Abstract
Increasing cases of infections caused by methicillin-resistant Staphylococcus aureus (MRSA) strains in healthy individuals have raised concerns worldwide. MRSA strains are resistant to almost the entire family of β-lactam antibiotics due to the acquisition of an extra penicillin-binding protein, PBP2a. Studies have shown that spoVG is involved in oxacillin resistance, while the regulatory mechanism remains elusive. In this study, we have found that SpoVG plays a positive role in oxacillin resistance through promoting cell wall synthesis and inhibiting cell wall degradation in MRSA strain N315. Deletion of spoVG in strain N315 led to a significant decrease in oxacillin resistance and a dramatic increase in Triton X-100-induced autolytic activity simultaneously. Real-time quantitative reverse transcription-PCR revealed that the expression of 8 genes related to cell wall metabolism or oxacillin resistance was altered in the spoVG mutant. Electrophoretic mobility shift assay indicated that SpoVG can directly bind to the putative promoter regions of lytN (murein hydrolase), femA, and lytSR (the two-component system). These findings suggest a molecular mechanism in which SpoVG modulates oxacillin resistance by regulating cell wall metabolism in MRSA.
INTRODUCTION
Staphylococcus aureus is a versatile and dangerous pathogen in humans, especially while the frequency of staphylococcal infections caused by methicillin-resistant Staphylococcus aureus (MRSA) increases steadily (1). S. aureus has four penicillin-binding proteins (PBPs) which are involved in the last stages of peptidoglycan synthesis (2). MRSA strains have acquired an extra PBP (PBP2a), encoded by the mecA gene on the staphylococcal cassette chromosome mec (SCCmec), which has a remarkably lower affinity for β-lactams than does PBP2 and is responsible for resisting these drugs (3). Besides the expression of PBP2a, which is responsible for a higher level of β-lactam resistance, S. aureus has another primary β-lactam resistance mechanism: the expression of β-lactamase enzymes, encoded by the blaZ gene, which hydrolyze β-lactams such as penicillin. In addition, several additional native genes, such as vraSR, pbp4, and fem (factor essential for methicillin resistance), have been identified as being essential to the full expression of oxacillin resistance (4–6). Inactivation of femA and pbp4 genes in the presence of an intact mecA gene usually results in strains with a low resistance to oxacillin. The VraS/VraR two-component regulatory system regulates the genes which are associated with cell wall biosynthesis, such as pbp2, sgtB, and murZ (7).
Most MRSA and glycopeptide-intermediately resistant Staphylococcus aureus (GISA) isolates are resistant to Triton X-100-induced autolysis, while instances of reduced resistance have been observed with concomitant increases of the autolysis rate (8–11). The expression of autolytic enzymes, including Atl, LytM, LytN, and Sle1 (12–15), is controlled by pleiotropic regulators, such as MgrA, SarV, SarA, LytSR, and ArlSR (autolysis-related locus) (16–20). Downstream of the lytSR operon is the lrgAB locus, the expression of which is regulated by LytSR (17). The cidABC and lrgAB operons may have an important role in the control of staphylococcal murein hydrolase activity and encode proteins analogous to the bacteriophage-encoded holins and antiholins (21–23).
SpoVG was initially identified in Bacillus subtilis and is involved in an unknown mechanism in sporulation (24). In nonsporulating bacteria, the σB-controlled spoVG affects capsule synthesis, expression of virulence factors, and antibiotic resistance (25–27). SpoVG is considered a site-specific DNA-binding protein (28). Except that spoVG is under the control of σB, a small RNA SprX negatively regulates spoVG expression by direct antisense pairings at the internal translation initiation signals of spoVG, without affecting YabJ translation (29). SpoVG is one of the target genes under the control of SprX, which is involved in glycopeptide resistance. However, the molecular mechanism of how spoVG participates in antibiotic resistance was largely unknown. In this study, we have investigated the role of spoVG in autolysis and resistance to oxacillin in MRSA strain N315.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were cultivated with shaking (220 rpm) in lysogeny broth (LB) medium (Oxoid) or on lysogeny broth agar (LA) at 37°C. S. aureus strains were grown with shaking (220 rpm) in tryptic soy broth (TSB) medium (Difco) or on tryptic soy agar (TSA) at 37°C. When required, the media were supplemented with 100 μg/ml of ampicillin or 50 μg/ml of kanamycin for E. coli and 15 μg/ml of chloromycetin for S. aureus strains.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| S. aureus strains | ||
| RN4220 | 8325-4, r−, initial recipient for modification of plasmids which are introduced into S. aureus from E. coli | NARSAb |
| N315 | HA-MRSA, SCCmec type II | NARSA |
| N315ΔspoVG | N315 strain deletion of spoVG | This study |
| E. coli strains | ||
| Trans1-T1 | Clone host strain; F− Φ80 (lacZ) ΔM15ΔlacX74 hsdR (rK− mK+) ΔrecA1398 endA1 tonA | TransGen |
| BL21(DE3) | Express strain; F− ompT hsdSB (rB− mB−) gal dcm (DE3) | TransGen |
| Plasmids | ||
| pBTs | Shuttle vector, temp sensitive, Ampr Chlr | |
| pBTsΔspoVG | pBTs derivative, for spoVG deletion in strain N315, Ampr Chlr | This study |
| pET28a(+) | Expression vector with a hexahistidine tag, Kanr | Novagen |
| pETSpoVG | pET28a(+) derivative, with ORF of spoVG, Kanr | This study |
| pLI50 | Shuttle vector, Ampr Chlr | Addgene |
| pLIspoVG | pLI50 derivative, harboring ORF of spoVG and its promoter, Ampr Chlr | This study |
r−, restriction system negative; Kanr, kanamycin resistant; Ampr, ampicillin resistant; Chlr, chloramphenicol resistant.
