Abstract
Most patients failing antiretroviral treatment in Uganda continue to fail their treatment regimen even if a dominant drug-resistant HIV-1 genotype is not detected. In a recent retrospective study, we observed that approximately 30% of HIV-infected individuals in the Joint Clinical Research Centre (Kampala, Uganda) experienced virologic failure with a susceptible HIV-1 genotype based on standard Sanger sequencing. Selection of minority drug-resistant HIV-1 variants (not detectable by Sanger sequencing) under antiretroviral therapy pressure can lead to a shift in the viral quasispecies distribution, becoming dominant members of the virus population and eventually causing treatment failure. Here, we used a novel HIV-1 genotyping assay based on deep sequencing (DeepGen) to quantify low-level drug-resistant HIV-1 variants in 33 patients failing a first-line antiretroviral treatment regimen in the absence of drug-resistant mutations, as screened by standard population-based Sanger sequencing. Using this sensitive assay, we observed that 64% (21/33) of these individuals had low-frequency (or minority) drug-resistant variants in the intrapatient HIV-1 population, which correlated with treatment failure. Moreover, the presence of these minority HIV-1 variants was associated with higher intrapatient HIV-1 diversity, suggesting a dynamic selection or fading of drug-resistant HIV-1 variants from the viral quasispecies in the presence or absence of drug pressure, respectively. This study identified low-frequency HIV drug resistance mutations by deep sequencing in Ugandan patients failing antiretroviral treatment but lacking dominant drug resistance mutations as determined by Sanger sequencing methods. We showed that these low-abundance drug-resistant viruses could have significant consequences for clinical outcomes, especially if treatment is not modified based on a susceptible HIV-1 genotype by Sanger sequencing. Therefore, we propose to make clinical decisions using more sensitive methods to detect minority HIV-1 variants.
INTRODUCTION
To date, 28 antiretroviral drugs from six drug classes have been approved for treatment of individuals infected with human immunodeficiency virus type 1 (HIV-1). Combinations of specific antiretroviral drugs are the basis for an effective therapy that suppresses viral replication, leading to partial immune reconstitution and considerable reduction in morbidity and mortality (1, 2). Together with prevention and educational efforts, antiretroviral treatment (ART) has been responsible for a worldwide reduction in AIDS-related deaths, as well as a 4-fold reduction in mother-to-child HIV-1 transmissions compared to the pretreatment era in Africa (3). Unfortunately, use of antiretroviral drugs in high-income countries (HICs) has also led to the emergence of HIV-1 drug resistance in many treated individuals (1). With high prevalence of HIV-1 drug resistance in the late 1990s/early 2000s, at least 10% of new infections were established by drug-resistant HIV-1 strains in HICs (4–6). In low- to middle-income countries (LMICs), poor access to clinical care, intermittent supply of antiretroviral drugs, and costs of travel to reach care providers result in suboptimal ART adherence, so that treatment failures and emergence of HIV-1 drug resistance have doubled in the last 10 years (6). With increased treatment access and high frequencies of treatment failures, HIV-1 with primary drug resistance is already found in 2% to 10% of treatment-naive individuals in Uganda, an increase of less than 1% to as high as 6.5% over the past 10 years (7–11).
Similar to other sub-Saharan countries, Uganda still has a high prevalence of people living with HIV-1 (approximately 2.1 million) (3), with over 750,000 HIV-infected adults receiving combination antiretroviral therapy (cART) in 2014 (12). Ugandan patients have access to first-line cART, consisting of a combination of two nucleoside/nucleotide reverse transcriptase (RT) inhibitors (NRTIs) and one nonnucleoside reverse transcriptase inhibitor (NNRTI), commonly tenofovir (TDF) or zidovudine (AZT) plus lamivudine (3TC) or emtricitabine (FTC) and efavirenz (EFV) or nevirapine (NVP) (13). Individuals experiencing virologic failure may have limited access to second- and third-line cART regimens due to limited drug availability and high costs (14), highlighting the need to identify the reason(s) for treatment failure. Unfortunately, treatment monitoring using plasma HIV RNA (viral) load measurements is limited in resource-limited settings, and regimen switching is typically guided by CD4+ T-cell counts and various clinical criteria. Access to HIV-1 genotyping to detect drug resistance is increasing but is often reserved for clinical studies and is rarely used as the standard of care (11, 15).
HIV-1 genotypic (antiretroviral) testing based on population (Sanger) sequencing is currently the most common method to manage patients infected with HIV-1 (1, 16–18); however, Sanger sequencing can detect only HIV-1 variants present at frequencies above 15% to 20% of the viral quasispecies (19–23) and thus fails to quantify low levels of HIV-1 drug-resistant variants (18, 24). These variants are usually present as minority members of the virus population, which can be selected to dominate over drug-susceptible variants under drug pressure (25–27). For this reason, a series of ultrasensitive HIV-1-genotyping assays, based on deep sequencing (next-generation sequencing [NGS]), have been developed to detect drug-resistant HIV-1 variants at levels below 20% of the viral population in an infected individual (24, 28–31). Several studies have associated early detection of these minority HIV-1 drug-resistant variants with subsequent treatment failure (32–37); however, with the advent of single-pill once-a-day (QD) cART regimens, treatment failures in HICs are rare, and the relevance of minority members of the viral population to the ART outcome is still under debate (32, 38–41).
Aside from differences in socioeconomic status between LMICs and HICs, patients in most LMICs in sub-Saharan Africa are infected with subtype A, C, D, and CRF02_AG HIV-1 strains that differ up to 10% in the Pol amino acid sequence, i.e., the target of most antiretroviral drugs. Infection with different HIV-1 subtypes can impact the treatment outcome (9, 42), due in part to differences in the pattern and frequency of drug-resistant mutations and/or difference in virus fitness and evolutionary rates (43, 44). For example, the frequencies of treatment failure and drug resistance were significantly greater in patients infected with the more pathogenic subtype D than in patients infected with subtype A HIV-1 strains (9). Similarly, the TDF resistance K65R mutation and the NNRTI resistance V106M mutation are more common in patients infected with subtype C HIV-1 strains than in individuals infected with other subtypes (45, 46). Despite extensive studies on HIV-1 drug resistance in sub-Saharan Africa, few studies have screened for minority drug-resistant variants in treatment-naive or -experienced patients. However, minority NNRTI-resistant variants appeared to feature prominently in treatment failures for women in Africa who had received an NVP-based treatment regimen following single-dose NVP treatment to prevent mother-to-child transmission (47, 48). Interestingly, NVP selective pressure, rather than the natural occurrence of K103N and Y181C at low frequencies (>1%) in the intrapatient HIV-1 population, was related to failure of NNRTI-based treatment regimens (49).
In our 10-year retrospective study (9), genotypic HIV-1 drug resistance was observed in approximately 70% of treatment failures at the Joint Clinical Research Centre (JCRC) in Kampala, Uganda (based on >3,000 HIV-1-genotyping tests). Thus, as the standard of care, physicians are informed of infection with fully drug-susceptible HIV-1 for over 30% of treatment failures (9, 50). In settings like this, with limited availability of salvage therapies, cART regimens are rarely switched in patients infected with drug-susceptible HIV-1. In this study, we used a novel HIV-1-genotyping assay based on deep sequencing (DeepGen) (24) to detect and quantify low-level drug-resistant HIV-1 variants in patients failing a first-line treatment regimen in the absence of drug-resistant mutations, as screened by standard population-based Sanger sequencing in a WHO-certified regional drug resistance test site in Kampala, Uganda. We then determined the treatment outcomes of patients failing treatment without HIV-1 drug resistance (by Sanger sequencing) but with minority drug-resistant HIV-1 variants detected by DeepGen.
(This research was presented in part at the 8th International AIDS Conference on HIV Pathogenesis, Treatment, and Prevention, Vancouver, Canada, 19 to 22 July 2015.)
MATERIALS AND METHODS
Cohort analyses.
Anonymized patient databases containing clinical demographics and laboratory values were merged with HIV-1 drug resistance databases without patient identifiers by an offsite data manager according to the stipulations of the institutional review board (IRB; Joint Clinical Research Centre, Kampala, Uganda) approval (EM10-07). A total of 1,967 plasma HIV-1 RNA loads (viral load) and 3,468 CD4+ T-cell counts, prior to and after HIV-1 drug resistance testing, were analyzed to determine treatment outcomes in a cohort of 423 HIV-infected individuals.
Clinical samples.
RNA specimens from 65 (n = 65) plasma samples were retested using our deep-sequencing-based HIV-1-genotyping assay (DeepGen; see below). These samples are collected for routine drug resistance testing as the standard of care for HIV-infected individuals failing treatment at the JCRC (Kampala, Uganda). Clinical and virological data were obtained from the patient care database at the JCRC under IRB approval (EM10-07) for HIV-1 drug resistance testing. Four groups of patient samples were selected from a repository based on virologic failure during treatment with an RTI-based (2 NRTIs plus an NNRTI) regimen, i.e., defined as a viral load above 2,000 copies/ml and/or CD4+ T-cell counts below 250 cells/mm3. A Sanger sequencing-based HIV-1-genotyping test had been performed on all patient samples at the time of treatment failure (9). Group I (n = 27) consisted of patients who failed treatment in the presence of RTI resistance mutations detected by Sanger sequencing, group II (n = 21) comprised patients who failed treatment in the absence of RTI resistance mutations using Sanger sequencing, group III (n = 12) included patients who failed treatment in the absence of RTI resistance mutations based on Sanger and deep sequencing, and group IV (n = 5) consisted of patients who did not fail antiretroviral treatment (Table 1). It is important to note that samples in groups II and III were defined only following DeepGen analysis, where group III consisted of patients experiencing treatment failure in the absence of drug resistance mutations, detected by Sanger or deep sequencing, associated with their respective RTI-based therapy.
TABLE 1.
