 113–116. 10.1007/BF02670468 [DOI] [Google Scholar]
- Chung T., Phillips A. R., Vierstra R. D. (2010). ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 62, 483–493. 10.1111/j.1365-313X.2010.04166.x [DOI] [PubMed] [Google Scholar]
- Chung T., Suttangkakul A., Vierstra R. D. (2009). The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 149, 220–234. 10.1104/pp.108.126714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J. H., Walker J. M., Friedman E. M., Thompson A. R., Vierstra R. D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277, 33105–33114. 10.1074/jbc.M204630200 [DOI] [PubMed] [Google Scholar]
- Farmer L. M., Rinaldi M. A., Young P. G., Danan C. H., Burkhart S. E., Bartel B. (2013). Disrupting autophagy restores peroxisomes function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. Plant Cell 25, 4085–4100. 10.1105/tpc.113.113407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., He D., Yao Z., Klionsky D. J. (2014). The machinery of macroautophagy. Cell Res. 24, 24–41. 10.1038/cr.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y., Noda N. N., Fujii K., Yoshimoto K., Ohsumi Y., Inagaki F. (2008). In vitro reconstitution of plant Atg8 and Atg12 conjugation systems essential for autophagy. J. Biol. Chem. 283, 1921–1928. 10.1074/jbc.M706214200 [DOI] [PubMed] [Google Scholar]
- Guiboileau A., Avila-Ospina L., Yoshimoto K., Soulay F., Azzopardi M., Marmagne A., et al. (2013). Physiological and metabolic consequences of autophagy deficiency for the management of nitrogen and protein resources in Arabidopsis leaves depending on nitrate availability. New Phytol. 199, 683–694. 10.1111/nph.12307 [DOI] [PubMed] [Google Scholar]
- Guiboileau A., Yoshimoto K., Soulay F., Bataille M. P., Avice J. C., Masclaux-Daubresse C. (2012). Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol. 194, 732–740. 10.1111/j.1469-8137.2012.04084.x [DOI] [PubMed] [Google Scholar]
- Gutierrez M. G., Master S. S., Singh S. B., Taylor G. A., Colombo M. I., Deretic V. (2004). Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766. 10.1016/j.cell.2004.11.038 [DOI] [PubMed] [Google Scholar]
- Hanada T., Satomi Y., Takao T., Ohsumi Y. (2009). The amino-terminal region of Atg3 is essential for association with phosphatidylethanolamine in Atg8 lipidation. FEBS Lett. 583, 1078–1083. 10.1016/j.febslet.2009.03.009 [DOI] [PubMed] [Google Scholar]
- Hanamata S., Kurusu T., Kuchitsu K. (2014). Roles of autophagy in male reproductive development in plants. Front. Plant Sci. 5:457. 10.3389/fpls.2014.00457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., et al. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129, 1181–1193. 10.1104/pp.011024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillwig M. S., Contento A. L., Meyer A., Ebany D., Bassham D. C., Macintosh G. C. (2011). RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. Proc. Natl. Acad. Sci. U.S.A. 108, 1093–1098. 10.1073/pnas.1009809108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., et al. (2000). A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492. 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- Ishida H., Yoshimoto K., Izumi M., Reisen D., Yano Y., Makino A., et al. (2008). Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 148, 142–155. 10.1104/pp.108.122770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T., Voss C., Dimmock S. A., Thumm M., Hussey P. J. (2004). Arabidopsis homologues of the autophagy protein Atg8 are a novel family of microtubule binding proteins. FEBS Lett. 567, 302–306. 10.1016/j.febslet.2004.04.088 [DOI] [PubMed] [Google Scholar]
- Kim J., Lee H., Lee H. N., Kim S. H., Shin K. D., Chung T. (2013). Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell 25, 4956–4966. 10.1105/tpc.113.117960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., Cregg J. M., Dunn W. A., Jr., Emr S. D., Sakai Y., Sandoval I. V., et al. (2003). A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5, 539–545. 10.1016/S1534-5807(03)00296-X [DOI] [PubMed] [Google Scholar]
- Li C., Wang P., Wei Z., Liang D., Liu C., Yin L., et al. (2012). The mitigation effects of exogenous melatonin on salinity-induced stress in Malus hupehensis. J. Pineal Res. 53, 298–306. 10.1111/j.1600-079X.2012.00999.x [DOI] [PubMed] [Google Scholar]
