Abstract
Ceftazidime is one of the few cephalosporins with activity against Pseudomonas aeruginosa. Using whole-genome comparative analysis, we set out to determine the prevalent mechanism(s) of resistance to ceftazidime (CAZ) using a set of 181 clinical isolates. These isolates represented various multilocus sequence types that consisted of both ceftazidime-susceptible and -resistant populations. A presumptive resistance mechanism against ceftazidime was identified in 88% of the nonsusceptible isolates using this approach.
TEXT
Pseudomonas aeruginosa is an opportunistic pathogen associated with numerous nosocomial infections, where β-lactam antibiotics remain key in treatment (1, 2). One of the major antimicrobials used to fight P. aeruginosa infections is ceftazidime (CAZ), a well-known cephalosporin that acts primarily as a penicillin-binding protein 3 (PBP3) inhibitor (3, 4).
A significant proportion of ceftazidime-resistant isolates arise through the horizontal acquisition of β-lactamases or altered expression of the chromosomal drug-inducible wide-spectrum class C β-lactamase AmpC (reviewed in reference 5). The overproduction of AmpC can result from mutations affecting the peptidoglycan (PG) recycling process, where accumulation of cell wall intermediates ultimately induces ampC overexpression (5).
We focused our study on a panel of 181 clinical P. aeruginosa isolates, where comparative analysis between multiple isolates belonging to the same multilocus sequence types (MLSTs) allowed for identification of chromosomal gene variants unique to the ceftazidime-resistant population (6).
The initial MIC to ceftazidime was determined using frozen plates (Thermo Scientific) following the Clinical and Laboratory Standards Institute guidelines (7, 8). Of the 181 isolates in the analysis set, 99 (55%) were resistant to ceftazidime (MIC, ≥16 μg/ml), and 82 were susceptible (MIC, ≤8 μg/ml) (Table 1; see Table S1 in the supplemental material).
TABLE 1.
Summary of β-lactamase content and AmpC regulon polymorphisms identified in P. aeruginosa isolates by MLST
| MLST | No. of isolates: |
No. of PBP3 polymorphisms (target modification) | Polymorphism(s) identified among regulators of ampC expression (n) | ||||
|---|---|---|---|---|---|---|---|
| Total | Susceptible to CAZ (MIC, <8 μg/ml) | Nonsusceptible to CAZ (MIC, >16 μg/ml) | Total with acquired β-lactamase | Total with acquired β-lactamase attributed to mediation of nonsusceptibility to CAZ | |||
| 111 | 24 | 8 | 16 | 19 | 8 (VIM-2) | AmpD (6), DacB (2) | |
| 116 | 4 | 3 | 1 | 0 | 0 | ||
| 155 | 4 | 3 | 1 | 1 | 0 | ||
| 167 | 3 | 1 | 2 | 2 | 2 (IMP-15) | ||
| 175 | 9 | 4 | 5 | 0 | 0 | AmpD (3), AmpR (2) | |
| 179 | 7 | 4 | 3 | 2 | 2 (PER-1) | ||
| 233 | 8 | 1 | 7 | 7 | 7 (VIM-2) | ||
| 235 | 37 | 11 | 26 | 23 | 14 (1 GES-19, 1 GES-9, 1 IMP-4, 1 VIM-11 and OXA-17, 1 OXA-19, 1 OXA-11, 1 OXA-74 and PER-1, 1 VIM-4, 3 KPC-2, 1 VIM-1, 1 VIM-2, 1 OXA-17) | 1 | AmpD (8), AmpR (1) |
| 244 | 9 | 5 | 4 | 3 | 1 (OXA-2 [Y158S]) | AmpD (2) | |
| 277 | 6 | 1 | 5 | 4 | 4 (SPM-1) | AmpD (1) | |
| 298 | 6 | 4 | 2 | 3 | 0 | AmpD (2) | |
| 308 | 12 | 5 | 7 | 3 | 3 (1 GES-7, 2 VIM-2) | AmpD (2) | |