Clinical and virological parameters
| Group no. (description)a | Patient identifier | Ageb | Sexc | HIV-1RNAd | HIV-1 subtypee | Treatment historyf |
|---|---|---|---|---|---|---|
| I (treatment failure in the presence of drug resistance mutations detected by Sanger sequencing [n = 27]) | 0113-13 | 35 | M | ND | A | 3TC, EFV, AZT |
| 135-13 | 15 | F | ND | A | TDF, 3TC, LPV, AZT, EFV | |
| 137-13 | 56 | M | 5.08 | D | TDF, 3TC, LPV, FTC, AZT, FTC, d4T | |
| dr-83-13 | Infant | Unk | 3.37 | A | TDF, 3TC, LPV | |
| dr-92-13 | Adult | Unk | ND | D | Unk | |
| 96-13 | 9 | F | 4.88 | D | AZT, 3TC, LPV, NVP, d4T, | |
| 223-13 | 20 | F | 4.71 | D | TDF, 3TC, LPV, ABC, EFV, AZT | |
| 176-13 | 35 | M | 6.09 | A | Unk | |
| 200-13 | 40 | M | 5.52 | D | TDF, 3TC, EFV, AZT, d4T, NVP | |
| EN-7-13 | Adult | Unk | ND | D | Unk | |
| 42-13 | 32 | F | 4.13 | D | AZT, 3TC, NVP | |
| 143-12 | 18 | F | 3.03 | D | AZT, 3TC, EFV | |
| 186-12 | 75 | F | ND | A | Unk | |
| 65-12 | Adult | M | 6.15 | A | AZT, ETR, ATV, RAL, LPV, TDF, 3TC, NVP | |
| 70-12 | 32 | F | 4.23 | A | TDF, 3TC, LPV, RAL, DRV, ETV, FTC, d4T, NVP | |
| 106-12 | 33 | F | 3.69 | A | ABC, NVP, LPV, ddI, NVP, AZT, 3TC, TDF | |
| 109-12 | Adult | M | 4.60 | A | FTC, TDF, LPV, AZT, 3TC | |
| 123-12 | 42 | F | 3.10 | D | ATV, AZT, ABC, AZT, NVP, 3TC, TDF | |
| 124-12 | 52 | F | ND | A | Unk | |
| 134-12 | 43 | M | ND | D | Unk | |
| 193-12 | 38 | F | 5.98 | D | TDF, 3TC, LPV, d4T, 3TC, NVP | |
| 207-12 | Unk | F | 6.10 | A | TDF, 3TC, EFV | |
| DR-31-11 | 50 | M | ND | A | TDF, 3TC, LPV, FTC | |
| DR-18-11 | 16 | M | 3.75 | A | AZT, 3TC, NVP | |
| DR-286-08 | Adult | F | ND | A | FTC, TDF, LPV, AZT, 3TC, NVP | |
| DR-371-08 | Adult | F | ND | A | ABC, ddI, LPV, 3TC, ABC, ddI, d4T, TDF, AZT | |
| DR-30-11 | Adult | M | ND | A | AZT, 3TC, TDF | |
| II (treatment failure in the absence of drug resistance mutations detected by Sanger sequencing [n = 21]) | 109-13 | Adult | M | 6.16 | D | EFV, FTC, TDF, LPV, AZT, 3TC, NVP |
| 89-13 | Adult | Unk | 4.21 | A | Unk | |
| 106-13 | 46 | F | 5.52 | D | AZT, 3TC, EFV | |
| 156-13 | 59 | Unk | ND | D | Unk | |
| 214-13 | 54 | Unk | 3.44 | D | ABC, 3TC, LPV | |
| 218-13 | 30 | M | 5.99 | D | TDF, 3TC, LPV, DRV, EFV, NVP | |
| 81-13 | 18 | M | 3.83 | A | TDF, 3TC, LPV | |
| 177-13 | 30 | M | 5.21 | D | TDF, 3TC, LPV, AZT, FTC, | |
| EN-10-13 | Adult | Unk | 4.80 | A/D | LPV, RAL, TDF, 3TC, NVP, AZT | |
| 72-12 | 35 | M | 5.29 | A | Unk | |
| dr-82-12 | Unk | Unk | 4.10 | A | TDF, 3TC, LPV | |
| 110-12 | 40 | F | 4.98 | D | TDF, 3TC, LPV | |
| 111-12 | 44 | M | 1.76 | A | TDF, 3TC, LPV, AZT, EFV, FTC | |
| 135-12 | 22 | F | 5.45 | A | AZT, 3TC, TDF, LPV, d4T, NVP | |
| 170-12 | 45 | M | 5.19 | D | Unk | |
| 172-12 | Adult | M | 5.23 | A | ABC, ddI, LPV, NVP, AZT, 3TC | |
| dr-0036-08 | Unk | Unk | ND | D | Unk | |
| DR-0115-11 | 14 | F | ND | A | 3TC, EFV, TDF, AZT | |
| DR-193-11 | Adult | M | 5.55 | A | AZT, 3TC, EFV | |
| DR-269-08 | Adult | M | ND | D | TDF, 3TC, LPV, AZT, NVP | |
| DR-50-11 | 16 | M | ND | C | AZT, 3TC, NVP | |
| III (treatment failure in the absence of drug resistance mutations detected by Sanger or deep sequencing [n = 12]) | 216-13 | 42 | F | ND | A | AZT, LPV, 3TC, EFV, TDF |
| 38-13 | 21 | Unk | 4.44 | A | TDF, 3TC, LPV, d4T, NVP, | |
| 66-13 | 21 | F | 4.63 | A | RAL, EFV, DRV, TDF, 3TC, LPV, NVP | |
| 130-12 | 50 | F | 5.01 | A | 3TC, TDF, ATV, IDV | |
| 30-12 | Adult | Unk | ND | D | Unk | |
| 22-12 | Adult | Unk | ND | D | 3TC, AZT, NVP, TDF, FTC | |
| 14-12 | Unk | F | ND | A | Unk | |
| 64-12 | 9 | F | 4.36 | A | AZT, 3TC, NVP, ABC, ddI, LPV, | |
| 205-12 | 16 | M | 4.53 | A | AZT, 3TC, ATV, LPV, ddI | |
| dr-0043-11 | 11 | M | ND | A | AZT, 3TC, NVP, ddI, DLV | |
| DR-212-11 | 34 | F | 5.12 | D | TDF, 3TC, NVP, LPV, d4T | |
| DR-001-09 | 42 | M | ND | D | TDF, 3TC, LPV | |
| IV (treatment success [n = 5]) | 62-12 | 33 | F | <20 | A | 3TC, AZT, NVP |
| 27-12 | 42 | M | <20 | A | AZT, 3TC, NVP | |
| 23-12 | Adult | Unk | <20 | D | AZT, 3TC, EFV | |
| 197-12 | 47 | M | <20 | A | d4T, 3TC, EFV | |
| 215-12 | 35 | F | <20 | D | AZT, 3TC, NVP |
Group of patients clustered by their response to treatment with antiretroviral drugs in the presence or absence of drug resistance mutations detected using HIV-1 genotyping assays based on Sanger or deep sequencing (DeepGen [24]).
Age in years when known; in some cases, patients were classified as infant or adult.
M, male; F, female; Unk, unknown.
Plasma viral load (log10 copies per milliliter); ND, not determined.
HIV-1 subtype determined using HIV-1 reverse transcriptase (Sanger) sequencing, as described previously (9).
Antiretroviral treatment history: AZT, zidovudine; ddI, didanosine; d4T, stavudine; 3TC, lamivudine; ABC, abacavir; TDF, tenofovir; FTC, emtricitabine; NVP, nevirapine; DLV, delavirdine; EFV, efavirenz; ETR, etravirine; IDV, indinavir; LPV, lopinavir; ATV, atazanavir; DRV, darunavir; RAL, raltegravir; Unk, unknown.
HIV-1 genotyping based on Sanger sequencing.
RT-PCR products, corresponding to the HIV-1 protease (PR)- and reverse-transcriptase-coding regions of HIV-1, were sequenced in the Case Western Reserve University/Center for AIDS Research Uganda Laboratory at the JCRC as part of regular HIV-1 drug resistance testing (9). HIV-1 sequences were interpreted and drug resistance profiles were generated based on the HIVdb Program Genotypic Resistance Interpretation Algorithm from the Stanford University HIV Drug Resistance Database (http://hivdb.stanford.edu). Phylogenetic analysis was used to predict the HIV-1 subtype as described previously (9).
HIV-1 genotyping based on deep sequencing of the gag-p2/NCp7/p1/p6/pol-PR/RT/IN-coding region.
Two overlapping RT-PCR products corresponding to the gag-p2/NCp7/p1/p6/pol-PR/RT/IN (1,657-nucleotide [nt] and 2,002-nt fragments)-coding region of HIV-1 were sequenced using DeepGen (24). Briefly, the two amplicons were purified (Agencourt AMPure XP; Beckman Coulter) and quantified (2100 Bioanalyzer DNA 7500; Agilent Technologies) prior to using the Ion Xpress Fragment Library kit (Life Technologies, Carlsbad, CA) to construct a multiplexed library for shotgun sequencing on the Ion Personal Genome Machine (PGM) (Life Technologies). For that purpose, a mixture of the purified DNA amplicons was randomly fragmented, and blunt ends were repaired using the Ion Shear Plus reagent (Life Technologies), followed by DNA purification (Agencourt AMPure XP; Beckman Coulter). The P1 adapter and one of 65 barcodes were ligated to the repaired fragment ends prior to DNA purification (Agencourt AMPure XP; Beckman Coulter). The DNA fragments were then selected by size (i.e., 280 to 320 bp; Pippin Prep; Life Technologies), and each barcoded library was purified (Agencourt AMPure XP; Beckman Coulter) and normalized using the Ion Library Equalizer kit (Life Technologies). Barcoded DNA libraries, corresponding to 65 patient-derived amplicons plus the HIV-1NL4-3 control, were pooled in equimolar concentrations, and templates were prepared and enriched for sequencing on the Ion sphere particles (ISPs) using the Ion OneTouch 200 template kit v2 (Life Technologies) in the Ion OneTouch 2 system (Life Technologies). The templated ISPs were quantified (Qubit 2.0; Life Technologies) and loaded into two Ion 318 chips (Life Technologies) to be sequenced on the Ion PGM using the Ion PGM Sequencing 200 kit v2 (Life Technologies). Signal processing and base calling were performed with Torrent Analysis Suite version 3.4.2.
Read mapping, variant calling, and phylogenetic analysis.
Reads were mapped and aligned against sample-specific reference sequences constructed for the gag-p2/NCp7/p1/p6/pol-PR/RT/IN genomic region using the DeepGen Software Tool Suite as described previously (24). The frequency of each amino acid present in each HIV-1 genomic position was calculated and summarized in a graphical interface. A list of the amino acids at these positions and their frequencies was exported as a tabulated text file and used with the HIVdb program genotypic resistance interpretation algorithm from the Stanford University HIV Drug Resistance Database (http://hivdb.stanford.edu) to infer the levels of susceptibility to protease, reverse transcriptase, and integrase (INT) inhibitors. In addition, for each data set, reads spanning amino acid positions (i) 50 to 85 in the protease (HIV-1 strain HXB2 2400 to 2508), (ii) 180 to 215 in the RT (HXB2 3087 to 3195), and (iii) 130 to 165 in the integrase (HXB2 4617 to 4725) were extracted, truncated, and translated for phylogenetic analysis. Within each data set, only one representative of any identical variants was maintained, but the overall frequency was stored. All variants with a frequency of ≥10 within the population were aligned using ClustalW (51), and the phylogeny was reconstructed using the neighbor-joining statistical method as implemented within MEGA 6.06 (52). In this study, minority variants were defined as amino acid substitutions detected in ≥1% (based on the intrinsic error rate of the system, as described previously [24]) and <20% of the virus population, corresponding to the mutations that could not be determined using population sequencing (19–23).
Statistical analyses.
Descriptive results are expressed as median values, standard deviations, and confidence intervals. The nonparametric Kruskal-Wallis one-way analysis of variance (ANOVA) test was used to compare the mutations detected in the four different groups. A paired t test was used to compare longitudinal measurements of plasma viral loads and CD4+ T-cell counts. All differences with a P value of <0.05 were considered statistically significant. All statistical analyses were performed using GraphPad Prism v.6.0b (GraphPad Software, La Jolla, CA) unless otherwise specified.
Nucleotide sequence accession numbers.
gag-p2/NCp7/p1/p6/pol-PR/RT/IN nucleotide sequences obtained by deep sequencing in this study have been submitted to the Los Alamos National Laboratory HIV-DB Next Generation Sequence Archive (http://www.hiv.lanl.gov/content/sequence/HIV/NextGenArchive/Kyeyune2016.html).
RESULTS
Antiretroviral treatment outcomes following Sanger sequencing-based HIV-1 drug resistance testing in Uganda.
We have previously reported on 937 of more than 3,000 drug resistance tests performed on HIV-infected individuals experiencing cART failures at the JCRC (Kampala, Uganda) from 1999 to 2009 (9). Upon treatment failure, these tests were offered to approximately 20,000 HIV patients receiving treatment at the JCRC through the PEPFAR and WHO/UNAIDS treatment programs. In contrast to those observed in HICs, HIV-1 drug resistance genotypes were observed in roughly 70% of Ugandans failing cART (>85% treatment success in HICs). Thus, drug-susceptible HIV-1 genotypes were reported to physicians for approximately 30% of patients failing cART with viral loads of >2,000 copies/ml and/or CD4+ counts of <250 cells/mm3 on two consecutive visits. As described above, due to various socioeconomic pressures, clinic visits four times per year are common in Uganda, with laboratory tests often performed semiannually; however, patients still miss some of these visits. Between these widely spaced visits, a small fraction of patients usually stop cART (<2% of patients/year), leading to rebounds in viral loads and loss of CD4+ T-cell counts. Thus, patients with “typical” treatment failures, which occur at a rate of approximately 10% per year (9, 53, 54), are mixed with patients with defined cessations of cART. Both groups would have the laboratory parameters to trigger a request for HIV-1 drug resistance testing. As described below, both types of treatment failures may have dominant or minority drug-resistant variants in their HIV-1 populations, but only the dominant drug resistance would be reported and influence future treatment decisions.
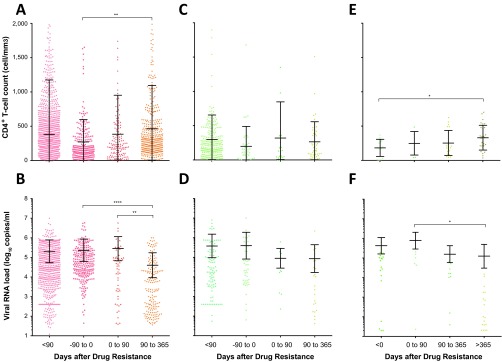
In Fig. 1, we show the treatment outcomes for patients with or without dominant drug-resistant HIV-1 genotypes. The vast majority of patients with drug resistance detected by Sanger sequencing switched treatment, whereas identification of a drug-susceptible virus led to at least a 3-month continuation of the ART regimen. Based on the criteria for drug resistance testing, CD4+ T-cell counts were at a mean of 259 ± 318 cells/mm3 of blood, with viral loads of 5.43 ± 5.92 RNA log10 copies/ml in plasma prior to drug resistance testing (data not shown). In patients with HIV-1 drug resistance, prior CD4+ T-cell counts (269 cells/mm3) and viral loads (5.38 log10 copies/ml) within the 0- to −90-day period analyses were not significantly different from those from all patients receiving drug resistance tests (resistant or susceptible). However, a significant gain of a mean of 83 CD4+ T cells/mm3 (P < 0.01; ANOVA and unpaired t tests) and a 5.5-fold drop in the mean viral load (P < 0.0001) were observed within a 3-month to 1-year window following detection of drug resistance and a switch in therapy (Fig. 1A and B).
FIG 1.