NARSA, Network on Antimicrobial Resistance in Staphylococcus aureus (https://www.beiresources.org/Collection/53/NARSA.aspx).
Construction of the spoVG mutant strain.
To create the spoVG mutant strain without extra genes introduced, the plasmid pBTs was used as previously described (30). The upstream and downstream regions of spoVG, which were amplified with primer pairs spoVG-mutant-up-F/spoVG-mutant-up-R and spoVG-mutant-down-F/spoVG-mutant-down-R (see Table S1 in the supplemental material), were ligated by SLiCE (31) to form an up-down fragment. Briefly, the two fragments were designed with a 20-bp overlap, the fragments (50 to 200 ng) were mixed at equimolar amounts, and then 1 μl of 1× SLiCE buffer (500 mM Tris-HCl, 100 mM MgCl2, 10 mM dithiothreitol [DTT]; pH 7.5), 1 μl of SLiCE extract, 1 μl of 10 mM ATP, and double-distilled water (ddH2O) were added to a final volume of 10 μl. The mixed solution was incubated at 37°C for 1 h and then amplified with primers spoVG-mutant-up-F/spoVG-mutant-down-R. The resultant fragment was digested with KpnI/EcoRI and then cloned into the KpnI/EcoRI-digested plasmid pBTs. The resulting plasmid, pBTsΔspoVG, was first electroporated into S. aureus strain RN4220 for modification and subsequently transformed into strain N315. An allelic replacement mutant was selected using a previously described method (32) and was further confirmed by PCR and sequencing.
Plasmid construction.
For the construction of complementary plasmid pLIspoVG, DNA fragments covering the open reading frame (ORF) of spoVG and the promoter of the yabJ-spoVG operon were amplified from N315 genomic DNA using primer pairs spoVG-complement-F/spoVG-complement-R and spoVG-mutant-up-F/PspoVG-R (see Table S1 in the supplemental material), respectively. The two DNA fragments were ligated by SLiCE and the resulting fragment was then cloned into the KpnI-digested plasmid pLI50 (33).
The protein expression plasmid pETSpoVG was constructed by amplifying the ORF of spoVG with primers SpoVG-exp-F and SpoVG-exp-R (see Table S1) from strain N315 chromosomal DNA. The PCR product was digested with NcoI and XhoI and then cloned into the expression vector pET28a (+) (Novagen). The recombinant plasmid was sequenced to confirm that spoVG was free of mutations and in frame with the hexahistidine tag.
Oxacillin susceptibility assay.
Determination of oxacillin MICs was performed by broth microdilution, as recommended by the Clinical and Laboratory Standards Institute (34). Population analysis profiles were established by plating appropriate dilutions of direct colony suspension on Mueller-Hinton agar with 2% NaCl containing increasing concentrations of oxacillin. The numbers of CFU were determined after 24 h of incubation at 37°C.
The growth experiments were performed as described elsewhere (20). In brief, overnight cultures of S. aureus were diluted into fresh TSB (Difco) with or without oxacillin to yield a starting optical density at 600 nm (OD600) of 0.05 and inoculated into 50-ml flasks in a final volume of 10 ml. Oxacillin was added at sub-MIC. The mixtures were then incubated with constant shaking (220 rpm) at 37°C, with the OD600 measured hourly for total 9 h using a spectrophotometer (DU 800; Beckman Coulter). These assays were repeated at least three times, with similar results.
Triton X-100-induced autolysis assay.
Triton X-100-stimulated autolysis was measured as described previously (35). Overnight-grown bacterial cells were diluted to an OD600 of 0.05 in TSB and allowed to grow to the early exponential (OD600 = 0.8) phase at 37°C with shaking (220 rpm). Cells were harvested, washed twice with 0.05 M Tris-HCl buffer (pH 7.5), resuspended in the original volume of Tris-HCl (0.05 M; pH 7.5) containing 0.05% (vol/vol) Triton X-100, incubated at 37°C with shaking, and checked for lysis by measuring the progressive decrease in absorbance (OD600) at half-hour intervals using a microplate reader (Elx800; Bio-Tek). The experiment was repeated at least three times, with similar results.