| 309 | 4 | 2 | 2 | 2 | 2 (1 GES-19, 1 VIM-2) | ||
| 313 | 5 | 3 | 2 | 0 | 0 | AmpD (2) | |
| 316 | 2 | 1 | 1 | 1 | 1 (GES-9) | ||
| 319 | 5 | 3 | 2 | 0 | 0 | AmpR (2) | |
| 348 | 8 | 5 | 3 | 2 | 1 (GES-1) | AmpD (2) | |
| 357 | 3 | 1 | 2 | 1 | 1 (VIM-5) | AmpD (1) | |
| 395 | 10 | 9 | 1 | 0 | 0 | AmpD (1) | |
| 446 | 2 | 1 | 1 | 0 | 0 | ||
| 500 | 3 | 2 | 1 | 0 | 0 | ||
| 560 | 3 | 1 | 2 | 0 | 0 | AmpD (2) | |
| 569 | 2 | 1 | 1 | 0 | 0 | 1 | |
| 606 | 2 | 1 | 1 | 0 | 0 | ||
| 1714 | 3 | 2 | 1 | 0 | 0 | ||
| Total | 181 | 82 | 99 | 73 | 46 | 2 | 39 |
Genomic analysis (see Table S1 in the supplemental material) was performed using CLC Genomic Workbench 7.0.4 (CLCBio). Unique sequence variants exclusive to the resistant population of each MLST group were flagged for further analysis as outlined in the summary column of Table S1. To account for resistance in the 99 ceftazidime-resistant isolates in a parsimonious manner, we followed a triage process—accounting first for resistance-inducing β-lactamases, second for mechanisms allowing for derepression of ampC, and third for other candidate causes of resistance.
Forty-six isolates had β-lactamases that have been reported to hydrolyze ceftazidime (Table 1). Analysis of the ampC regulon and additional cephalosporin targets was also completed for these isolates (see Table S1 in the supplemental material); however, resistance was attributed primarily to the presence of the β-lactamases, as clinically, detection of such an element would rule out treatment with ceftazidime. It was apparent that certain MLST lineages were enriched in β-lactamases, particularly sequence type 111 (ST111) and ST233, which were the predominant carriers of blaVIM-2. This is consistent with previous reports that these lineages represent global disseminators of the class B metallo-β-lactamases (9–12).
Comparative analysis of the ampC regulons from the remaining 53 ceftazidime-resistant isolates identified mutations in ampR, ampD, and dacB (Table 1; see Table S1 in the supplemental material). In the global transcriptional regulator ampR, which directly controls expression of the intrinsic β-lactamase ampC, unique amino acid changes were identified in five isolates of the resistant population. Of these isolates, three (AZPAE14890, AZPAE14909, and AZPAE15058) had a mutation that resulted in the D135N amino acid change. This mutation has previously been reported to affect the regulatory function of ampR, leading to derepression of ampC (13). Two of these isolates were obtained in France and were of different STs (ST175 and ST235) and 1 was from Spain (ST175), suggesting this mutation can be independently acquired by an isolate rather than being unique to a single lineage. Two other isolates (both ST319) had a G154R variation in AmpR. This change occurs within a region that has been purported to interact with the permease, AmpG (14). Reverse transcription-PCR (RT-PCR) analysis revealed a >20-fold increase in ampC expression in the presence of ceftazidime from these two isolates. As no other changes in the ampC regulon were apparent in these isolates, the increased expression level was attributed to the change in AmpR.