CD4 cell counts and viral loads prior to and following antiretroviral drug resistance tests in Kampala, Uganda. (A to D) CD4+ T-cell counts (CD4) (A and C) and viral loads (VL) (B and D) were analyzed for <90 days to 1 year, up to 90 days preceding, 0 to 90 days following, or 90 days to 1 year following the identification of a drug-resistant genotype (A and B) or having a drug-susceptible genotype (C and D). A total of 356 and 67 patients with 3,017 CD4/1,727 VL and 451 CD4/239 VL tests were analyzed for panels A and B and for panels C and D, respectively. (E and F) A subset of 33 patients (groups II and II) lacking HIV-1 drug resistance by Sanger sequencing were analyzed for CD4+ T-cell counts (n = 69) (E) and viral loads (n = 66) (F). *, P < 0.05; **, P < 0.01; ****, P < 0.0001; ANOVA and multiple-comparison tests. Mean values and standard deviations are indicated.
CD4+ T-cell and viral-load levels fluctuated but were not significantly different for the first year (Fig. 1C and D) following the report of a susceptible virus despite earlier evidence of treatment failure (<250 CD4+ T cells/mm3 and viral loads of >1,000 copies/ml at the time of testing). Interestingly, over the first year following an HIV-1 drug resistance genotype test, patients who maintained their current ART regimen with a susceptible virus had poor prognostic outcomes (with increasing viral loads and decreasing CD4+ T-cell counts), whereas patients with drug-resistant viruses who switched treatment regimens fared much better over the first year (Fig. 1). Most patients had 2 or 3 visits in the first year following drug resistance testing, but like every patient clinic (especially in Uganda and other African countries), patient visits did not follow a set monthly calendar, which is the reason for the >90 to 365 days required to determine treatment outcomes following drug resistance testing.
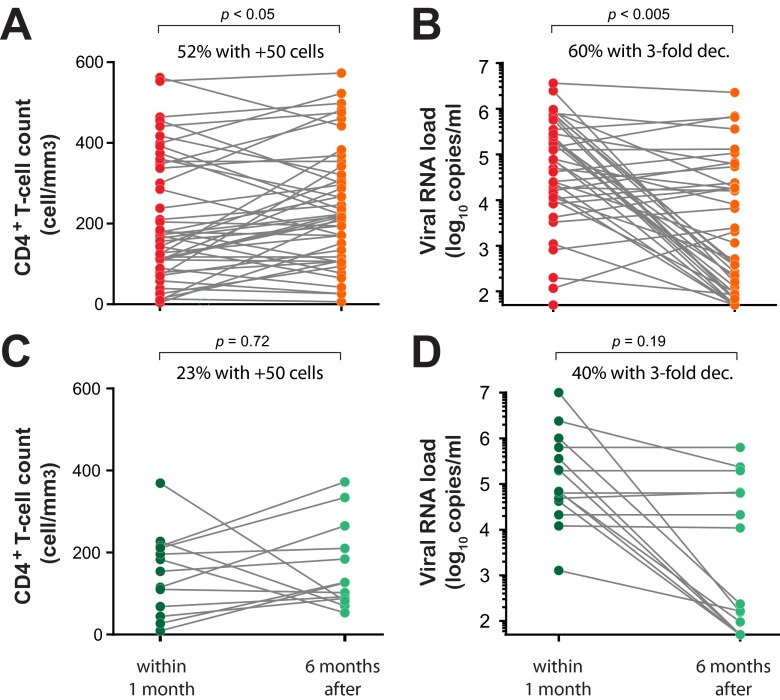
In paired analyses of plasma viral loads and CD4+ T-cell counts within 1 month before versus 6 months after HIV-1 drug resistance testing, a significant viral-load decrease and CD4+ T-cell count increase was observed only in the patients with a drug resistance genotype that led to a switch in treatment regimens (P < 0.005 and < 0.05, respectively; paired one-tailed t test). A 3-fold difference in plasma viral loads reflects the typical sensitivity and error in the Roche Amplicor 1.5 assay in our JCRC laboratory based on over 20,000 plasma viral-load determinations. The increase of >50 CD4 cells/ml was also determined to be significant by numerous clinical studies. Approximately 60% of the patients with drug-resistant genotypes had a 3-fold or greater drop in the viral load (Fig. 2B) compared to only 40% of the patients with this viral load drop (Fig. 2D) following a report of HIV-1 that was susceptible to the current cART. A >50-cell/mm3 increase in CD4+ T-cell counts (considered statistically significant in Fig. 1) was observed in 52% of the patients with a drug-resistant genotype (Fig. 2A) and only 23% of the patients with a drug-susceptible genotype (Fig. 2C). Based on viral-load decreases and CD4+ T-cell count decreases, a “better” response to cART in patients with a drug-resistant genotype versus a susceptible genotype was not significant based on Fisher's exact test. Based on these differences, we would require at least 200 patients with paired samples 1 month before and 6 months after drug resistance testing, which is difficult to control in an observational analysis involving over 12,000 treated patients with a treatment failure rate of approximately 5 to 10% per year and with highly variable patient visit schedules. These issues were the basis of the larger cohort analyses shown in Fig. 1, with reduced stringency on the timing of viral-load and CD4+ T-cell count testing. Based on our analysis of these clinical outcomes, we propose that low-frequency drug-resistant HIV-1 variants are present in the intrapatient viral quasispecies population that are not detected by Sanger sequencing methods and that they may impact antiretroviral treatment outcomes.
FIG 2.
Paired analyses of viral-load and CD4+ T-cell count changes before and after drug resistance testing. We performed paired analyses with CD4+ T-cell counts (A and C) and viral-RNA loads (B and D) and with samples taken prior to but closest to the drug resistance test date (within 1 month) and with the paired samples from the patients closest to 6 months after drug resistance testing. (A and B) A total of 46 patients were analyzed for CD4+ T-cell count pairs before and after detection of drug resistance by Sanger sequencing (A) and 42 patients for plasma viral-RNA loads before and after detection of drug resistance by Sanger sequencing (B). (C and D) Paired samples 1 month before and 6 months after drug resistance testing were also analyzed for patients with no detectable HIV drug resistance by Sanger sequencing, i.e., 13 patients/paired samples for CD4+ T-cell counts (C) and 15 patients/paired samples for plasma viral loads (D). The P values were calculated using paired one-tailed t tests.
Antiretroviral drug susceptibility determined using a standard HIV-1-genotyping assay based on Sanger sequencing.
Plasma samples were obtained from 60 HIV-infected individuals experiencing virologic failure and 5 patients responding to treatment while on a first-line RTI-based treatment regimen (2 NRTIs plus an NNRTI). The HIV-1 drug resistance genotypes of the viral RNA from these plasma samples were previously determined using standard (Sanger) population sequencing (9) (Table 1). Our Sanger sequencing-based drug resistance genotyping assay was performed in a facility and with an assay certified by the World Health Organization (WHO). Based on WHO (55) and European (56) guidelines, any nucleotide substitution (from the wild-type reference HIV-1 sequence) at a drug resistance-associated position that is identified on the chromatogram at a 15% to 20% or higher frequency over the background is reported as a drug resistance mutation that confers reduced susceptibility to a specific drug. However, this sequencing/genotyping cutoff was determined for a HIV-1 subtype B-infected population (19–23). Our experience in Sanger sequencing thousands of highly diverse non-B HIV-1 sequences leads us to believe that the threshold for detection of mixed nucleotides at drug resistance sites in the pol gene is approximately 30% in non-B HIV-1 strains.
Twenty-seven patients (group I) failed cART with viruses harboring multiple mutations in resistance to NRTIs (mean, 1.1 [range, 0 to 4] NRTI resistance mutations) and/or NNRTIs (mean, 1.6; [range, 0 to 6]). M184V and K103N were the most common NRTI and NNRTI resistance mutations, respectively. Based on Sanger sequencing, 33 subjects failed treatment in the absence of any primary mutations conferring resistance to reverse transcriptase inhibitors (groups II and III). It is important to note that groups II and III were defined following deep-sequencing analyses. For this study, we also obtained 20 samples from JCRC patients who responded to cART and who had undetectable plasma viral loads (<50 copies/ml), but we were successful in amplifying the p2-INT regions of only 5 patient samples, which were used as control group IV. A complete list of all amino acid substitutions in the RT-coding region, determined directly from plasma-derived amplicons using Sanger sequencing, is included in Table S1 in the supplemental material. No significant difference was observed in the distributions of age (9 to 75 years old), sex, plasma HIV-1 RNA load (<20 to 6.15 log10 copies/ml), HIV-1 subtype (37 subtype A, 27 D, and 1 C), or treatment history among patients in all four groups (Table 1).
Antiretroviral drug susceptibility determined using an HIV-1-genotyping assay based on deep sequencing.
Using our deep-sequencing method (DeepGen), all 65 patient samples were genotyped to obtain an average sequencing coverage of 10,795 reads at each nucleotide position, which varied with each sample and HIV-1 genomic region analyzed, i.e., protease (mean, 11,678; range, 3,603 to 15,314), reverse transcriptase (mean, 10,929; range, 1,108 to 20,792), and integrase (mean, 9,716; range, 3,589 to 13,847). Based on this sequencing depth, we detected 288 mutations at a frequency of 1% or greater associated with reduced susceptibility to antiretroviral drugs, i.e., 146 primary and 142 secondary or compensatory drug resistance mutations as defined by the Stanford University HIV Drug Resistance Database (http://hivdb.stanford.edu). As described below, most of these drug resistance mutations were detected by DeepGen in patients failing cART. In contrast to the results from Sanger sequencing, more than half of these drug resistance mutations (156/288) were detected at frequencies below our 20% limit of detection for the Sanger-based HIV-1-genotyping assay.
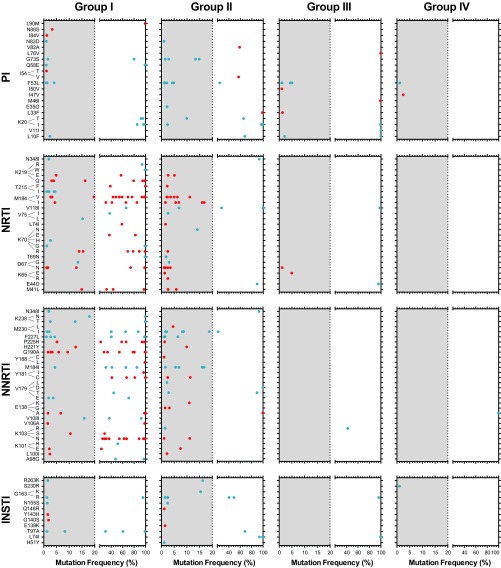
As described above, we split our 60 patients failing cART into 27 individuals who were failing treatment with drug resistance (group I) and 33 who were failing in the absence of drug resistance as determined by Sanger sequencing (groups II and III). A total of 75 mutations in positions associated with resistance to NRTIs or NNRTIs were detected by Sanger sequencing in the 27 patients from group I (see Table S1 in the supplemental material). HIV-1 variants from individuals failing treatment showed a high number of primary NRTI (45/60 [75%]) and NNRTI (45/76 [59.2%]) resistance mutations (group I), most of them as majority members of the intrapatient HIV-1 population, i.e., 87 of the 136 drug resistance mutations were detected at frequencies above 20%. M41L, D67N, K70R, M184V/I, and K219Q were among the most common primary NRTI resistance mutations, while K103N, Y181C, G190A, and P225H were the most common amino acid substitutions associated with resistance to NNRTIs in group I (Fig. 3). As expected, all the mutations identified by Sanger sequencing in group I were also detected by deep sequencing (DeepGen); however, 49 additional drug resistance mutations were identified only using DeepGen, and most at a low frequency (Fig. 3). The number of drug resistance mutations in the PR-, RT-, and INT-coding regions not only reflected the antiretroviral treatment history of each patient, but also correlated with the RTI-based therapy regimen at the time the plasma samples were collected. For example, few mutations associated with resistance to protease inhibitors (PIs) or integrase strand transfer inhibitors (INSTIs) were identified in the four groups, with most corresponding to compensatory mutations (40/52 [76.9%] and 23/27 [85.2%] in PR and IN, respectively), and many of these were detected as minority mutations, i.e., 59.6% (31/52) in PR and 59.3% (16/27) in IN (Fig. 3). A V82A mutation found at a 59% frequency and I54V at a 54% frequency in the HIV-1 populations of two different patients in group I may reflect initial transmission with PI-resistant virus or, more likely, treatment with a PI that was not recorded or was provided by another clinic. The L33F mutation was found at 99% in the HIV-1 population of a patient from group II infected with a subtype C virus; however, this mutation confers PI resistance only when combined with other primary mutations (57). Although some primary PI resistance mutations were detected at high frequency within the population, all primary INSTI resistance mutations were detected as minority members of the quasispecies, e.g., Y143H (1.6%) or Q148R (1.1%), and likely reflect natural polymorphisms at these IN sites.
FIG 3.