Total RNA isolation and real-time quantitative reverse transcription-PCR (qRT-PCR).
Overnight cultures of S. aureus were diluted 1:100 in TSB and cultivated to early exponential (OD600 = 0.5), mid-logarithmic (OD600 = 2.5), and early stationary (OD600 = 6) phases. Cells were harvested and processed with 1 ml of RNAiso plus (TaKaRa) in combination with 0.1-mm-diameter silica beads in a FastPrep-24 automated system (MP Biomedicals). Residual DNA was removed with RNase-free DNase I (TaKaRa).
For the reverse transcription, the cDNAs were synthesized using a PrimeScript 1st Strand cDNA synthesis kit (TaKaRa) and qRT-PCR was performed with SYBR Premix Ex Taq (TaKaRa) using the StepOne real-time PCR system (Applied Biosystems). The quantity of cDNA measured by real-time PCR was normalized to the abundance of hu cDNA (36). All qRT-PCR assays were repeated at least three times.
Purification of SpoVG-His6.
Expression and purification of the recombinant SpoVG-His6 protein were conducted as described previously, with modification (28). E. coli BL21(DE3) was transformed with the protein expression plasmid pETSpoVG, and the transformant was cultivated in LB at 37°C to an OD600 of 0.5 and induced with 0.5 mM isopropyl-β-d-1-thiogalactopyranoside at 37°C for an additional 3 h. The cells were harvested and lysed by sonication in lysis buffer (50 mM Tris-HCl [pH 8.0], 300 mM NaCl). The hexahistidine-tagged SpoVG protein was purified with a nickel-nitrilotriacetic acid-agarose solution (Qiagen) by following the manufacturer's recommendation. The bound protein was eluted with an elution buffer (200 mM imidazole, 50 mM Tris-HCl [pH 8.0], 300 mM NaCl). The imidazole in the eluent was removed using a Centrifuge Biomax-5 column (Millipore), and the protein was stored with storage buffer (50 mM Tris-HCl, 25 mM KCl, 100 nM DTT, 10% [vol/vol] glycerol, 0.01% [vol/vol] Tween 20) at −80°C until use. Protein purity and concentration were assessed via SDS-PAGE and the bicinchoninic acid assay with bovine serum albumin as the standard. The final concentration of purified SpoVG was 30 μM.
EMSA.
To determine whether the purified SpoVG binds to lytN, lytSR, and femA promoters, the biotin-labeled DNA fragments containing the putative promoter regions of lytN (254 bp), lytSR (253 bp), or femA (273 bp) were obtained by PCR with primers listed in Table S1 in the supplemental material. For electrophoretic mobility shift assay (EMSA), the biotin-labeled promoter regions of lytN (1 fmol), lytS (1 fmol), or femA (2 fmol) were incubated at 25°C for 30 min with various amounts of SpoVG in 10 μl of incubation buffer [50 mM Tris-HCl (pH 7.5), 1 mM DTT, 150 nM EDTA, 50 ng/ml of poly(dI-dC)]. After incubation, the mixtures were electrophoresed in a 5% native polyacrylamide gel in 1× Tris-borate-EDTA (TBE) buffer and then transferred to a nylon membrane in 0.5× TBE buffer. The band shifts were detected and analyzed according to the manufacturer's instructions. The images were obtained using ImageQuant LAS 4000 (GE). The unlabeled fragments of each promoter were added to the labeled fragments at a ratio of approximately 100:1 as specific competitors. The unlabeled DNA fragment of the pta ORF (100-fold or 200-fold) was added as a nonspecific competitor.
RESULTS
Influence of spoVG on oxacillin resistance in MRSA strain N315.
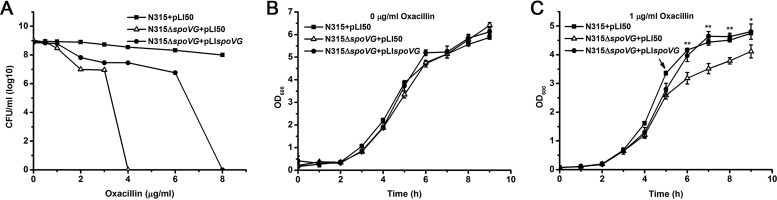
Either deletion of the yabJ-spoVG operon or inhibition of spoVG expression reduces antibiotic resistance in Staphylococcus aureus (25, 29). To investigate the regulatory mechanism in which SpoVG is involved in antibiotic resistance, we deleted spoVG in MRSA strain N315. Oxacillin resistance in the spoVG mutant was significantly reduced and the complementary strain exhibited a partial effect as shown by population analysis profiles (Fig. 1A). We also tested cell growth in TSB broth without antibiotic or exposed to 1 μg/ml of oxacillin (representing approximately one-quarter MIC for the mutant strain) at the 5th hour. No significant difference was observed when cells were grown in TSB broth without oxacillin (Fig. 1B), whereas the spoVG mutant exhibited a clear growth defect compared to the wild type in the medium containing oxacillin (Fig. 1C). In addition, there is no significant difference in vancomycin and teicoplanin resistance between the spoVG mutant and wide-type strains as shown by population analysis profiles (see Fig. S1 in the supplemental material). Ceftizoxime resistance in the spoVG mutant was significantly reduced, and the complementary strain also had a partial complementary action (see Fig. S1).