The most common sequence variations identified within the ampC regulon were located in the 1,6-anhydro-N-acetylmuramyl-[scapi]l-alanine amidase, AmpD, with 32 isolates having unique sequence changes. Mutations of ampD were easily identified in 14/32 isolates as they introduced early stop codons, frameshifts, or in-frame deletions known to result in inactivation of ampD. The remaining isolates had unique amino acid variations within AmpD. These differences were aligned with the sequence of AmpD from Citrobacter freundii, for which a structure (PDB accession no. 1J3G) has been determined (15), to deduce changes that may affect the activity of this enzyme (Fig. 1). Of the 18 P. aeruginosa isolates with unique variations, 6 were deduced to affect the activity of this enzyme in C. freundii (15). Another 6/18 isolates had changes that have been experimentally noted to affect the activity of AmpD in multiple Enterobacteriaceae (16). Additional unique sequence variants were identified among the remaining 6 isolates (Fig. 1), and RT-PCR was used to determine the impact of these variations on the level of ampC expression (Table 2). Briefly, isolates were grown to the mid-log phase in Mueller-Hinton broth II (MHB II) at 37°C with shaking (200 rpm). The culture was split into two aliquots, where one was exposed to 1/2 the respective MIC of ceftazidime for 15 min and the other was treated as an unexposed control. Samples were treated with RNAprotect cell reagent (Qiagen), and RNA was prepared using a Maxwell 16 LEV simplyRNA purification kit (Promega). A total of 5 ng RNA was used in an RT-PCR assay using a QuantiTect SYBR green RT-PCR kit (Qiagen) with a Bio-Rad CFX96 instrument. The level of expression of rpsL was used for normalization, and the relative quantification of ampC expression was performed using a comparative threshold cycle (CT) method. The oligonucleotides used to quantify transcript expression for ampC and rpsL were obtained from previous publications (17, 18).
FIG 1.
Changes identified within AmpD responsible for derepression of ampC. Alignment of the protein sequence of Citrobacter freundii and P. aeruginosa PAO1 for comparative purposes. Red boxes above the sequence of C. freundii denote residues identified from structural studies and mutational analysis as being important in the activity and functionality of AmpD (15, 31). Blue dots below indicate residues that had changes exclusive to the ceftazidime-resistant population and which previously have been shown to be important in increasing ampC expression in other Enterobacteriaceae (16). Yellow dots indicate newly identified changes that were exclusive to the ceftazidime-resistant P. aeruginosa population.
TABLE 2.
Summary of AmpD variants identified among the clinical population of P. aeruginosaa
| Variant type | Isolate | MLST | MIC of CAZ (μg/ml) | Allelic variationb | Δ−CT: |
|
|---|---|---|---|---|---|---|
| Relative to PAO1c | Induced/uninduced ratiod | |||||
| Control | PAO1 | 1 | 1.00 | 1.91 | ||
| With structurally important residue identified in C. freundii crystal structure | AZPAE14403 | 175 | 16 | P41S (39) | 0.56 | 3.69 |
| AZPAE14892 | 313 | 64 | P41S (39) | 3.61 | 7.52 | |
| AZPAE14860 | 308 | 32 | A96T (94) | 54.16 | 1.13 | |
| AZPAE15054 | 298 | 16 | A96T (94) | 49.38 | 6.06 | |
| AZPAE14886 | 111 | 16 | R164S (161) | 0.05 | 4.18 | |
| AZPAE14983 | 111 | 16 | R164S (161) | 0.18 | 5.94 | |
| With structurally important residue identified in Enterobacteriaceae | AZPAE14394 | 175 | 64 | R82C (80) | 3.96 | 70.67 |
| AZPAE15006 | 235 | 16 | G84D (82) | 3.50 | 6.82 | |
| AZPAE14842 | 235 | 16 | G84D (82) | 24.18 | 2.01 | |
| AZPAE14422 | 235 | 32 | G84D (82) | 5.24 | 2.63 | |
| AZPAE14979 | 235 | 32 | G84D (82) | 28.56 | 4.82 | |
| AZPAE14843 | 235 | 32 | G84D (82) | 13.99 | 6.35 | |
| With unique changes identified among isolates | AZPAE14722 | 175 | 32 | H77Y (75) | 2.33 | 16.00 |
| AZPAE14730 | 235 | 64 | F89S (87) | 3.96 | 12.45 | |
| AZPAE15015 | 235 | 32 | C92Y (90) | 6.60 | 2.00 | |
| AZPAE14987 | 298 | 32 | G121R (119) | 12.26 | 1.47 | |
| AZPAE15035 | 560 | 16 | T139A (137) | 1.26 | 2.29 | |
| AZPAE14710 | 235 | 32 | P162L (159) | 0.69 | 2.28 | |
Presented are mutations that have been shown to be important based upon the C. freundii structure, those identified to be important from the study of other Enterobacteriaceae, and those changes that were unique and identified among isolates in this study. RT-PCR values for ampC are provided for these isolates to confirm overexpression of the intrinsic β-lactamase.