Numbers and frequencies of HIV-1 drug resistance mutations in all 65 patients quantified using DeepGen. Primary and secondary/compensatory drug resistance mutations, defined by the Stanford University HIV Drug Resistance Database (http://hivdb.stanford.edu), are indicated by red and blue dots, respectively. Amino acid substitutions (mutations) associated with resistance to PIs, NRTIs, NNRTIs, and INSTIs identified in any of the 65 patients are shown. The gray shading depicts the range of minority HIV-1 mutations (i.e., ≥1% to <20%) identified by DeepGen but usually not detected by standard Sanger sequencing.
Groups II and III comprised 33 patients experiencing virologic failure with a susceptible HIV-1 genotype based on Sanger sequencing who were genotyped using DeepGen. Interestingly, in group II, we identified minority HIV-1 variants resistant to NRTIs (29/33 [87.9%]) and/or NNRTIs (29/34 [85.3%]), with many primary mutations associated with resistance to NRTIs (24/29) or NNRTIs (13/29). For example, M41L was identified in two patients (at frequencies of 2.6% and 5.8% of the intrapatient HIV-1 population), D67N in four individuals (ranging from 1.1% to 2.5%), K103N in two patients (1.1% and 11.2%), Y181C in two patients (2.5% and 11.3%), M184I in five individuals (ranging from 1.6% to 16.8%), and M184V in six patients (ranging from 2.1% to 11.1%) (Fig. 3). Conversely, viruses from patients in groups III and IV had very limited numbers of RTI resistance mutations, even using the most sensitive DeepGen test, i.e., only five (two of them minority HIV-1 variants) and one, respectively (Fig. 3). Moreover, the few minority (primary or compensatory) mutations (<20% frequency) detected in group III were not linked to reduced susceptibility to the antiretroviral drugs included in the RTI-based treatment of any of the individuals in the group. Therefore, group II was defined as patients failing cART with a susceptible HIV-1 genotype as determined by Sanger sequencing but having low-frequency drug resistance mutations in the intrapatient HIV-1 population as determined by DeepGen. It is important to note the 20 HIV-1 RNA copies was likely the maximum number of RNA templates derived from patients in group IV, i.e., cART-treated patients with undetectable viral loads. Thus, only mutations detected above a 5% frequency in group IV samples should be considered. The IRB approvals did not permit collection of large blood volumes (>200 ml) to increase the sensitivity of the assay to as low as 1% drug resistance mutations in this group of patients. A complete list of all amino acid substitutions in the protease-, RT-, and integrase-coding regions, determined directly from plasma-derived amplicons using deep sequencing, is included in Table S2 in the supplemental material.
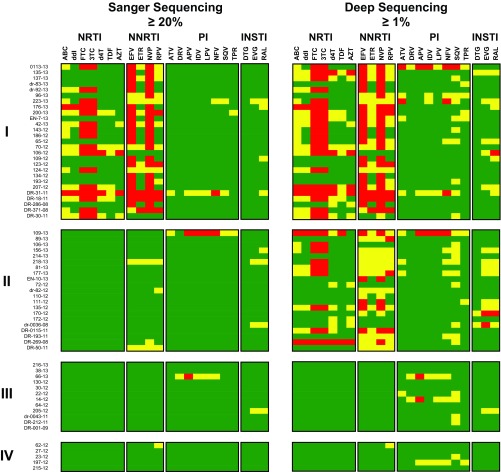
As described above, the drug resistance mutational patterns in the entire PR and RT sequences were analyzed with the HIVdb Program Genotypic Resistance Interpretation Algorithm to compare the results obtained by deep sequencing with standard (Sanger) HIV-1-genotyping data. When assuming equal impacts of >1% (by DeepGen) and >20% (by Sanger) mutation frequencies on predicted drug resistance, all 27 patients in group I showed intermediate to high-level resistance to several NRTIs or NNRTIs, which correlated with the drugs employed in the current RTI-based treatment regimen (Fig. 4). For example, many viruses were highly resistant to 3TC and FTC (16/27) and/or to NNRTIs, such as NVP (24/27), EFV (21/27), rilpivirine (RPV) (5/7), or etravirine (ETR) (1/27). On the other hand, the Sanger HIV-1 genotype for samples in groups II, III, and IV was essentially susceptible to all antiretroviral drugs, with a few minor exceptions of intermediate resistance associated with the presence of secondary or compensatory drug resistance mutations (Fig. 4). Interestingly, some of these drug resistance profiles changed when the minority HIV-1 variants detected by DeepGen were taken into account, e.g., more viruses in group I were now resistant to 3TC and FTC (20/27), as well as to additional NRTIs or NNRTIs. More importantly, although the resistance pattern was not considerably modified by using deep-sequencing-based HIV-1 genotyping in patients from groups III and IV, all 21 viruses from patients failing treatment in the absence of RTI resistance mutations using Sanger sequencing (group II) now showed an intermediate to high-level reduction in susceptibility to NRTIs (12/21) or NNRTIs (17/21) when minority HIV-1 variants were used in the genotypic resistance interpretation (Fig. 4). In summary, in this study, 55% (33/60) of the Ugandan patients failing treatment at the JCRC in Kampala, Uganda, were infected with drug-susceptible HIV-1 as determined by Sanger sequencing (groups II and III) (Table 1). Our more sensitive DeepGen assay revealed that 21 (group II) of the 33 patients harbored drug-resistant viruses at low frequency (1% to 18%), leaving only 12 of the patients experiencing treatment failure in the absence of drug-resistant HIV-1 variants (group III). That is, 80% (48/60) of the HIV-infected Ugandans failing cART in this study (groups I and II) harbored some proportion of drug-resistant HIV-1, which may be prognostic of their future treatment success.
FIG 4.
HIV-1 genotypic resistance interpretation based on Sanger or deep sequencing. A list of all the amino acid substitutions was used with the HIVdb Program Genotypic Resistance Interpretation Algorithm from the Stanford University HIV Drug Resistance Database (http://hivdb.stanford.edu) to infer the levels of susceptibility to protease, reverse transcriptase, and integrase inhibitors. High-level and intermediate resistance profiles are indicated in red and yellow, respectively, while a susceptible genotype is depicted in green. All 65 HIV-infected individuals classified in groups I, II, III, and IV, as described in Table 1, are indicated. NRTIs (ABC, ddI, FTC, 3TC, stavudine [d4T], TDF, and AZT), NNRTIs (EFV, ETR, NVP, and RPV), PIs (atazanavir [ATV], darunavir [DRV], amprenavir [APV], indinavir [IDV], lopinavir [LPV], nelfinavir [NFV], saquinavir [SQV], and tipranavir [TPV]), and INSTIs (dolutegravir [DTG], elvitegravir [EVG], and raltegravir [RAL]) are shown.
Phylogenetic and diversity analysis using deep sequencing.
As described above, our HIV-1-genotyping and coreceptor tropism assay (DeepGen) is based on deep sequencing of viral RNA extracted from plasma samples and optimized to accurately detect minority HIV-1 variants above a 1% level in the HIV-1 population (24). In addition to identifying low-level drug-resistant viruses otherwise not detected by Sanger sequencing, this methodology is capable of generating over 10,000 HIV-1 sequences (reads) per patient to analyze intra- and interpatient HIV-1 genetic diversity (58, 59). All 65 patient samples were genotyped, generating a total of 7,979,986 quality reads with an average read length of 171 bp. The high viral loads in most of these patients (ranging from 3.03 to 6.16 log10 copies/ml) and average sequencing coverage (a minimum of 1,000 reads per nucleotide position) ensured accurate detection of minor variants present at a 1% or higher level in the intrapatient HIV-1 population (60).
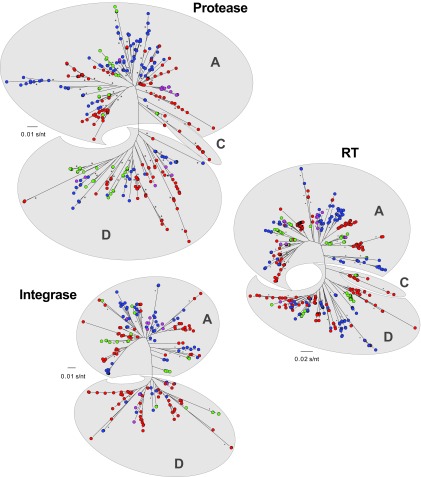
Deep-sequencing reads corresponding to 105-nt fragments in the protease-, RT-, and integrase-coding regions and present at a frequency of ≥10% of the overall intrapatient HIV-1 population were aligned and used to construct neighbor-joining phylogenetic trees and to calculate intra- and interpatient genetic diversity (Fig. 5). A total of 358 unique protease (mean, 5.1 [range, 1 to 24] per patient sample), 422 unique reverse transcriptase (5.9 [1 to 28]), and 211 unique integrase (3.1 [1 to 16]) sequence reads were included in each phylogenetic analysis. “Twigs,” or branch points (shown as colored dots in Fig. 5), on a single branch represent the unique reads within a patient sample. As expected, multiple unique reads were identified per patient, which branched together in each of the three HIV-1 genomic regions and clustered based on the HIV-1 subtype identification, i.e., A, C, or D (Fig. 5).
FIG 5.
Neighbor-joining phylogenetic trees were constructed using reads with a frequency of ≥10 corresponding to 105-bp fragments from the protease, RT, and integrase regions. Each color-coded dot represents a unique variant (frequency not depicted) in each patient corresponding to groups I (blue), II (red), III (green), and IV (purple). HIV-1 subtype-specific clusters are depicted by clouds labeled for each subtype, i.e., A, C, or D. Bootstrap resampling (1,000 data sets) of the multiple alignments tested the statistical robustness of the trees, with percentages above 75% indicated by asterisks. s/nt, substitutions per nucleotide.