FIG 1.
Oxacillin susceptibility assay. (A) Deletion of spoVG reduced oxacillin resistance in strain N315. The oxacillin resistance levels were evaluated by population analysis profiles as recommended by the Clinical and Laboratory Standards Institute. The colonies were counted after incubation at 37°C for 24 h. (B and C) Growth of the wild-type (N315+pLI50), the spoVG mutant (N315ΔspoVG+pLI50), and the spoVG complementary (N315ΔspoVG+pLIspoVG) strains in TSB broth at 37°C containing 0 or 1 μg/ml of oxacillin at the 5th hour. Statistically significant differences, calculated by the unpaired two-tailed Student t test, are indicated as follows: NS, not significant (P > 0.05); *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
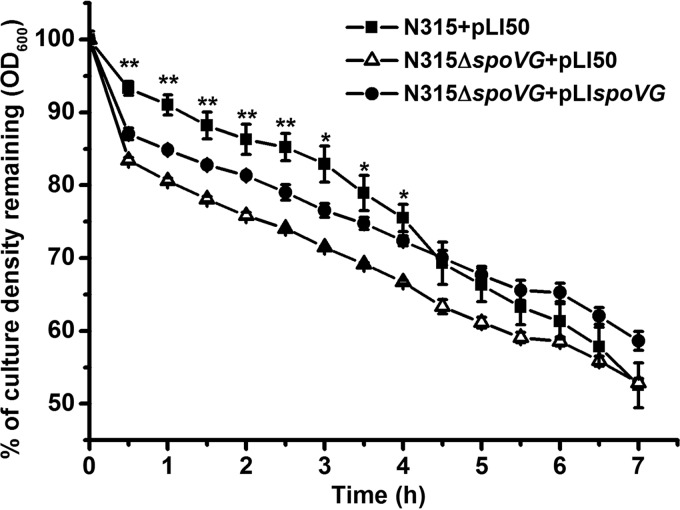
Effect of the spoVG mutation on Triton X-100-induced autolysis.
Autolysis is commonly associated with the killing mechanism of penicillin and β-lactams. We examined Triton X-100-induced autolytic activity in the wild-type and spoVG mutant strains. The spoVG mutant exhibited increased autolysis rates compared with those of the wild type. The phenotype was restored by introducing the complementary plasmid pLIspoVG (Fig. 2), indicating that cell wall turnover was modulated by SpoVG.
FIG 2.
Triton X-100-induced autolysis. Autolysis of the wild-type (N315+pLI50), the spoVG mutant (N315ΔspoVG+pLI50), and the spoVG complementary (N315ΔspoVG+pLIspoVG) strains at 37°C in Tris-HCl buffer containing 0.05% Triton X-100 was determined by measuring changes in optical densities upon exposure to the detergent. *, P < 0.05; **, P < 0.01.
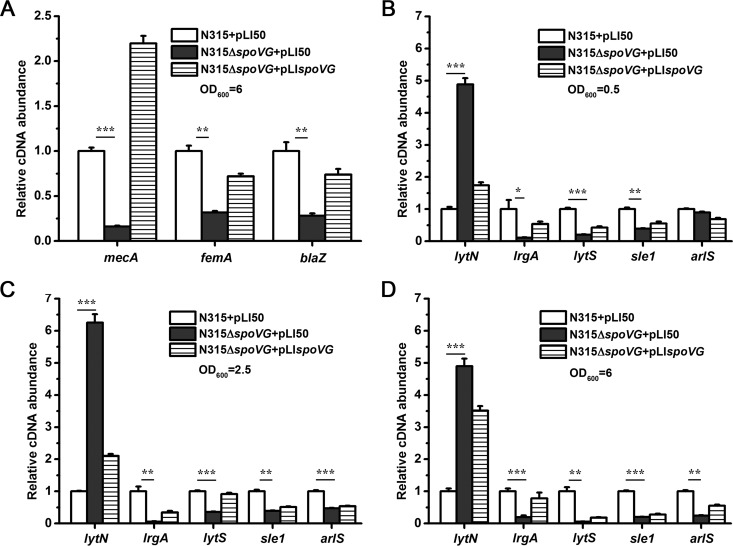
Transcriptional analysis by qRT-PCR.