The positions listed are numbered according to the sequence of the AmpD from P. aeruginosa PAO1. Positions provided in parentheses are for the corresponding position in AmpD of C. freundii.
Results are representative of 3 independent experiments. The Δ−CT ratio is calculated relative to the RT-PCR result for the housekeeping gene rpsL. The values listed represent the ratio of the Δ−CT of the isolate in MHB II relative to that of strain PAO1.
The values listed represent the ratio of the Δ−CT of the isolate in MHB II in the presence of 1/2 the MIC of ceftazidime for 15 min in the log phase compared to that in MHB II at the same point in time.
Isolates were examined for constitutive expression of ampC in MHB II and derepressed ampC expression in the presence of sub-MICs of ceftazidime (Table 2). Elevated constitutive expression (>4-fold) of ampC relative to the sensitive control strain PAO1 was evident in 8/18 isolates, and elevated induced expression was present in the remainder.
Additional analysis of the data set identified two isolates of the ST111 lineage (AZPAE14727 and AZPAE14728) with the same unique variation in the dacB gene. DacB is a nonessential low-molecular-weight PBP that is involved in maintaining PG composition and mediates β-lactam resistance through increased expression of AmpC and the two-component system, BrlAB, also known to mediate resistance (19, 20). RT-PCR of ampC expression from these isolates also indicated derepression of ampC (ampC/rpsL ratio of >10-fold; ampC expression relative to PAO1, 4.82-fold).
No unique changes to the ampR/ampC promoter region were identified in the resistant population, and examination of additional genes of the amp regulon did not identify variants unique to the ceftazidime-resistant population (see Table S1 in the supplemental material). It is interesting to note that the comparative analysis of alleles by MLST grouping showed almost identical sequence profiles, with the exception of genes encoding the lytic transglycosylases, which were quite diverse. Further studies on the structure of these enzymes and the effect of changes in the mature protein are needed to understand the genetic diversity and potential impact of these changes in the lytic transglycosylases.
Mutations within or near the active sites of the essential PBPs (PBP3, PBP1a, and PBP1b) may mediate decreased susceptibility to ceftazidime in P. aeruginosa (21). Unique changes to the PBP3 sequence were identified in two of the clinical isolates (AZPAE13850 and AZPAE12156). Both had the same PBP3 mutation resulting in the change of R504C. This residue is part of an important hinge region of the PBP (22) and may cause interference with ceftazidime binding. These isolates were of different STs from India and the United States, strongly indicating independent acquisition of this amino acid variation. Examination of PBP1a and PBP1b, which can both be inhibited by ceftazidime at high concentrations (4), did not identify any sequence variations unique to the resistant population.
Overexpression of efflux components has also been implicated in reduced susceptibility to ceftazidime (23, 24). Although 3 (AZPAE12150, AZPAE13876, and AZAPE14872) of the remaining 12 ceftazidime-resistant isolates did have mutations in efflux regulatory components (nalD and mexZ), examination of the whole population showed that they were not exclusive to the ceftazidime-resistant population. However, it is likely that these mutations contribute to the overall resistance or reduced susceptibility of the organisms.
Additional alleles associated with resistance, including genes identified in studies with transposon libraries (25), mutator-associated genes (26), and quorum sensing genes (26, 27), were also evaluated (see Table S2 in the supplemental material). Analysis of these alleles did not reveal any variants that were unique to the resistant isolates (data not shown).