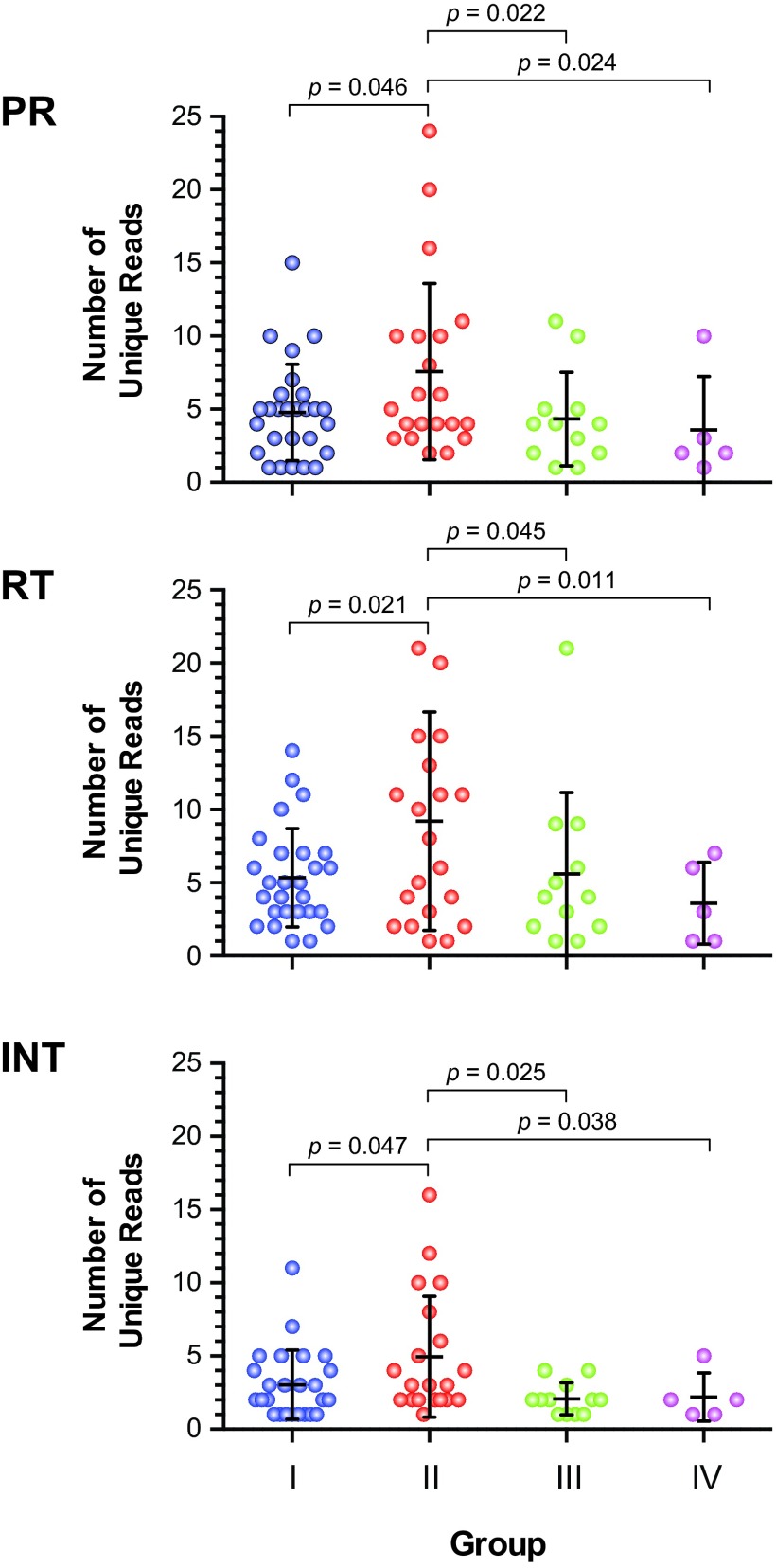
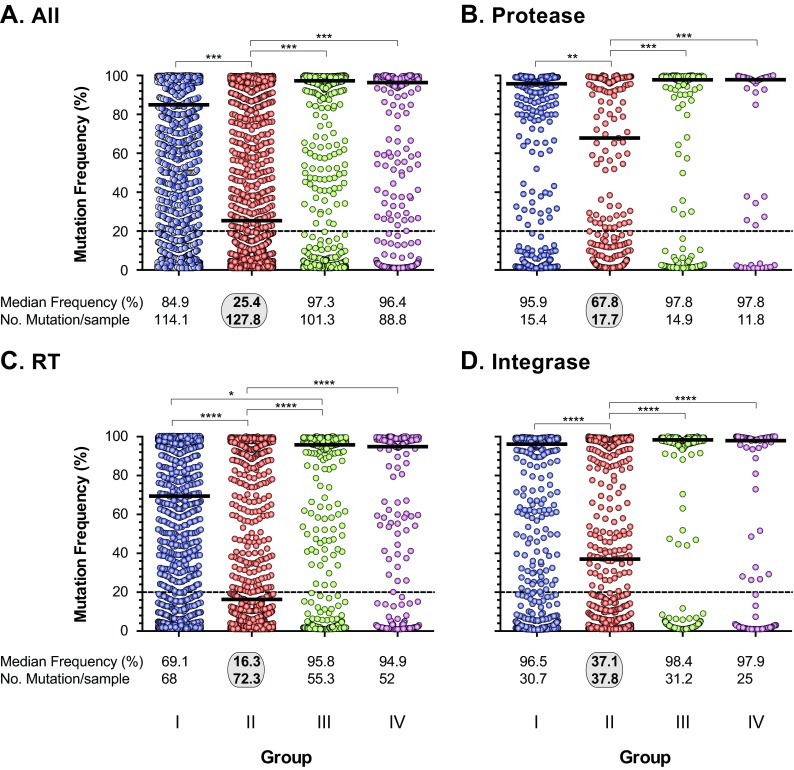
Although not statistically significant, the mean interpatient genetic distances in the two groups failing treatment with HIV-1 drug-resistant viruses (groups I and II) were higher than the genetic distances in groups III and IV, with susceptible viruses (in all three HIV-1 genomic regions) (data not shown). Intrapatient HIV-1 diversity, based on the number of unique PR, RT, or IN reads per patient, was significantly higher in patients from group II than in subjects in groups I, III, and IV, i.e., mean numbers of unique reads of 7.6 versus 4.7, 4.3, and 3.6, respectively, in PR (P value range, 0.022 to 0.046; unpaired t test); 9.2 versus 5.3, 5.6, and 3.6 in RT (0.011 to 0.045); and 4.95 versus 3.0, 2.1, and 2.2 in IN (0.025 to 0.047) (Fig. 6). Increased numbers of unique reads corresponded to higher numbers of overall mutations across the pol gene, particularly in the RT-coding region, which were also significantly higher in viruses from group II patients than in group I, II, and IV patients, i.e., a mean number of pol mutations of 127.8 versus 114.1, 101.3, and 88.8, respectively (P value range, 0.018 to 0.041; unpaired t test) and a mean number of RT mutations of 72.3 versus 68, 55.3, and 52, respectively (P value range, 0.024 to 0.043; unpaired t test) (Fig. 7). Finally, the number of minority HIV-1 variants was significantly higher in patients from group II versus group I, III, and IV patients based on an overall mutation frequency of 25.4% versus 84.9%, 97.3%, and 96.4%, respectively, in the pol gene (P < 0.001; Kruskal-Wallis ANOVA test) (Fig. 7A). Similar results were observed when the PR-, RT-, and INT-coding regions were individually analyzed (Fig. 7B, C, and D).
FIG 6.
Intrapatient HIV-1 genetic-diversity analysis was performed by quantifying the unique deep-sequencing reads with a frequency of ≥10, corresponding to 105-bp fragments from the PR-, RT-, and INT-coding regions for each patient. Groups I, II, III, and IV were defined as shown in Table 1. Means ± standard deviations and statistically significant differences (unpaired t test; P values) are indicated.
FIG 7.
Numbers and frequencies of all amino acid substitutions (mutations) detected at ≥1% in the entire HIV-1 pol gene (A), protease (B), reverse transcriptase (C), and integrase (D) coding regions using deep sequencing (DeepGen) in all 65 HIV-infected individuals. The median frequency and the mean number of mutations per sample in each group of patients are indicated. Statistically significant differences (Kruskal-Wallis ANOVA test) are marked: ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05.
Clinical outcome and association with HIV-1 drug susceptibility by DeepGen.
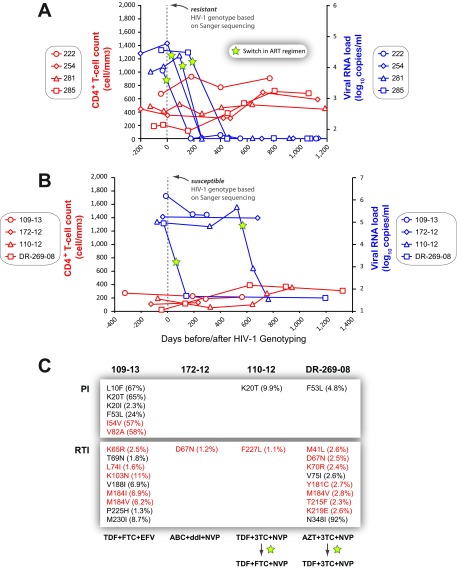
Group I patients were infected with drug-resistant HIV-1 variants that dominate the intrapatient HIV-1 population as detected by Sanger sequencing (and by DeepGen); however, minority HIV-1 drug-resistant variants in group II patients were detected only by DeepGen. The cumulative findings shown in Fig. 6 and 7 suggest that group II patients have an increased number of unique HIV-1 sequence reads (i.e., variants) bearing drug-resistant HIV-1 mutations that either are in the process of selection due to ongoing treatment or have faded in the intrapatient HIV-1 population due to intermittent treatment. HIV-1-genotyping reports (based on Sanger sequencing) were provided to the physicians treating all 60 patients from groups I, II, and III at the JCRC in Kampala, Uganda. To evaluate the potential contribution of minority HIV-1 drug-resistant variants, we examined the clinical outcomes for 38 (of 65) patients up to 3 years following the HIV-1-genotyping reports. As expected, antiretroviral drugs were switched in the ART regimens of the 27 patients from group I upon detection of drug resistance by Sanger sequencing. Patients in group I (as in the larger population shown in Fig. 1A and B) with a drug-resistant genotype and a switch in treatment had better treatment outcomes based on increased CD4+ T-cell counts and decreasing viral loads (data not shown). However, no significant change was observed in CD4+ T-cell counts and viral loads in the first year for individuals in group II (a subset of the JCRC patient population in Fig. 1E and F), who remained on the same antiretroviral treatment with a drug-susceptible HIV-1 genotype based on Sanger sequencing. After 1 year, the outcomes for the group II subset improved, with increasing CD4+ T-cell counts and lower viral loads in these patients due to an eventual switch in antiretroviral treatment (Fig. 1F). Figure 2C shows significant decreases in CD4+ T-cell counts and increases in viral loads for the paired patient analyses of the subset. In Fig. 8A, we provide specific examples of patients in group I where evidence of drug resistance by Sanger sequencing led to a change in cART, a subsequent drop to undetectable viral loads, and slow increases in CD4+ T-cell counts. Three of four patients in group II who were “susceptible” to the ongoing cART (109-13, 172-12, and 110-12) showed no decline in viral loads and no appreciable increases in CD4+ T-cell counts while remaining under the same treatment regimens (Fig. 8B). In Fig. 8C, we show that these patients from group II (109-13, 172-12, 110-12, and DR-269-08) had low-frequency HIV-1 drug resistance mutations conferring resistance to RTIs. Patient 109-13 continued on ATRIPLA (TDF plus FTC plus EFV) despite having a series of mutations associated with resistance to this cART, e.g., K65R (2.5%), M814V (6.2%), and K130N (11%), undetected by the standard HIV-1 genotype test. Patients 172-12 and 110-12 remained on abacavir (ABC) plus didanosine (ddI) plus NVP and TDF plus 3TC plus NVP, respectively. Due to issues with drug tolerance, patient 110-12 switched 3TC for FTC but retained TDF and NVP in the regimen. A drop in the viral load was then observed over a 200-day period. Finally, patient DR-269-08 carried viral variants with thymidine analog mutations at low frequency, e.g., M41L (2.6%), D67N (2.5%), K70R (2.4%), and T215F (2.3%), in addition to Y181C (2.7%) and M184V (2.8%), associated with reduced susceptibility to NVP and 3TC, respectively (Fig. 8C). Fortunately, this patient switched to TDF plus 3TC plus NVP shortly after the drug-resistant-genotype test, which resulted in a drop to undetectable viral loads within 150 days, most likely due to the potent activity of TDF.
FIG 8.
(A and B) Specific treatment outcomes in patients who switched (A) or remained on current (B) antiretroviral treatment regimens following the identification of drug resistance (A) or a drug-susceptible genotype (B) after a Sanger sequencing-based HIV-1-genotyping test. Viral-RNA levels (copies per milliliter of plasma) and CD4+ T-cell counts (cells per cubic millimeter of blood) were monitored over a 1,000- to 1,200-day period following a drug resistance test. (C) Frequencies of all amino acid substitutions (mutations) associated with resistance to PIs or RTIs detected at ≥1% using DeepGen. Primary (red) and secondary/compensatory (black) drug resistance mutations, defined by the Stanford University HIV Drug Resistance Database (http://hivdb.stanford.edu), are indicated. cART regimens for each patient are included.
Observation of clinical management following HIV drug resistance testing.
As discussed below and highlighted throughout, this observational and retrospective study could analyze only the clinical data from patient visits that did not follow a routine schedule (typical in a clinical trial). Furthermore, we did not or could not randomize patients to maintain or switch treatment regimens in cases of treatment failure in the absence of HIV drug resistance detected by Sanger sequencing. Although we cannot produce exact numbers, the majority of physicians treating patients with cART failure in Uganda usually request an HIV drug resistance test, which is relatively inexpensive by HIC standards (i.e., approximately $100 per test) but still costly for patient management in Uganda. We typically report HIV drug resistance by Sanger sequencing in over 75% to 80% of patients failing treatment (i.e., with plasma viral loads of >2,000 copies/ml and/or CD4+ T-cell counts of <250 cells/mm3 on two consecutive visits). In cases of clear HIV drug resistance, patients are started on a new cART regimen. However, we fail to detect HIV drug resistance in approximately 20% of these patients. Due in part to stipulations by the various international funding groups, these patients are usually maintained or restarted on the same treatment regimen. From the cohort analyses, patients failing treatment without HIV drug resistance by Sanger sequencing and maintaining the same treatment regimen fare significantly worse than those who had HIV drug resistance detected by Sanger sequencing. However, with observational studies and without randomization, our physicians have the patients' well-being in mind, and all patients are eventually switched to salvage treatment within the first 1 to 2 years of experiencing continued treatment failure (as illustrated in Fig. 8). Due to the cost, this switch rarely involves a second HIV drug resistance test. From our subset analyses, we know that the majority of the treatment failures in Uganda, without HIV drug resistance detected by Sanger sequencing, will have drug resistance mutations detected by deep sequencing. As discussed below, the significant observation of continued treatment failure 1 year following detection of drug-susceptible virus by Sanger sequencing (i.e., approximately 20% of the cART failures in Uganda) and the significant detection of low-frequency drug-resistant virus in a subset of these patients (64%) have prompted the immediate adoption of DeepGen for HIV drug resistance genotyping at the JCRC. Further studies from our group will compare the treatment outcomes of patients who switched to a salvage regimen based on dominant HIV drug resistance with those of individuals carrying low-frequency drug-resistant viruses and those with a lack of drug resistance.