The data presented above clearly indicate that SpoVG participates in the control of oxacillin resistance as well as in autolysis. Many genes involved in cell wall synthesis or cell wall degradation can influence methicillin resistance or autolysis. To determine whether the expression of these genes was altered in the spoVG mutant, we performed qRT-PCR to examine the mRNA levels of 17 potential target genes, among which 8 genes were associated with cell wall synthesis or oxacillin resistance (femA, mecA, blaZ, pbp1, pbp2, pbp3, pbp4, and vraS) and 9 genes were implicated in cell wall degradation (atl, lytN, lrgA, arlS, sle1, lytS, cidA, lytM, and sarA). The transcriptional levels of mecA, femA, and blaZ were decreased in the spoVG mutant during the early stationary phase of growth but not markedly altered at early exponential and mid-logarithmic phases (Fig. 3A). In addition to these genes, other target genes involved in methicillin resistance (pbp1, pbp2, pbp3, pbp4, and vraS) were not significantly changed in the mutant strain (see Fig. S2 in the supplemental material). A significant increase in the transcript level of lytN, encoding a murein hydrolase, was observed in the spoVG mutant compared with those in the wild-type and complemented strains in the different growth stages (Fig. 3B to D). In contrast, the mRNA levels of lrgA (antiholin) and sle1 (murein hydrolase) were significantly reduced in the spoVG mutant (Fig. 3B to D). The transcriptional levels of lytS and arlS were also found to be decreased in the mutant strain (Fig. 3B to and D). The transcript levels of atl, lytM, cidA, mgrA, and sarA were not significantly affected in the spoVG mutant (see Fig. S2). These data indicate that SpoVG controls regulatory genes as well as target genes involved in cell wall metabolism.
FIG 3.
Transcriptional analysis of cell wall-related genes. The relative transcription levels of several cell wall biosynthesis- and hydrolysis-related genes in the wild-type (N315+pLI50), the spoVG mutant (N315ΔspoVG+pLI50), and the spoVG complementary (N315ΔspoVG+pLIspoVG) strains are shown. (A) Relative transcription levels of femA, mecA, and blaZ genes in early stationary phase of growth. (B to D) Relative mRNA levels of target genes (lytN, lrgA, and sle1) and regulatory loci (lytSR and arlSR) involved in autolysis in early exponential (OD600 = 0.5), mid-logarithmic (OD600 = 2.5), and early stationary (OD600 = 6) phases. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
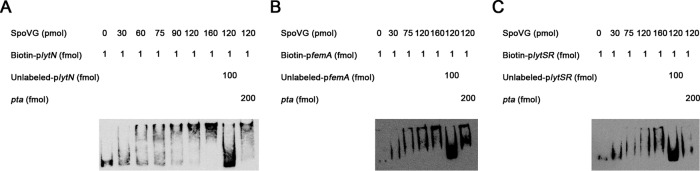
Binding of SpoVG to the putative promoter regions of lytN, femA, and lytSR.
To test whether the target genes are under the direct control of SpoVG, EMSA was performed with biotin-labeled putative promoter regions and the recombinant SpoVG (SpoVG-His6). The results showed that SpoVG can retard the mobility of the lytN promoter in a dose-dependent manner (Fig. 4A). This shifted band disappeared in the presence of an approximately 100-fold excess of unlabeled lytN promoter DNA, but not in the presence of a 200-fold excess of an unlabeled coding sequence DNA of pta. EMSA was also performed using the promoter regions of femA and lytSR, and similar band shift patterns were observed with the femA and lytSR promoters (Fig. 4B and C). These data suggest that SpoVG can regulate the expression of lytN, femA, and lytSR by directly binding to their promoter regions.
FIG 4.
Electrophoretic mobility shift assay of the purified SpoVG with the biotin-labeled DNA fragments containing the putative promoter regions of lytN (254 bp), femA (273 bp), or lytSR (253 bp). Increasing concentrations of purified SpoVG and 1 or 2 fmol of the biotin-labeled probe were used in the reactions. The specific competitor concentration was 100-fold that of the labeled promoter, and the concentration of the nonspecific competitor (pta) was 100-fold or 200-fold.