The 12 isolates and all other strains belonging to the same STs were mapped to the reference strain P. aeruginosa PAO1 in an attempt to identify common polymorphisms unique to this population; however, no single target gene was identified from this analysis. In part, this may be due to the small number of isolates spread across a diverse genetic background. Additionally, this is not unexpected as resistance can occur singularly or in a multifaceted manner through direct target changes, expression-level changes of numerous factors, as well as changes to membrane permeability, to name but a few. Indeed, 6 of these 12 isolates had a MIC to ceftazidime (16 μg/ml) 1 doubling dilution higher than the nonsusceptible breakpoint. This level of elevation could easily be due to the combinatorial changes in several factors, as opposed to a single predominant factor.
It may also be prudent to consider the pathogenic/disease association of the isolates. Two of the isolates for which a mechanism of resistance was not clearly defined were highly resistant to ceftazidime (MIC, 128 μg/ml) and were collected from cystic fibrosis (CF) patients. Isolates associated with CF are often multidrug resistant due to phenotypic traits that change and develop with adaptation to the lung environment (28). For example, the overproduction of alginate may affect the susceptibility of an isolate as it provides another barrier to antibiotic entry (29). A mutation in mucA, an anti-sigma factor that controls alginate production (30), was identified in isolate AZPAE12416 and may be one of many contributory factors to resistance of this isolate.
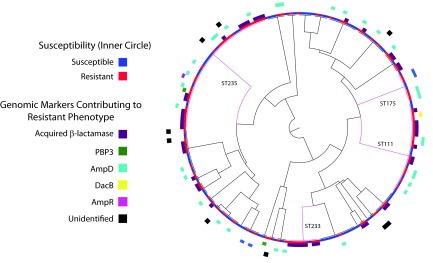
Using a comparative genomic approach with alleles previously associated with ceftazidime resistance in P. aeruginosa, we were able to identify the probable factor(s) mediating resistance in 88% of the 99 resistant isolates in our data set (Fig. 2). This type of analysis provides a real depiction of the probable mutations that are mediating resistance among a relevant population and is invaluable in aiding our understanding of resistance mechanisms and designing new antimicrobials that evade these pathways.
FIG 2.
Summary of resistance mechanisms among ceftazidime-resistant clinical isolates of P. aeruginosa by MLST. Global lineages are highlighted on the MLST tree in pink and labeled accordingly. The 12 isolates for which a genomic marker for resistance could not be determined are highlighted on the outer circle with black boxes.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03113-15.
REFERENCES
- 1.Mesaros N, Nordmann P, Plesiat P, Roussel-Delvallez M, Van Eldere J, Glupczynski Y, Van Laethem Y, Jacobs F, Lebecque P, Malfroot A, Tulkens PM, Van Bambeke F. 2007. Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin Microbiol Infect 13:560–578. doi: 10.1111/j.1469-0691.2007.01681.x. [DOI] [PubMed] [Google Scholar]
- 2.Zervosen A, Sauvage E, Frere JM, Charlier P, Luxen A. 2012. Development of new drugs for an old target: the penicillin binding proteins. Molecules 17:12478–12505. doi: 10.3390/molecules171112478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liao X, Hancock RE. 1997. Susceptibility to beta-lactam antibiotics of Pseudomonas aeruginosa overproducing penicillin-binding protein 3. Antimicrob Agents Chemother 41:1158–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayes MV, Orr DC. 1983. Mode of action of ceftazidime: affinity for the penicillin-binding proteins of Escherichia coli K12, Pseudomonas aeruginosa and Staphylococcus aureus. J Antimicrob Chemother 12:119–126. doi: 10.1093/jac/12.2.119. [DOI] [PubMed] [Google Scholar]