DISCUSSION
Over 40% of HIV-1-infected patients around the world are now receiving treatment compared to less than 1% in 2000 (http://www.unaids.org/en/resources/documents/2015/AIDS_by_the_numbers_2015). Initiating cART earlier in disease is obviously beneficial to patient outcome, but only if the patient remains adherent to the cART with full viral suppression (61). Full viral suppression on first-line cART is achieved in >85% of treatment-naive HIV-infected individuals/year in HICs (62–64), but in LMICs, these rates are less than 80% per year (64–66). Antiretroviral treatment failure is largely due to poor adherence in sub-Saharan Africa, but purely behavior-based decisions to stop medications are rare, and with access to medication, adherence rates are similar to those in HICs (64, 66). Thus, poor adherence in LMICs is generally related to limited access to clinical centers, high travel costs, intermittent antiretroviral drug supply, and interruptions of funding programs that provide cART (67). In Uganda, it is not uncommon for treating physicians to have limited knowledge of patient-specific adherence issues, and poor adherence is rarely noted in patient charts.
At the Joint Clinical Research Centre in Uganda, we provide HIV-1 drug resistance genotype testing for any patient with a viral loads of >1,000 copies/ml and/or CD4+ T-cell counts below 250 cells/mm3 on two consecutive visits. Since 1999, over 5,000 HIV-1 drug resistance genotype tests (based on Sanger sequencing) have been performed in the JCRC's WHO-designed laboratory. At the JCRC, a HIV-1 drug-resistant genotype with mutations in primary resistance to at least one antiretroviral drug is observed in approximately 76% of patients failing treatment (9). In a prospective study of over 2,000 patients, Hamers et al. (65) found a 10% failure rate after 1 year on first-line cART, and approximately 70% of these treatment failures harbored drug-resistant genotypes. Despite similar drug resistance rates (70% to 76%) for first-line treatment failures in these prospective (65) and cross-sectional (9) cohort analyses, several smaller cross-sectional clinical studies in Uganda have indicated that nearly all first-line treatment failures were associated with the appearance of HIV-1 drug-resistant genotypes (68, 69). The concordance between first-line treatment failures and drug resistance in HICs is generally greater than 95% (65). Hamers et al. (65) suggested that close monitoring of viral loads and CD4+ T-cell counts over the first year identified treatment failures prior to the emergence of dominant HIV-1 drug resistance. In contrast, Kyeyune et al. (9) reported that treatment failures in this cross-sectional cohort were frequently identified in patients with prolonged times between clinic visits. Unlike many clinics in HICs, where samples are stored during viral-load testing and where drug resistance testing is performed on the same sample with a detectable plasma viral load, the JCRC requests HIV drug resistance testing only when the previous visit (commonly 3 months earlier) showed a high viral load or the patient had repeated CD4+ T-cell counts below 200 cells/mm3. Many patients stop taking their cART during the intervening period between a positive viral load and subsequent drug resistance testing, which likely accounts for the low-level HIV drug resistance being undetectable by Sanger sequencing. As described below, we suspect that both studies are correct in their assumptions, in that (i) primary HIV-1 drug resistance mutations can contribute to treatment failures prior to the dominance of drug-resistant HIV-1 variants in the intrapatient population and (ii) drug-resistant HIV-1 strains quickly fade in the intrapatient HIV-1 population in the absence of drug pressure (i.e., poor adherence).
Based on the hypotheses of Hamer et al. (65) and Kyeyune et al. (9), we proposed that Uganda HIV patients failing first-line antiretroviral treatment were likely to carry drug-resistant HIV-1 as minority variants in their intrapatient virus population. Furthermore, if the same treatment regimens (as the first line) were resumed, we proposed that treatment failure would arise or worsen if the minority drug-resistant HIV-1 variants were present. Counter to our hypothesis, Gupta et al. (70) reported that 27% of all first-line treatment failures (n = 70) in the Development of Anti-Retroviral Therapy in Africa (DART) study showed resuppression of the viral load when the same first-line regimen was continued. Resuppression was even observed in 7 of 10 patients failing treatment in the presence of a drug-resistant genotype (70). In our study, we observed resuppression based on a drop in the viral load in only 40% of the patients failing treatment but lacking Sanger-detected drug resistance. Our standard-of-care HIV-1 genotype testing in the JCRC clinic led to a switch to second-line ART regimens if resistance to the first-line treatment was detected. Here, we analyzed 33 patients (groups II and III) experiencing treatment failure in the absence of HIV-1 drug resistance detected by Sanger sequencing. These patients resumed their treatment, and there was no significant change in their viral loads or CD4+ T-cell counts 6 months before and 6 months after drug resistance testing. Despite being a “susceptible virus” in our Sanger-based HIV-1 drug resistance report, resumption of the first-line treatment regimen led to only 40% of patients having a 3-fold or greater decrease in the viral load over the next 6 months. In contrast, detection of a virus resistant to the first-line treatment and “susceptible” to the second-line treatment led to over 60% of the patients having significant drops in the viral load over the first 6 months of the second-line treatment. With a second-line treatment regimen, over 20% reached complete viral suppression in 6 months and another 24% in 12 months. Thus, our data suggest that despite a “drug-susceptible” HIV-1 genotype (based on Sanger sequencing) and intense adherence counseling, resumption of the first-line treatment regimen provides only a weak virologic response (in 40% of the patients), suggesting that low-frequency HIV-1 drug resistance could be responsible for the continued treatment failure.
Emergence of a dominant drug-resistant virus from the low-frequency variant during continued treatment failure would provide absolute proof of the impact of low-frequency HIV drug resistance on treatment outcomes in Uganda. However, physicians typically switch to a salvage treatment regimen if treatment failure continues and HIV drug resistance was not detected by the Sanger-based test on the previous visit sample. Based on (i) the strong correlations between the detection of low-frequency HIV drug resistance mutations and continued treatment failure (here) and (ii) previous studies in Ugandan women showing that low-frequency NVP mutations led to failure of NNRTI-containing cART (49, 71), our ethics committee decided against a study in which the same cART was continued when low-frequency HIV drug resistance mutations were detected at >5%. More importantly, we have also adopted DeepGen as the preferred HIV drug resistance genotyping test in the JCRC.
The impact of low-frequency drug-resistant HIV-1 variants on treatment outcomes in patients is a matter of great debate (32, 34, 38–41, 72). Several studies suggest that preexisting minority drug-resistant variants in treatment-naive patients have minimal to no effect on the treatment outcome even if the treatment involves drugs associated with the drug resistance pattern (e.g., treatment with NVP when a patient has 3% NVP-resistant variants) (32, 49). However, the impact of these drug-resistant minority variants in treatment-naive patients is also highly dependent on the level of resistance conferred by the specific mutation, cross-resistance to other drugs in the regimen, and the replicative fitness of the HIV-1-resistant strain. Unfortunately, there have been few research studies examining the impact of low-frequency drug-resistant HIV-1 variants in treatment-experienced patients due to the current success of cART in HICs; very low failure rates; and, so far, limited availability of clinical tests aimed at quantifying low-frequency HIV-1 variants (24, 73). The OCTANE/A5208 study by the AIDS Clinical Trial Group provides our best indication that low-frequency mutations can impact treatment outcomes (49, 71, 74, 75). Women who had previously received a single-dose of NVP to prevent mother-to-child transmission had a 30% rate of treatment failure if randomized to receive an NVP-containing regimen versus a <10% failure rate with a ritonavir-boosted lopinavir (LPV/r)-containing regimen of FTC plus TDF. Preexisting dominant NVP mutations were a major risk factor for failure. Subsequent studies also showed that a high risk of treatment failure was also observed in women who had received single-dose NVP and had NVP-resistant HIV-1 variants at low frequency (49, 71).
Most studies on the impact of low-frequency drug-resistant viruses have been limited by clinical-trial design and patient numbers. With the accumulation of over 3,000 drug-resistant genotypes from treatment failures in Uganda, we used a deep-sequencing-based HIV-1-genotyping assay (DeepGen) on 21 (of approximately 900) and 27 (of approximately 2,100) patients failing treatment without (group II) or with (group I) HIV-1 drug resistance as determined by Sanger sequencing. In 27 patients from group I, we obtained strong concordance between the dominant mutations detected by both sequencing methods, but our DeepGen test identified 66 additional drug-resistant mutations at a low frequency. Despite the lack of drug resistance by Sanger sequencing, all 21 patients in group II had drug resistance mutations detected in between 1% and 20% of the intrapatient HIV-1 population. These findings suggest that new drug-resistant HIV-1 variants were about to emerge in these patients or that high-frequency HIV-1 drug-resistant variants had been replaced by more fit wild-type variants with higher replicative capacity due to poor treatment adherence. Interestingly, the presence of drug-resistant minority variants was also related to higher genetic diversity in the patient's virus, whereas dominant drug-resistant HIV-1 had low diversity in treatment failures. Since the physicians received a report of drug-susceptible HIV-1 based on Sanger sequencing, the treatment regimen was continued, and treatment failure persisted. This continued treatment failure correlated with the presence of the minority drug-resistant HIV-1 variants in the patient samples (tested retrospectively with DeepGen). In contrast, viral suppression was achieved when the treatment regimen was switched to drugs that lacked any patterns of cross-resistance to previous treatment drugs.
In summary, we have shown that most patients failing cART in Uganda would continue to fail their treatment regimen even if a dominant drug-resistant HIV-1 variant was not detected. At least 25% to 30% of treatment failure cases in Uganda occur in the absence of dominant HIV-1 drug resistance (9, 65). Unfortunately, when informed of a drug-susceptible HIV-1 genotype, treating physicians typically maintained the existing treatment regimen and only switched cART upon retesting for HIV-1 drug resistance or after observing continued virologic failure at the next visit (typically 4 to 12 months later). Our analyses of a subset of samples using the more sensitive DeepGen assay revealed that most patients with treatment failure had low-frequency (or minority) drug-resistant variants in the intrapatient HIV-1 population, which correlated with the continued treatment failure. For that reason, we have now adopted the DeepGen assay in place of a Sanger sequencing-based HIV-1-genotyping assay to report HIV-1 drug resistance at a dominant (>20%) or subdominant (1% to 20%) level for patients failing treatment at the JCRC in Kampala, Uganda. Implementing DeepGen or another deep-sequencing-based HIV-1-genotyping assay as the standard of care in Uganda (and possibly in the rest of sub-Saharan Africa) is related to a higher likelihood that drug resistance exists as a minority variant in patients in LMICs than in patients in HICs. As described above, patients in LMICs—specifically in Uganda—have irregular and infrequent clinic visits; incomplete clinical data related to viral-RNA loads and CD4+ T-cell counts; and, perhaps more relevant, poor adherence. Again, it is important to stress that breaks in drug treatment or poor adherence are more related to socioeconomic factors, such as drug shortages, costs of both drugs and travel to obtain drugs, and stigma than to various behavioral factors. Due to the rapid selection and/or decline of drug-resistant HIV-1 variants from the complex intrapatient HIV-1 population, we should not base clinical decisions on an arbitrary detection of drug-resistant HIV-1 set to 20% in the virus population (i.e., the empirical limits of Sanger sequencing) but should use more sensitive methods to detect these HIV-1 variants at low levels.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dane Winner and Christine Sayir from the University Hospitals Translational Laboratory (Cleveland, OH) for their help with the deep-sequencing experiments.
F.K. was funded by a scholarship from NIH Fogarty International Center grants D43-TW000011 and D43-TW009780. E.J.A. was funded by NIAID/NIH AI49170 and holds the Canada Research Chair in HIV-1 Pathogenesis and Viral Control. M.E.Q.-M. was partially supported by the CWRU/UH Center for AIDS Research (P30 AI036219) and by funding from University Hospitals Case Medical Center (UHCMC) for the University Hospitals Translational Laboratory (UHTL).
R.M.G. and M.E.Q.-M. developed the novel HIV-1-genotyping and coreceptor tropism assay, DeepGenHIV. There are no further patents, products in development, or marketed products to declare.
F.K., E.J.A., and M.E.Q,-M, designed the study, collected and assembled the data, and drafted and wrote the manuscript. F.K. and R.M.G. performed all sequencing experiments, while I.N., S.M., and E.N. performed all molecular and Sanger sequencing experiments. F.K. and M.E.Q.-M. contributed to the overall analysis of the data. R.A.S., C.N.K., and P.M. provided clinical material and data and critical review and advice. We all read and approved the final manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00038-16.