DISCUSSION
Previous studies have shown that SpoVG is the major factor of the yabJ-spoVG operon required in S. aureus for antibiotic resistance. In this study, we have also found that spoVG deletion can reduce oxacillin resistance in MRSA strain N315. SpoVG can directly regulate fmtB, which is one of the fem or aux genes (26, 28). It has been reported that femA, also one of fem or aux genes, is involved in the synthesis of pentaglycin cross-links (5). Our studies indicate that SpoVG acts as a positive regulator for femA. A decrease of the mRNA level of femA in the spoVG mutant implies that the synthesis process of peptiglycan cross-links may be damaged, thus reducing oxacillin resistance. With the normal function of PBP2, the decreased expression of mecA is likely to be associated with the lower resistance to oxacillin. In addition, the decrease in the transcript level of blaZ in the spoVG mutant suggests that the ability to hydrolyze β-lactams decreased. PBP2 is the only native PBP which is capable of both transpeptidation and transglycosylation of the peptidoglycan in S. aureus. PBP2a is capable of replacing the transpeptidase domain of PBP2, and the transglycosylase domain of PBP2 is necessary for resistance (37, 38). PBP4 has a low affinity for most β-lactam antibiotics and possesses transpeptidase and carboxypeptidase activities. The deletion of pbp4 in MRSA strain MW2 affects the transcription of pbp2 in cells challenged with oxacillin (6). Disruption of pbp4 also caused increased sensitivity to ceftizoxime in the ZOX3 mutant, which has a decreased affinity of its PBP2 variant for ceftizoxime (39). Therefore, two native S. aureus transpeptidases (PBP2 and PBP4) and an acquired transpeptidase (PBP2A) are likely to cooperate functionally in the biosynthesis of peptidoglycan and susceptibility to antimicrobial agents. How the activities of PBP1, which is involved in the enhancement of the activity of daptomycin against MRSA in the presence of β-lactam antibiotics, and the “nonessential” PBP3 of S. aureus interact in this cooperative system remains to be explored (40, 41). The VraS/VraR system regulates many cell wall biosynthesis-related genes, and the inactivation of it yielded significant decrease of resistance against teicoplanin, β-lactams, bacitracin, and fosfomycin (7). However, the transcript levels of pbp1, pbp2, pbp3, pbp4, and vraS were not affected by SpoVG in strain N315. In addition, our results indicate that there were no dramatic differences in vancomycin and teicoplanin resistances between the spoVG mutant and wild-type strains. This result is not in agreement with the previous reports (25, 29), perhaps due to the diverse genetic backgrounds of strains.
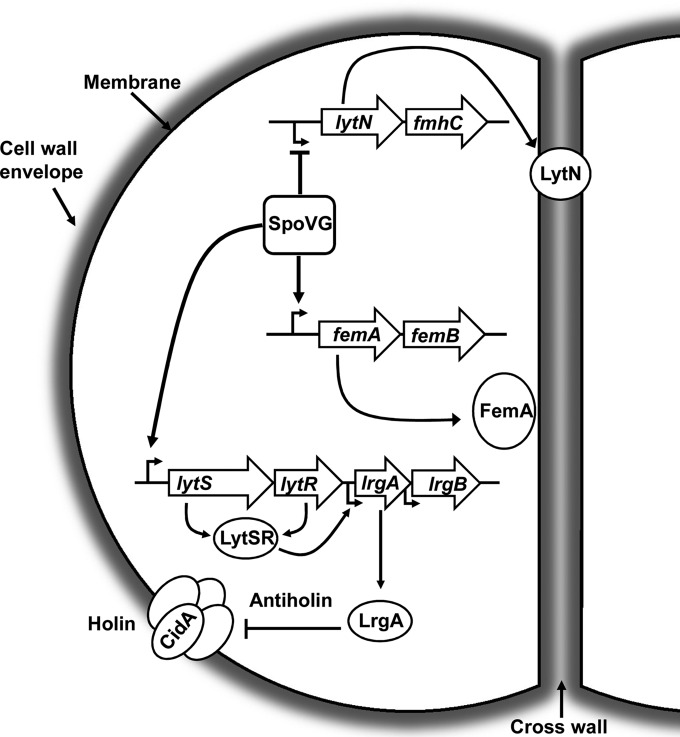
Since our data in this study were consistent with the previous report that reduced resistance was observed with concomitant increases of autolysis rate in S. aureus isolates, we further investigated the mRNA levels of cell wall degradation-related genes by qRT-PCR. Two regulatory systems, LytSR and ArlRS, have been shown to modulate autolytic activity negatively (17, 22). LytSR positively regulates lrgA and lrgB, a set of genes downstream of lytSR. Our studies indicate that SpoVG acts as a positive regulator for lytSR and, in turn, activates lrgA and leads to the repression of murein hydrolase activity. LytN, a murein hydrolase, is negatively regulated by SpoVG, which is consistent with the hypothesis that SpoVG is a negative regulator of autolysis in S. aureus. Our data suggest that SpoVG indirectly affects the expressions of arlRS and sle1. Sle1 is a peptidoglycan hydrolase which has been implicated in cell separation of S. aureus. But, unlike Atl (a major autolysin), Sle1 is not directly involed in autolysis of S. aureus (14). In summary, these results indicate that SpoVG as a transcriptional factor regulates the expression of lytN, femA, and lytS genes involved in cell wall metabolism (Fig. 5). Even though we identified several genes involved in oxacillin resistance and autolysis under the control of SpoVG, it cannot be excluded that other genes or small regulatory RNAs may be involved in the regulatory process.
FIG 5.
SpoVG is involved in the control of cell wall metabolism and oxacillin resistance in Staphylococcus aureus strain N315. SpoVG positively regulates the expression of femA, which is involved in the synthesis of pentaglycin cross-links. LytN, a murein hydrolase, is negatively regulated by SpoVG, which is consistent with the hypothesis that SpoVG is a negative regulator of autolysis in S. aureus. Besides, SpoVG acts as a positive regulator for lytSR and, in turn, activates lrgA, leading to the repression of murein hydrolase activity.