- 5.Fisher JF, Mobashery S. 2014. The sentinel role of peptidoglycan recycling in the beta-lactam resistance of the Gram-negative Enterobacteriaceae and Pseudomonas aeruginosa. Bioorg Chem 56:41–48. doi: 10.1016/j.bioorg.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kos VN, Deraspe M, McLaughlin RE, Whiteaker JD, Roy PH, Alm RA, Corbeil J, Gardner H. 2015. The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob Agents Chemother 59:427–436. doi: 10.1128/AAC.03954-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically—ninth edition. Approved standard M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing: twenty-third informational supplement M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 9.Turton JF, Wright L, Underwood A, Witney AA, Chan YT, Al-Shahib A, Arnold C, Doumith M, Patel B, Planche TD, Green J, Holliman R, Woodford N. 2015. High-resolution analysis by whole-genome sequencing of an international lineage (sequence type 111) of Pseudomonas aeruginosa associated with metallo-carbapenemases in the United Kingdom. J Clin Microbiol 53:2622–2631. doi: 10.1128/JCM.00505-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zafer MM, Al-Agamy MH, El-Mahallawy HA, Amin MA, El Din Ashour S. 2015. Dissemination of VIM-2 producing Pseudomonas aeruginosa ST233 at tertiary care hospitals in Egypt. BMC Infect Dis 15:122. doi: 10.1186/s12879-015-0861-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perez F, Hujer AM, Marshall SH, Ray AJ, Rather PN, Suwantarat N, Dumford D III, O'Shea P, Domitrovic TN, Salata RA, Chavda KD, Chen L, Kreiswirth BN, Vila AJ, Haussler S, Jacobs MR, Bonomo RA. 2014. Extensively drug-resistant Pseudomonas aeruginosa isolates containing blaVIM-2 and elements of Salmonella genomic island 2: a new genetic resistance determinant in northeast Ohio. Antimicrob Agents Chemother 58:5929–5935. doi: 10.1128/AAC.02372-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samuelsen O, Toleman MA, Sundsfjord A, Rydberg J, Leegaard TM, Walder M, Lia A, Ranheim TE, Rajendra Y, Hermansen NO, Walsh TR, Giske CG. 2010. Molecular epidemiology of metallo-beta-lactamase-producing Pseudomonas aeruginosa isolates from Norway and Sweden shows import of international clones and local clonal expansion. Antimicrob Agents Chemother 54:346–352. doi: 10.1128/AAC.00824-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caille O, Zincke D, Merighi M, Balasubramanian D, Kumari H, Kong KF, Silva-Herzog E, Narasimhan G, Schneper L, Lory S, Mathee K. 2014. Structural and functional characterization of Pseudomonas aeruginosa global regulator AmpR. J Bacteriol 196:3890–3902. doi: 10.1128/JB.01997-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong KF, Aguila A, Schneper L, Mathee K. 2010. Pseudomonas aeruginosa beta-lactamase induction requires two permeases, AmpG and AmpP BMC Microbiol 10:328. doi: 10.1186/1471-2180-10-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liepinsh E, Genereux C, Dehareng D, Joris B, Otting G. 2003. NMR structure of Citrobacter freundii AmpD, comparison with bacteriophage T7 lysozyme and homology with PGRP domains. J Mol Biol 327:833–842. doi: 10.1016/S0022-2836(03)00185-2. [DOI] [PubMed] [Google Scholar]