REFERENCES
- 1.Paredes R, Clotet B. 2010. Clinical management of HIV-1 resistance. Antiviral Res 85:245–265. doi: 10.1016/j.antiviral.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 2.Zolopa AR. 2010. The evolution of HIV treatment guidelines: current state-of-the-art of ART. Antiviral Res 85:241–244. doi: 10.1016/j.antiviral.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 3.UNAIDS. 2013. Global update on HIV treatment 2013: results, impact, and opportunities. UNAIDS, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/85327/1/WHO_HIV_2013.9_eng.pdf. [Google Scholar]
- 4.Tang JW, Pillay D. 2004. Transmission of HIV-1 drug resistance. J Clin Virol 30:1–10. doi: 10.1016/j.jcv.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Geretti AM. 2007. Epidemiology of antiretroviral drug resistance in drug-naive persons. Curr Opin Infect Dis 20:22–32. doi: 10.1097/QCO.0b013e328013caff. [DOI] [PubMed] [Google Scholar]
- 6.Pham QD, Wilson DP, Law MG, Kelleher AD, Zhang L. 2014. Global burden of transmitted HIV drug resistance and HIV-exposure categories: a systematic review and meta-analysis. AIDS 28:2751–2762. doi: 10.1097/QAD.0000000000000494. [DOI] [PubMed] [Google Scholar]
- 7.Ndembi N, Hamers RL, Sigaloff KC, Lyagoba F, Magambo B, Nanteza B, Watera C, Kaleebu P, Rinke de Wit TF. 2011. Transmitted antiretroviral drug resistance among newly HIV-1 diagnosed young individuals in Kampala. AIDS 25:905–910. doi: 10.1097/QAD.0b013e328346260f. [DOI] [PubMed] [Google Scholar]
- 8.Nazziwa J, Njai HF, Ndembi N, Birungi J, Lyagoba F, Gershim A, Nakiyingi-Miiro J, Nielsen L, Mpendo J, Nanvubya A, Debont J, Grosskurth H, Kamali A, Seeley J, Kaleebu P. 2013. Short communication: HIV type 1 transmitted drug resistance and evidence of transmission clusters among recently infected antiretroviral-naive individuals from Ugandan fishing communities of Lake Victoria. AIDS Res Hum Retrovir 29:788–795. doi: 10.1089/aid.2012.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kyeyune F, Nankya I, Metha S, Akao J, Ndashimye E, Tebit DM, Rodriguez B, Kityo C, Salata RA, Mugyenyi P, Arts E. 2013. Treatment failure and drug resistance is more frequent in HIV-1 subtype D versus subtype A-infected Ugandans over a 10-year study period. AIDS 27:1899–1909. doi: 10.1097/QAD.0b013e3283610ec7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhee SY, Blanco JL, Jordan MR, Taylor J, Lemey P, Varghese V, Hamers RL, Bertagnolio S, Rinke de Wit TF, Aghokeng AF, Albert J, Avi R, Avila-Rios S, Bessong PO, Brooks JI, Boucher CA, Brumme ZL, Busch MP, Bussmann H, Chaix ML, Chin BS, D'Aquin TT, De Gascun CF, Derache A, Descamps D, Deshpande AK, Djoko CF, Eshleman SH, Fleury H, Frange P, Fujisaki S, Harrigan PR, Hattori J, Holguin A, Hunt GM, Ichimura H, Kaleebu P, Katzenstein D, Kiertiburanakul S, Kim JH, Kim SS, Li Y, Lutsar I, Morris L, Ndembi N, Ng KP, Paranjape RS, Peeters M, Poljak M, Price MA, Ragonnet-Cronin ML, Reyes-Teran G, Rolland M, Sirivichayakul S, Smith DM, Soares MA, Soriano VV, Ssemwanga D, Stanojevic M, Stefani MA, Sugiura W, Sungkanuparph S, Tanuri A, Tee KK, Truong HH, van de Vijver DA, Vidal N, Yang C, Yang R, Yebra G, Ioannidis JP, Vandamme AM, Shafer RW. 2015. Geographic and temporal trends in the molecular epidemiology and genetic mechanisms of transmitted HIV-1 drug resistance: an individual-patient- and sequence-level meta-analysis. PLoS Med 12:e1001810. doi: 10.1371/journal.pmed.1001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ssemwanga D, Lihana RW, Ugoji C, Abimiku A, Nkengasong J, Dakum P, Ndembi N. 2015. Update on HIV-1 acquired and transmitted drug resistance in Africa. AIDS Rev 17:3–20. [PubMed] [Google Scholar]
- 12.UNAIDS. 2014. The HIV and AIDS Uganda country progess report 2014. UNAIDS, Geneva, Switzerland: http://www.unaids.org/sites/default/files/country/documents/UGA_narrative_report_2015.pdf. [Google Scholar]
- 13.WHO. 2013. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. WHO, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/85321/1/9789241505727_eng.pdf?ua=1. [PubMed] [Google Scholar]
- 14.Boender TS, Sigaloff KC, McMahon JH, Kiertiburanakul S, Jordan MR, Barcarolo J, Ford N, Rinke de Wit TF, Bertagnolio S. 2015. Long-term virological outcomes of first-line antiretroviral therapy for HIV-1 in low- and middle-income countries: a systematic review and meta-analysis. Clin Infect Dis 61:1453–1461. doi: 10.1093/cid/civ556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boender TS, Hoenderboom BM, Sigaloff KC, Hamers RL, Wellington M, Shamu T, Siwale M, Labib Maksimos EE, Nankya I, Kityo CM, Adeyemo TA, Akanmu AS, Mandaliya K, Botes ME, Ondoa P, Rinke de Wit TF. 2015. Pretreatment HIV drug resistance increases regimen switches in sub-Saharan Africa. Clin Infect Dis 61:1749–1758. doi: 10.1093/cid/civ656. [DOI] [PubMed] [Google Scholar]
- 16.Grant PM, Zolopa AR. 2009. The use of resistance testing in the management of HIV-1-infected patients. Curr Opin HIV AIDS 4:474–480. doi: 10.1097/COH.0b013e328331c14f. [DOI] [PubMed] [Google Scholar]
- 17.Lessells RJ, Avalos A, de Oliveira T. 2013. Implementing HIV-1 genotypic resistance testing in antiretroviral therapy programs in Africa: needs, opportunities, and challenges. AIDS Rev 15:221–229. [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson RM, Schmotzer CL, Quinones-Mateu ME. 2014. Next-generation sequencing to help monitor patients infected with HIV: ready for clinical use? Curr Infect Dis Rep 16:401. doi: 10.1007/s11908-014-0401-5. [DOI] [PubMed] [Google Scholar]
- 19.Larder BA, Kohli A, Kellam P, Kemp SD, Kronick M, Henfrey RD. 1993. Quantitative detection of HIV-1 drug resistance mutations by automated DNA sequencing. Nature 365:671–673. doi: 10.1038/365671a0. [DOI] [PubMed] [Google Scholar]
- 20.Church JD, Jones D, Flys T, Hoover D, Marlowe N, Chen S, Shi C, Eshleman JR, Guay LA, Jackson JB, Kumwenda N, Taha TE, Eshleman SH. 2006. Sensitivity of the ViroSeq HIV-1 genotyping system for detection of the K103N resistance mutation in HIV-1 subtypes A, C, and D. J Mol Diagn 8:430–432. doi: 10.2353/jmoldx.2006.050148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halvas EK, Aldrovandi GM, Balfe P, Beck IA, Boltz VF, Coffin JM, Frenkel LM, Hazelwood JD, Johnson VA, Kearney M, Kovacs A, Kuritzkes DR, Metzner KJ, Nissley DV, Nowicki M, Palmer S, Ziermann R, Zhao RY, Jennings CL, Bremer J, Brambilla D, Mellors JW. 2006. Blinded, multicenter comparison of methods to detect a drug-resistant mutant of human immunodeficiency virus type 1 at low frequency. J Clin Microbiol 44:2612–2614. doi: 10.1128/JCM.00449-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leitner T, Halapi E, Scarlatti G, Rossi P, Albert J, Fenyo EM, Uhlen M. 1993. Analysis of heterogeneous viral populations by direct DNA sequencing. Biotechniques 15:120–127. [PubMed] [Google Scholar]
- 23.Grant RM, Kuritzkes DR, Johnson VA, Mellors JW, Sullivan JL, Swanstrom R, D'Aquila RT, Van Gorder M, Holodniy M, Lloyd RM Jr, Reid C, Morgan GF, Winslow DL. 2003. Accuracy of the TRUGENE HIV-1 genotyping kit. J Clin Microbiol 41:1586–1593. doi: 10.1128/JCM.41.4.1586-1593.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibson RM, Meyer AM, Winner D, Archer J, Feyertag F, Ruiz-Mateos E, Leal M, Robertson DL, Schmotzer CL, Quinones-Mateu ME. 2014. Sensitive deep sequencing-based HIV-1 genotyping assay to simultaneously determine susceptibility to protease, reverse transcriptase, integrase, and maturation inhibitors, as well as HIV-1 coreceptor tropism. Antimicrob Agents Chemother 58:2167–2185. doi: 10.1128/AAC.02710-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinones-Mateu ME, Arts EJ. 2002. Fitness of drug resistant HIV-1: methodology and clinical implications. Drug Resist Updat 5:224–233. doi: 10.1016/S1368-7646(02)00123-1. [DOI] [PubMed] [Google Scholar]
- 26.Quinones-Mateu ME, Arts EJ. 2001. HIV-1 fitness: implications for drug resistance, disease progression, and global epidemic evolution, p 134–170. In Kuiken C, Foley B, Hahn B, Marx P, McCutchan F, Mellors J, Wolinsky S, Korber B (ed), HIV sequence compendium 2001. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM. [Google Scholar]
- 27.Quinones-Mateu ME, Arts EJ. 2006. Virus fitness: concept, quantification, and application to HIV population dynamics. Curr Top Microbiol Immunol 299:83–140. [DOI] [PubMed] [Google Scholar]
- 28.Avidor B, Girshengorn S, Matus N, Talio H, Achsanov S, Zeldis I, Fratty IS, Katchman E, Brosh-Nissimov T, Hassin D, Alon D, Bentwich Z, Yust I, Amit S, Forer R, Vulih Shultsman I, Turner D. 2013. Evaluation of a benchtop HIV ultradeep pyrosequencing drug resistance assay in the clinical laboratory. J Clin Microbiol 51:880–886. doi: 10.1128/JCM.02652-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang MW, Oliveira G, Yuan J, Okulicz JF, Levy S, Torbett BE. 2013. Rapid deep sequencing of patient-derived HIV with ion semiconductor technology. J Virol Methods 189:232–234. doi: 10.1016/j.jviromet.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dudley DM, Chin EN, Bimber BN, Sanabani SS, Tarosso LF, Costa PR, Sauer MM, Kallas EG, O'Connor DH. 2012. Low-cost ultra-wide genotyping using Roche/454 pyrosequencing for surveillance of HIV drug resistance. PLoS One 7:e36494. doi: 10.1371/journal.pone.0036494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lapointe HR, Dong W, Lee GQ, Bangsberg DR, Martin JN, Mocello AR, Boum Y, Karakas A, Kirkby D, Poon AF, Harrigan PR, Brumme CJ. 2015. HIV drug resistance testing by high-multiplex “wide” sequencing on the MiSeq instrument. Antimicrob Agents Chemother 59:6824–6833. doi: 10.1128/AAC.01490-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li JZ, Kuritzkes DR. 2013. Clinical implications of HIV-1 minority variants. Clin Infect Dis 56:1667–1674. doi: 10.1093/cid/cit125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gianella S, Richman DD. 2010. Minority variants of drug-resistant HIV. J Infect Dis 202:657–666. doi: 10.1086/655397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li JZ, Paredes R, Ribaudo HJ, Svarovskaia ES, Metzner KJ, Kozal MJ, Hullsiek KH, Balduin M, Jakobsen MR, Geretti AM, Thiebaut R, Ostergaard L, Masquelier B, Johnson JA, Miller MD, Kuritzkes DR. 2011. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA 305:1327–1335. doi: 10.1001/jama.2011.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swenson LC, Mo T, Dong WW, Zhong X, Woods CK, Thielen A, Jensen MA, Knapp DJ, Chapman D, Portsmouth S, Lewis M, James I, Heera J, Valdez H, Harrigan PR. 2011. Deep V3 sequencing for HIV type 1 tropism in treatment-naive patients: a reanalysis of the MERIT trial of maraviroc. Clin Infect Dis 53:732–742. doi: 10.1093/cid/cir493. [DOI] [PubMed] [Google Scholar]
- 36.Swenson LC, Mo T, Dong WW, Zhong X, Woods CK, Jensen MA, Thielen A, Chapman D, Lewis M, James I, Heera J, Valdez H, Harrigan PR. 2011. Deep sequencing to infer HIV-1 co-receptor usage: application to three clinical trials of maraviroc in treatment-experienced patients. J Infect Dis 203:237–245. doi: 10.1093/infdis/jiq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodman DD, Zhou Y, Margot NA, McColl DJ, Zhong L, Borroto-Esoda K, Miller MD, Svarovskaia ES. 2011. Low level of the K103N HIV-1 above a threshold is associated with virological failure in treatment-naive individuals undergoing efavirenz-containing therapy. AIDS 25:325–333. doi: 10.1097/QAD.0b013e3283427dcb. [DOI] [PubMed] [Google Scholar]