Previous studies have shown that the 5′ noncoding regions of cap5, fmtB, esxA, and lukED all contain at least two 5′-TAATTT/A-3′ sequences to which SpoVG binds with high affinity and specificity (28). In our study, we have also found at least two 5′-TAATTT/A-3′ sequences existing in the 5′ noncoding regions of lytN, femA, and lytSR. Whether this motif or other surrounding DNA sequences or structures contribute to the binding of SpoVG to target genes remains to be determined.
In conclusion, our findings reveal that SpoVG modulates β-lactam antibiotic resistance through promoting cell wall synthesis and inhibiting cell wall degradation in the presence of an intact mecA gene. While the molecular mechanism of mecA expression regulated by mecR1-mecI has been widely recognized, antibiotic resistance caused by auxiliary factors deserves further study.
Supplementary Material
ACKNOWLEDGMENT
This study was supported by the National Natural Science Foundation of China (81371850).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00026-16.
REFERENCES
- 1.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Ghuysen JM. 1991. Serine β-lactamases and penicillin-binding proteins. Annu Rev Microbiol 45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 3.Hartman BJ, Tomasz A. 1984. Low-affinity penicillin-binding protein associated with β-lactam resistance in Staphylococcus aureus. J Bacteriol 158:513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle-Vavra S, Yin S, Daum RS. 2006. The VraS/VraR two-component regulatory system required for oxacillin resistance in community-acquired methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett 262:163–171. doi: 10.1111/j.1574-6968.2006.00384.x. [DOI] [PubMed] [Google Scholar]
- 5.Maidhof H, Reinicke B, Blumel P, Bergerbachi B, Labischinski H. 1991. femA, which encodes a factor essentia for expression of methicillin resistance, affects glycine content of peptidoglycan in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J Bacteriol 173:3507–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Memmi G, Filipe SR, Pinho MG, Fu Z, Cheung A. 2008. Staphylococcus aureus PBP4 is essential for β-lactam resistance in community-acquired methicillin-resistant strains. Antimicrob Agents Chemother 52:3955–3966. doi: 10.1128/AAC.00049-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol 49:807–821. [DOI] [PubMed] [Google Scholar]
- 8.Renzoni A, Barras C, Francois P, Charbonnier Y, Huggler E, Garzoni C, Kelley WL, Majcherczyk P, Schrenzel J, Lew DP, Vaudaux P. 2006. Transcriptomic and functional analysis of an autolysis-deficient, teicoplanin-resistant derivative of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 50:3048–3061. doi: 10.1128/AAC.00113-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sieradzki K, Tomasz A. 1997. Inhibition of cell wall turnover and autolysis by vancomycin in a highly vancomycin-resistant mutant of Staphylococcus aureus. J Bacteriol 179:2557–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maki H, Yamaguchi T, Murakami K. 1994. Cloning and characterization of a gene affecting the methicillin resistance level and the autolysis rate in Staphylococcus aureus. J Bacteriol 176:4993–5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyle-Vavra S, Challapalli M, Daum RS. 2003. Resistance to autolysis in vancomycin-selected Staphylococcus aureus isolates precedes vancomycin-intermediate resistance. Antimicrob Agents Chemother 47:2036–2039. doi: 10.1128/AAC.47.6.2036-2039.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramadurai L, Jayaswal RK. 1997. Molecular cloning, sequencing, and expression of lytM, a unique autolytic gene of Staphylococcus aureus. J Bacteriol 179:3625–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oshida T, Sugai M, Komatsuzawa H, Hong YM, Suginaka H, Tomasz A. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-L-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc Natl Acad Sci U S A 92:285–289. doi: 10.1073/pnas.92.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kajimura J, Fujiwara T, Yamada S, Suzawa Y, Nishida T, Oyamada Y, Hayashi I, Yamagishi JI, Komatsuzawa H, Sugai M. 2005. Identification and molecular characterization of an N-acetylmuramyl-L-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus. Mol Microbiol 58:1087–1101. doi: 10.1111/j.1365-2958.2005.04881.x. [DOI] [PubMed] [Google Scholar]
- 15.Frankel MB, Hendrickx APA, Missiakas DM, Schneewind O. 2011. LytN, a murein hydrolase in the cross-wall compartment of Staphylococcus aureus, is involved in proper bacterial growth and envelope assembly. J Biol Chem 286:32593–32605. doi: 10.1074/jbc.M111.258863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manna AC, Ingavale SS, Maloney M, van Wamel W, Cheung AL. 2004. Identification of sarV (SA2062), a new transcriptional regulator, is repressed by SarA and MgrA (SA0641) and involved in the regulation of autolysis in Staphylococcus aureus. J Bacteriol 186:5267–5280. doi: 10.1128/JB.186.16.5267-5280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunskill EW, Bayles KW. 1996. Identification of LytSR-regulated genes from Staphylococcus aureus. J Bacteriol 178:5810–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingavale SS, Van Wamel W, Cheung AL. 2003. Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol Microbiol 48:1451–1466. doi: 10.1046/j.1365-2958.2003.03503.x. [DOI] [PubMed] [Google Scholar]