- Li D. D., Shi W., Deng X. X. (2002). Agrobacterium-mediated transformation of embryogenic calluses of Ponkan mandarin and the regeneration of plants containing the chimeric ribonuclease gene. Plant Cell Rep. 21, 153–156. 10.1007/s00299-002-0492-6 [DOI] [Google Scholar]
- Li F., Chung T., Pennington J. G., Federico M., Kaeppler H., Kaeppler S. W., et al. (2015). Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 27, 1389–1408. 10.1105/tpc.15.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Chung T., Vierstra R. D. (2014). AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 26, 788–807. 10.1105/tpc.113.120014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Vierstra R. D. (2012). Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 17, 526–537. 10.1016/j.tplants.2012.05.006 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler K., Wellburn A. R. (1983). Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 11, 591–592. 10.1042/bst0110591 [DOI] [Google Scholar]
- Liu Y., Bassham D. C. (2012). Autophagy: pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63, 215–237. 10.1146/annurev-arplant-042811-105441 [DOI] [PubMed] [Google Scholar]
- Marshall R. S., Li F., Gemperline D. C., Book A. J., Vierstra R. D. (2015). Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/Ubiquitin receptor RPN10 in Arabidopsis. Mol. Cell 58, 1053–1066. 10.1016/j.molcel.2015.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux C., Valadier M. H., Brugiere N., Morot-Gaudry J. F., Hirel B. (2000). Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta 211, 510–518. 10.1007/s004250000310 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B. (2010). Autophagy in mammalian development and differentiation. Nat. Cell Biol. 12, 823–830. 10.1038/ncb0910-823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., Ohsumi Y. (2011). The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132. 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- Modgil M., Mahajan K., Chakrabarti S. K., Sharma D. R., Sobti R. C. (2005). Molecular analysis of genetic stability in micro propagated apple rootstock MM 106. Sci. Hortic. 104, 151 10.1016/j.scienta.2004.07.009 [DOI] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- Nakagawa I., Amano A., Mizushima N., Yamamoto A., Yamaguchi H., Kamimoto T., et al. (2004). Autophagy defends cells against invading group A Streptococcus. Science 306, 1037–1040. 10.1126/science.1103966 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. (2009). Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467. 10.1038/nrm2708 [DOI] [PubMed] [Google Scholar]
- Noda N. N., Inagaki F. (2015). Mechanisms of autophagy. Annu. Rev. Biophys. 44, 101–122. 10.1142/S0192415X16500282 [DOI] [PubMed] [Google Scholar]
- Noda N. N., Satoo K., Fujioka Y., Kumeta H., Ogura K., Nakatogawa H., et al. (2011). Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol. Cell 44, 462–475. 10.1016/j.molcel.2011.08.035 [DOI] [PubMed] [Google Scholar]
- Oh S. A., Lee S. Y., Chung I. K., Lee C. H., Nam H. G. (1996). A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol. Biol. 30, 739–754. 10.1007/BF00019008 [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. (2014). Historical landmarks of autophagy research. Cell Res. 24, 9–23. 10.1038/cr.2013.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips A. R., Suttangkakul A., Vierstra R. D. (2008). The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178, 1339–1353. 10.1534/genetics.107.086199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V., Dotsch V., Johansen T., Kirkin V. (2014). Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell 53, 167–178. 10.1016/j.molcel.2013.12.014 [DOI] [PubMed] [Google Scholar]
- Rose T. L., Bonneau L., Der C., Marty-Mazars D., Marty F. (2006). Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol. Cell 98, 53–67. 10.1042/BC20040516 [DOI] [PubMed] [Google Scholar]