- 38.Stekler JD, Ellis GM, Carlsson J, Eilers B, Holte S, Maenza J, Stevens CE, Collier AC, Frenkel LM. 2011. Prevalence and impact of minority variant drug resistance mutations in primary HIV-1 infection. PLoS One 6:e28952. doi: 10.1371/journal.pone.0028952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li JZ, Paredes R, Ribaudo HJ, Kozal MJ, Svarovskaia ES, Johnson JA, Geretti AM, Metzner KJ, Jakobsen MR, Hullsiek KH, Ostergaard L, Miller MD, Kuritzkes DR. 2013. Impact of minority nonnucleoside reverse transcriptase inhibitor resistance mutations on resistance genotype after virologic failure. J Infect Dis 207:893–897. doi: 10.1093/infdis/jis925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li JZ, Paredes R, Ribaudo HJ, Svarovskaia ES, Kozal MJ, Hullsiek KH, Miller MD, Bangsberg DR, Kuritzkes DR. 2012. Relationship between minority nonnucleoside reverse transcriptase inhibitor resistance mutations, adherence, and the risk of virologic failure. AIDS 26:185–192. doi: 10.1097/QAD.0b013e32834e9d7d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Codoner FM, Pou C, Thielen A, Garcia F, Delgado R, Dalmau D, Alvarez-Tejado M, Ruiz L, Clotet B, Paredes R. 2011. Added value of deep sequencing relative to population sequencing in heavily pre-treated HIV-1-infected subjects. PLoS One 6:e19461. doi: 10.1371/journal.pone.0019461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soares EA, Santos AF, Sousa TM, Sprinz E, Martinez AM, Silveira J, Tanuri A, Soares MA. 2007. Differential drug resistance acquisition in HIV-1 of subtypes B and C. PLoS One 2:e730. doi: 10.1371/journal.pone.0000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cornelissen M, van den Zorgdrager BRF, Lukashov V, Goudsmit J. 1997. pol gene diversity of five human immunodeficiency virus type 1 subtypes: evidence for naturally occurring mutations that contribute to drug resistance, limited recombination patterns, and common ancestry for subtypes B and D. J Virol 71:6348–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quinones-Mateu ME, Albright JL, Mas A, Soriano V, Arts EJ. 1998. Analysis of pol gene heterogeneity, viral quasispecies, and drug resistance in individuals infected with group O strains of human immunodeficiency virus type 1. J Virol 72:9002–9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Derache A, Wallis CL, Vardhanabhuti S, Bartlett J, Kumarasamy N, Katzenstein D. 2016. Phenotype, genotype, and drug resistance in subtype C HIV-1 infection. J Infect Dis 213:250–256. doi: 10.1093/infdis/jiv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skhosana L, Steegen K, Bronze M, Lukhwareni A, Letsoalo E, Papathanasopoulos MA, Carmona SC, Stevens WS. 2015. High prevalence of the K65R mutation in HIV-1 subtype C infected patients failing tenofovir-based first-line regimens in South Africa. PLoS One 10:e0118145. doi: 10.1371/journal.pone.0118145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fisher RG, Smith DM, Murrell B, Slabbert R, Kirby BM, Edson C, Cotton MF, Haubrich RH, Kosakovsky Pond SL, Van Zyl GU. 2015. Next generation sequencing improves detection of drug resistance mutations in infants after PMTCT failure. J Clin Virol 62:48–53. doi: 10.1016/j.jcv.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samuel R, Paredes R, Parboosing R, Moodley P, Gordon M. 2014. Minority HIV-1 drug-resistant mutations and prevention of mother-to-child transmission: perspectives for resource-limited countries. AIDS Rev 16:187–198. [PubMed] [Google Scholar]
- 49.Boltz VF, Bao Y, Lockman S, Halvas EK, Kearney MF, McIntyre JA, Schooley RT, Hughes MD, Coffin JM, Mellors JW. 2014. Low-frequency nevirapine (NVP)-resistant HIV-1 variants are not associated with failure of antiretroviral therapy in women without prior exposure to single-dose NVP. J Infect Dis 209:703–710. doi: 10.1093/infdis/jit635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richard N, Juntilla M, Abraha A, Demers K, Paxinos E, Galovich J, Petropoulos C, Whalen CC, Kyeyune F, Atwine D, Kityo C, Mugyenyi P, Arts EJ. 2004. High prevalence of antiretroviral resistance in treated Ugandans infected with non-subtype B human immunodeficiency virus type 1. AIDS Res Hum Retroviruses 20:355–364. doi: 10.1089/088922204323048104. [DOI] [PubMed] [Google Scholar]
- 51.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 52.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hosseinipour MC, Gupta RK, Van Zyl G, Eron JJ, Nachega JB. 2013. Emergence of HIV drug resistance during first- and second-line antiretroviral therapy in resource-limited settings. J Infect Dis 207(Suppl 2):S49–S56. doi: 10.1093/infdis/jit107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.WHO. 2014. Surveillance of HIV drug resistance in adults receiving ART (acquired HIV drug resistance). Concept note. WHO, Geneva, Switzerland. [Google Scholar]
- 55.WHO. 2012. HIV drug resistance report. HIV/AIDS Prorgramme. WHO, Geneva, Switzerland. [Google Scholar]
- 56.Vandamme AM, Camacho RJ, Ceccherini-Silberstein F, de Luca A, Palmisano L, Paraskevis D, Paredes R, Poljak M, Schmit JC, Soriano V, Walter H, Sonnerborg A. 2011. European recommendations for the clinical use of HIV drug resistance testing: 2011 update. AIDS Rev 13:77–108. [PubMed] [Google Scholar]
- 57.Rhee SY, Taylor J, Fessel WJ, Kaufman D, Towner W, Troia P, Ruane P, Hellinger J, Shirvani V, Zolopa A, Shafer RW. 2010. HIV-1 protease mutations and protease inhibitor cross-resistance. Antimicrob Agents Chemother 54:4253–4261. doi: 10.1128/AAC.00574-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gibson RM, Weber J, Winner D, Miller MD, Quinones-Mateu ME. 2014. Contribution of human immunodeficiency virus type 1 minority variants to reduced drug susceptibility in patients on an integrase strand transfer inhibitor-based therapy. PLoS One 9:e104512. doi: 10.1371/journal.pone.0104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quinones-Mateu ME, Avila S, Reyes-Teran G, Martinez MA. 2014. Deep sequencing: becoming a critical tool in clinical virology. J Clin Virol 61:9–19. doi: 10.1016/j.jcv.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang C, Mitsuya Y, Gharizadeh B, Ronaghi M, Shafer RW. 2007. Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance. Genome Res 17:1195–1201. doi: 10.1101/gr.6468307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fox MP, Rosen S. 2015. Retention of adult patients on antiretroviral therapy in low- and middle-income countries: systematic review and meta-analysis 2008-2013. J Acquir Immune Defic Syndr 69:98–108. doi: 10.1097/QAI.0000000000000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Braitstein P, Brinkhof MW, Dabis F, Schechter M, Boulle A, Miotti P, Wood R, Laurent C, Sprinz E, Seyler C, Bangsberg DR, Balestre E, Sterne JA, May M, Egger M. 2006. Mortality of HIV-1-infected patients in the first year of antiretroviral therapy: comparison between low-income and high-income countries. Lancet 367:817–824. doi: 10.1016/S0140-6736(06)68337-2. [DOI] [PubMed] [Google Scholar]
- 63.Cescon AM, Cooper C, Chan K, Palmer AK, Klein MB, Machouf N, Loutfy MR, Raboud J, Rachlis A, Ding E, Lima VD, Montaner JS, Rourke SB, Smieja M, Tsoukas C, Hogg RS. 2011. Factors associated with virological suppression among HIV-positive individuals on highly active antiretroviral therapy in a multi-site Canadian cohort. HIV Med 12:352–360. doi: 10.1111/j.1468-1293.2010.00890.x. [DOI] [PubMed] [Google Scholar]
- 64.Pillay P, Ford N, Shubber Z, Ferrand RA. 2013. Outcomes for efavirenz versus nevirapine-containing regimens for treatment of HIV-1 infection: a systematic review and meta-analysis. PLoS One 8:e68995. doi: 10.1371/journal.pone.0068995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hamers RL, Sigaloff KC, Wensing AM, Wallis CL, Kityo C, Siwale M, Mandaliya K, Ive P, Botes ME, Wellington M, Osibogun A, Stevens WS, Rinke de Wit TF, Schuurman R. 2012. Patterns of HIV-1 drug resistance after first-line antiretroviral therapy (ART) failure in 6 sub-Saharan African countries: implications for second-line ART strategies. Clin Infect Dis 54:1660–1669. doi: 10.1093/cid/cis254. [DOI] [PubMed] [Google Scholar]
- 66.Amoroso A, Etienne-Mesubi M, Edozien A, Ojoo S, Sheneberger R, Obiefune M, Hossain MB, Stafford K, Redfield RR. 2012. Treatment outcomes of recommended first-line antiretroviral regimens in resource-limited clinics. J Acquir Immune Defic Syndr 60:314–320. doi: 10.1097/QAI.0b013e31824e5256. [DOI] [PubMed] [Google Scholar]
- 67.Ramadhani HO, Thielman NM, Landman KZ, Ndosi EM, Gao F, Kirchherr JL, Shah R, Shao HJ, Morpeth SC, McNeill JD, Shao JF, Bartlett JA, Crump JA. 2007. Predictors of incomplete adherence, virologic failure, and antiviral drug resistance among HIV-infected adults receiving antiretroviral therapy in Tanzania. Clin Infect Dis 45:1492–1498. doi: 10.1086/522991. [DOI] [PubMed] [Google Scholar]
- 68.Sigaloff KC, Kayiwa J, Musiime V, Calis JC, Kaudha E, Mukuye A, Matama C, Nankya I, Nakatudde L, Dekker JT, Hamers RL, Mugyenyi P, Rinke De Wit TF, Kityo C. 2013. Short communication: high rates of thymidine analogue mutations and dual-class resistance among HIV-infected Ugandan children failing first-line antiretroviral therapy. AIDS Res Hum Retrovir 29:925–930. doi: 10.1089/aid.2012.0218. [DOI] [PubMed] [Google Scholar]
- 69.Reynolds SJ, Laeyendecker O, Nakigozi G, Gallant JE, Huang W, Hudelson SE, Quinn TC, Newell K, Serwadda D, Gray RH, Wawer MJ, Eshleman SH. 2012. Antiretroviral drug susceptibility among HIV-infected adults failing antiretroviral therapy in Rakai, Uganda. AIDS Res Hum Retrovir 28:1739–1744. doi: 10.1089/aid.2011.0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gupta RK, Goodall RL, Ranopa M, Kityo C, Munderi P, Lyagoba F, Mugarura L, Gilks CF, Kaleebu P, Pillay D. 2014. High rate of HIV resuppression after viral failure on first-line antiretroviral therapy in the absence of switch to second-line therapy. Clin Infect Dis 58:1023–1026. doi: 10.1093/cid/cit933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boltz VF, Zheng Y, Lockman S, Hong F, Halvas EK, McIntyre J, Currier JS, Chibowa MC, Kanyama C, Nair A, Owino-Ong'or W, Hughes M, Coffin JM, Mellors JW. 2011. Role of low-frequency HIV-1 variants in failure of nevirapine-containing antiviral therapy in women previously exposed to single-dose nevirapine. Proc Natl Acad Sci U S A 108:9202–9207. doi: 10.1073/pnas.1105688108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simen BB, Simons JF, Hullsiek KH, Novak RM, Macarthur RD, Baxter JD, Huang C, Lubeski C, Turenchalk GS, Braverman MS, Desany B, Rothberg JM, Egholm M, Kozal MJ. 2009. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis 199:693–701. doi: 10.1086/596736. [DOI] [PubMed] [Google Scholar]
- 73.Kagan RM, Johnson EP, Siaw M, Biswas P, Chapman DS, Su Z, Platt JL, Pesano RL. 2012. A genotypic test for HIV-1 tropism combining Sanger sequencing with ultradeep sequencing predicts virologic response in treatment-experienced patients. PLoS One 7:e46334. doi: 10.1371/journal.pone.0046334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lockman S, Hughes M, Sawe F, Zheng Y, McIntyre J, Chipato T, Asmelash A, Rassool M, Kimaiyo S, Shaffer D, Hosseinipour M, Mohapi L, Ssali F, Chibowa M, Amod F, Halvas E, Hogg E, Alston-Smith B, Smith L, Schooley R, Mellors J, Currier J. 2012. Nevirapine- versus lopinavir/ritonavir-based initial therapy for HIV-1 infection among women in Africa: a randomized trial. PLoS Med 9:e1001236. doi: 10.1371/journal.pmed.1001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lockman S, Hughes MD, McIntyre J, Zheng Y, Chipato T, Conradie F, Sawe F, Asmelash A, Hosseinipour MC, Mohapi L, Stringer E, Mngqibisa R, Siika A, Atwine D, Hakim J, Shaffer D, Kanyama C, Wools-Kaloustian K, Salata RA, Hogg E, Alston-Smith B, Walawander A, Purcelle-Smith E, Eshleman S, Rooney J, Rahim S, Mellors JW, Schooley RT, Currier JS. 2010. Antiretroviral therapies in women after single-dose nevirapine exposure. N Engl J Med 363:1499–1509. doi: 10.1056/NEJMoa0906626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.