- 16.Petrosino JF, Pendleton AR, Weiner JH, Rosenberg SM. 2002. Chromosomal system for studying AmpC-mediated beta-lactam resistance mutation in Escherichia coli. Antimicrob Agents Chemother 46:1535–1539. doi: 10.1128/AAC.46.5.1535-1539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dumas JL, van Delden C, Perron K, Kohler T. 2006. Analysis of antibiotic resistance gene expression in Pseudomonas aeruginosa by quantitative real-time-PCR. FEMS Microbiol Lett 254:217–225. doi: 10.1111/j.1574-6968.2005.00008.x. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Martinez JM, Poirel L, Nordmann P. 2009. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:4783–4788. doi: 10.1128/AAC.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moya B, Dotsch A, Juan C, Blazquez J, Zamorano L, Haussler S, Oliver A. 2009. Beta-lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog 5:e1000353. doi: 10.1371/journal.ppat.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zamorano L, Moya B, Juan C, Oliver A. 2010. Differential beta-lactam resistance response driven by ampD or dacB (PBP4) inactivation in genetically diverse Pseudomonas aeruginosa strains. J Antimicrob Chemother 65:1540–1542. doi: 10.1093/jac/dkq142. [DOI] [PubMed] [Google Scholar]
- 21.Moya B, Beceiro A, Cabot G, Juan C, Zamorano L, Alberti S, Oliver A. 2012. Pan-beta-lactam resistance development in Pseudomonas aeruginosa clinical strains: molecular mechanisms, penicillin-binding protein profiles, and binding affinities. Antimicrob Agents Chemother 56:4771–4778. doi: 10.1128/AAC.00680-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sainsbury S, Bird L, Rao V, Shepherd SM, Stuart DI, Hunter WN, Owens RJ, Ren J. 2011. Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms. J Mol Biol 405:173–184. doi: 10.1016/j.jmb.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castanheira M, Mills JC, Farrell DJ, Jones RN. 2014. Mutation-driven beta-lactam resistance mechanisms among contemporary ceftazidime-nonsusceptible Pseudomonas aeruginosa isolates from U.S. hospitals. Antimicrob Agents Chemother 58:6844–6850. doi: 10.1128/AAC.03681-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laudy AE, Osinska P, Namyslowska A, Zajac O, Tyski S. 2015. Modification of the susceptibility of Gram-negative rods producing ESbetaLS to beta-lactams by the efflux phenomenon. PLoS One 10:e0119997. doi: 10.1371/journal.pone.0119997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarez-Ortega C, Wiegand I, Olivares J, Hancock RE, Martinez JL. 2011. The intrinsic resistome of Pseudomonas aeruginosa to beta-lactams. Virulence 2:144–146. doi: 10.4161/viru.2.2.15014. [DOI] [PubMed] [Google Scholar]
- 26.Karatuna O, Yagci A. 2010. Analysis of quorum sensing-dependent virulence factor production and its relationship with antimicrobial susceptibility in Pseudomonas aeruginosa respiratory isolates. Clin Microbiol Infect 16:1770–1775. doi: 10.1111/j.1469-0691.2010.03177.x. [DOI] [PubMed] [Google Scholar]
- 27.D'Argenio DA, Wu M, Hoffman LR, Kulasekara HD, Deziel E, Smith EE, Nguyen H, Ernst RK, Larson Freeman TJ, Spencer DH, Brittnacher M, Hayden HS, Selgrade S, Klausen M, Goodlett DR, Burns JL, Ramsey BW, Miller SI. 2007. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol 64:512–533. doi: 10.1111/j.1365-2958.2007.05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sousa AM, Pereira MO. 2014. Pseudomonas aeruginosa diversification during infection development in cystic fibrosis lungs—a review. Pathogens 3:680–703. doi: 10.3390/pathogens3030680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratchik EM, Viswanathan V. 1975. GLC determination of saccharin in pharmaceutical products. J Pharm Sci 64:133–135. doi: 10.1002/jps.2600640131. [DOI] [PubMed] [Google Scholar]
- 30.Pulcrano G, Iula DV, Raia V, Rossano F, Catania MR. 2012. Different mutations in mucA gene of Pseudomonas aeruginosa mucoid strains in cystic fibrosis patients and their effect on algU gene expression. New Microbiol 35:295–305. [PubMed] [Google Scholar]
- 31.Stapleton P, Shannon K, Phillips I. 1995. DNA sequence differences of ampD mutants of Citrobacter freundii. Antimicrob Agents Chemother 39:2494–2498. doi: 10.1128/AAC.39.11.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.