- Sakuraba Y., Lee S. H., Kim Y. S., Park O. K., Hortensteiner S., Paek N. C. (2014). Delayed degradation of chlorophylls and photosynthetic proteins in Arabidopsis autophagy mutants during stress-induced leaf yellowing. J. Exp. Bot. 65, 3915–3925. 10.1093/jxb/eru008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong E. S., Song K. J. (2008). Factors affecting the early gene transfer step in the development of transgenic ‘Fuji’ apple plants. Plant Growth Regul. 54, 89–95. 10.1007/s10725-007-9231-x [DOI] [Google Scholar]
- Shibata M., Oikawa K., Yoshimoto K., Kondo M., Mano S., Yamada K., et al. (2013). Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell 25, 4967–4983. 10.1105/tpc.113.116947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavikova S., Ufaz S., Avin-Wittenberg T., Levanony H., Galili G. (2008). An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J. Exp. Bot. 59, 4029–4043. 10.1093/jxb/ern244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taherbhoy A. M., Tait S. W., Kaiser S. E., Williams A. H., Deng A., Nourse A., et al. (2011). Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway. Mol. Cell 44, 451–461. 10.1016/j.molcel.2011.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H. (2013). Senescence, ageing and death of the whole plant. New Phytol. 197, 696. 10.1111/nph.12047 [DOI] [PubMed] [Google Scholar]
- Thompson A. R. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138, 2097–2110. 10.1104/pp.105.060673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A. R., Vierstra R. D. (2005). Autophagic recycling: lessons from yeast help define the process in plants. Curr. Opin. Plant Biol. 8, 165–173. 10.1016/j.pbi.2005.01.013 [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M., Egner R., Koch B., Schlumpberger M., Straub M., Veenhuis M., et al. (1994). Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 349, 275–280. 10.1016/0014-5793(94)00672-5 [DOI] [PubMed] [Google Scholar]
- Tsukada M., Ohsumi Y. (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169–174. 10.1016/0014-5793(93)80398-E [DOI] [PubMed] [Google Scholar]
- van der Graaff E., Schwacke R., Schneider A., Desimone M., Flugge U. I., Kunze R. (2006). Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 141, 776–792. 10.1104/pp.106.079293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S., Ishida H., Izumi M., Yoshimoto K., Ohsumi Y., Mae T., et al. (2009). Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 149, 885–893. 10.1104/pp.108.130013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Sun X., Yue Z., Liang D., Wang N., Ma F. (2014). Isolation and characterization of MdATG18a, a WD40-repeat AuTophaGy-related gene responsive to leaf senescence and abiotic stress in Malus. Sci. Hortic. Amsterdam 165, 51–61. 10.1016/j.scienta.2013.10.038 [DOI] [Google Scholar]
- Xia T., Xiao D., Liu D., Chai W., Gong Q., Wang N. N. (2012). Heterologous expression of ATG8c from soybean confers tolerance to nitrogen deficiency and increases yield in Arabidopsis. PLoS ONE 7:e37217. 10.1371/journal.pone.0037217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Li S., Zhang R., Zhao J., Chen Y., Zhao Q., et al. (2012). The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apple. Plant Cell Environ. 35, 1884–1897. 10.1111/j.1365-3040.2012.02523.x [DOI] [PubMed] [Google Scholar]
- Xiong Y., Contento A. L., Bassham D. C. (2005). AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42, 535–546. 10.1111/j.1365-313XX.2005.02397.x [DOI] [PubMed] [Google Scholar]
- Yang X., Bassham D. C. (2015). Chapter one – New insights into the mechanism and function of autophagy in plant cells. Int. Rev. Cell Mol. Biol. 320, 1–40. 10.1016/bs.ircmb.2015.07.005 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., et al. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16, 2967–2983. 10.1105/tpc.104.025395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K., Jikumaru Y., Kamiya Y., Kusano M., Consonni C., Panstruga R., et al. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21, 2914–2927. 10.1105/tpc.109.068635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S. S., Niu Q., Chua N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. 10.1038/nprot.2006.97 [DOI] [PubMed] [Google Scholar]
- Zhou J., Wang J., Cheng Y., Chi Y. J., Fan B., Yu J. Q., et al. (2013). NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 9:e1003196. 10.1371/journal.pgen.1003196 [DOI] [PMC free article] [PubMed] [Google Scholar]