- 19.Memmi G, Nair DR, Cheung A. 2012. Role of ArlRS in autolysis in methicillin-sensitive and methicillin-resistant Staphylococcus aureus strains. J Bacteriol 194:759–767. doi: 10.1128/JB.06261-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trotonda MP, Xiong YQ, Memmi G, Bayer AS, Cheung AL. 2009. Role of mgrA and sarA in methicillin-resistant Staphylococcus aureus autolysis and resistance to cell wall-active antibiotics. J Infect Dis 199:209–218. doi: 10.1086/595740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A 104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groicher KH, Firek BA, Fujimoto DF, Bayles KW. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J Bacteriol 182:1794–1801. doi: 10.1128/JB.182.7.1794-1801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadykov MR, Bayles KW. 2012. The control of death and lysis in staphylococcal biofilms: a coordination of physiological signals. Curr Opin Microbiol 15:211–215. doi: 10.1016/j.mib.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuno K, Sonenshein AL. 1999. Role of SpoVG in asymmetric septation in Bacillus subtilis. J Bacteriol 181:3392–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulthess B, Meier S, Homerova D, Goerke C, Wolz C, Kormanec J, Berger-Bachi B, Bischoff M. 2009. Functional characterization of the sigma(B)-dependent yabJ-spoVG operon in Staphylococcus aureus: role in methicillin and glycopeptide resistance. Antimicrob Agents Chemother 53:1832–1839. doi: 10.1128/AAC.01255-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulthess B, Bloes DA, Francois P, Girard M, Schrenzel J, Bischoff M, Berger-Bachi B. 2011. The sigma(B)-dependent yabJ-spoVG operon is involved in the regulation of extracellular nuclease, lipase, and protease expression in Staphylococcus aureus. J Bacteriol 193:4954–4962. doi: 10.1128/JB.05362-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meier S, Goerke C, Woiz C, Seidl K, Homerova D, Schulthess B, Kormanec J, Berger-Baechi B, Bischoff M. 2007. sigma(B) and the sigma(B)-dependent arlRS and yabJ-spoVG loci affect capsule formation in Staphylococcus aureus. Infect Immun 75:4562–4571. doi: 10.1128/IAI.00392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jutras BL, Chenail AM, Rowland CL, Carroll D, Miller MC, Bykowski T, Stevenson B. 2013. Eubacterial SpoVG homologs constitute a new family of site-specific DNA-binding proteins. PLoS One 8(6):e66683. doi: 10.1371/journal.pone.0066683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eyraud A, Tattevin P, Chabelskaya S, Felden B. 2014. A small RNA controls a protein regulator involved in antibiotic resistance in Staphylococcus aureus. Nucleic Acids Res 42:4892–4905. doi: 10.1093/nar/gku149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu J, Zhang X, Liu X, Chen C, Sun B. 2015. Mechanism of reduced vancomycin susceptibility conferred by walK mutation in community-acquired methicillin-resistant Staphylococcus aureus strain MW2. Antimicrob Agents Chemother 59:1352–1355. doi: 10.1128/AAC.04290-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Werling U, Edelmann W. 2012. SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res 40(8):e55. doi: 10.1093/nar/gkr1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 33.Lee CY, Buranen SL, Ye ZH. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101–105. doi: 10.1016/0378-1119(91)90399-V. [DOI] [PubMed] [Google Scholar]
- 34.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, approved standard, 7th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 35.Sun H, Yang Y, Xue T, Sun B. 2013. Modulation of cell wall synthesis and susceptibility to vancomycin by the two-component system AirSR in Staphylococcus aureus NCTC8325. BMC Microbiol 13:286. doi: 10.1186/1471-2180-13-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valihrach L, Demnerova K. 2012. Impact of normalization method on experimental outcome using RT-qPCR in Staphylococcus aureus. J Microbiol Methods 90:214–216. doi: 10.1016/j.mimet.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Pinho MG, Filipe SR, De Lencastre H, Tomasz A. 2001. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus. J Bacteriol 183:6525–6531. doi: 10.1128/JB.183.22.6525-6531.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinho MG, de Lencastre H, Tomasz A. 2001. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc Natl Acad Sci U S A 98:10886–10891. doi: 10.1073/pnas.191260798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Łeski TA, Tomasz A. 2005. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J Bacteriol 187:1815–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinho MG, De Lencastre H, Tomasz A. 2000. Cloning, characterization, and inactivation of the gene pbpC, encoding penicillin-binding protein 3 of Staphylococcus aureus. J Bacteriol 182:1074–1079. doi: 10.1128/JB.182.4.1074-1079.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berti AD, Sakoulas G, Nizet V, Tewhey R, Rose WE. 2013. β-Lactam antibiotics targeting PBP1 selectively enhance daptomycin activity against methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 57:5005–5012. doi: 10.1128/AAC.00594-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.