Abstract
The rapid emergence of drug-resistant malaria parasites during the course of an infection remains a major challenge for providing accurate treatment guidelines. This is particularly important in cases of malaria treatment failure. Using a previously well-characterized case of malaria treatment failure, we show the utility of using next-generation sequencing for early detection of the rise and selection of a previously reported atovaquone-proguanil (malarone) drug resistance-associated mutation.
TEXT
Development of resistance to antimalarial drugs has become a major challenge. Molecular tools that can detect resistant alleles have been useful in population-based studies in tracking resistant parasites in countries where malaria is endemic. The detection of molecular markers of resistance has historically involved various PCR-based genotyping methods, such as sequence-specific PCR amplification, pyrosequencing, and Sanger sequencing of a genomic region containing one or more resistance mutations (1–4). One major limitation of these conventional genotyping methods is their detection limit of minor allele frequencies (MAFs). Some studies have reported that the detection of MAFs is limited to 15% to 30% of the population by Sanger sequencing or pyrosequencing (3, 5). This is particularly important in the context of treatment failures, where treatment with an antimalarial drug can exert selection pressure on minor populations of drug-resistant parasites. The inability of Sanger and/or pyrosequencing to detect minor alleles below certain thresholds can be overcome by the use of next-generation sequencing (NGS) methods, which can detect mutations with an MAF as low as 1% (6).
In this report, we investigated the use of an NGS method for early detection of drug resistance mutations in an imported case of malaria in the United States that was ultimately shown to be a well-characterized case of atovaquone-proguanil treatment failure (7).
A patient from the United States with a history of travel to Nigeria was diagnosed with uncomplicated Plasmodium falciparum malaria by microscopy and treated with the full 3-day treatment course of the drug combination atovaquone-proguanil (four 250/100-mg tablets once per day) on two occasions, once starting on day 0 (D0) and then again on D11. Four blood samples were obtained, on D0 prior to treatment, on D11 during a regularly scheduled follow-up visit, and on D34 and D35 when the patient presented with a recrudescent P. falciparum infection. DNA was extracted from the four whole-blood samples (D0, D11, D34, and D35) using the QIAamp DNA blood kit (catalog number 51104, Qiagen, USA). The cytochrome b gene (AIJ28900.1) from the P. falciparum mitochondrial genome was amplified using a high-fidelity polymerase (catalog number M530L, New England BioLabs, USA) with the forward primer 5′-CTATTAATTTAGTTAAAGCACAC-3′ and reverse primer 5′-ACAGAATAATCTCTAGCACCA-3′. These are the same primers that were used in the previous Sanger-sequencing protocol (7). Final amplicon products were cleaned using Agencourt AMPure XP beads (catalog number A63880, Beckman Coulter, USA) and visualized using gel electrophoresis. Sequencing-library preparation was performed using the Nextera XT v2 kit (catalog number FC-131-1024, Illumina, USA). Final amplicon purity and expected size were analyzed using the Bioanalyzer 2100 (Agilent, USA) prior to pooling the samples. Last, NGS was performed on an Illumina HiSeq 2500 using the SBSv2 kit. Final data quality filtering and analysis were performed using the Geneious R8 Pro v8.1.6 software. Only sequence reads with >200 bp, a Q20 score of >98%, and a minimum number (30) of low-quality bases were mapped to cytochrome b (AIJ28900.1), and polymorphisms were called only if they exceeded 2% of the total reads (MAF ≤ 2%). Polymorphisms were considered only if they were found on both the forward and reverse paired reads. The average coverage for the entire cytochrome b gene for all six samples ranged from 270,000 to 900,000 reads.
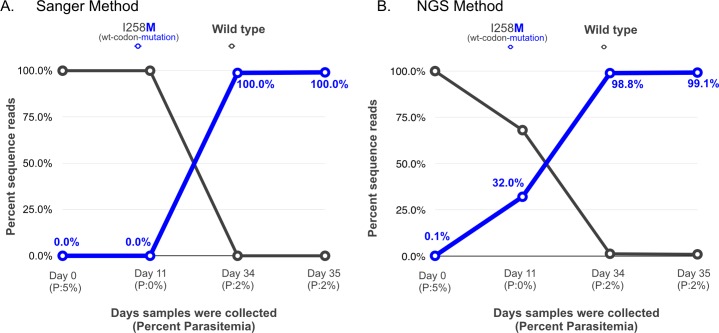
Sanger sequencing of the cytochrome b gene, encoding the molecular target of atovaquone, confirmed P. falciparum infection and revealed a wild-type cytochrome b sequence on D0 and D11 (only a single chromatographic trace was present on these days) and a previously unreported nonsynonymous mutation, I258M, on D34 and D35 (7). In contrast, using NGS, the I258M mutation was found at 0.1% as early as D0, at 32.0% on D11, at 98.8% on D34, and at 99.1% on D35 (Fig. 1), demonstrating the advantage of this method for the early detection of resistant alleles.
FIG 1.
Comparison of results between (A) Sanger sequencing and (B) next-generation sequencing of D0, D11, D34, and D35 patient samples. Percentage parasitemia is shown below each day (P:n%). Percentage sequence reads contributing to the specific atovaquone-proguanil resistance-associated loci in cytochrome b are shown on the y axis, and pretreatment (D0) and day of treatment failure samples (D11 to D35) are shown on the x axis.
This finding shows that NGS allowed for the detection of low-frequency mutations much earlier than that with conventional sequencing methods. While conventional methods such as Sanger sequencing provide longer read lengths (600 to 1,000 bp) than those provided by the latest Illumina sequencing platforms (250 to 350 bp), only one or two sequences are obtained per sequencing run, limiting the detection of minor alleles in a population. Next-generation sequencing is becoming more affordable and, on a per-sequence basis, is substantially cheaper than Sanger sequencing. The application of NGS in the context of treatment failures can detect drug resistance mutations before they are detectable by Sanger sequencing. While we detected the novel I258M mutation at 0.1% on D0, it was within the known 1% error margin of the Illumina sequencing platform and thus was not considered a real mutation (8, 9). Mutations were considered only at an MAF of greater than 2% and if found on both the forward and reverse reads. Therefore, it is most likely that the I258M mutation arose de novo. Antimalarials, such as atovaquone, have been shown to frequently give rise to de novo mutations during the course of treatment (10–13). This case illustrates how NGS techniques can provide a high-resolution picture of both major and minor alleles in clinical specimens. This is particularly applicable to malaria, in which phenotype-based approaches are impractical to implement in a typical clinical microbiology laboratory. Due to decreasing costs and an increasing list of applications, including multiplexing with multiple pathogens, NGS may soon be routine in such laboratories to manage common infections, such as Clostridium difficile or HIV infections (14). While analyzing for malaria resistance markers would not usually be the main focus in clinical microbiology laboratories, especially in countries with no malaria transmission, the flexibility of the NGS technology might allow easy incorporation of this additional capability (15). For example, the MiSeqDx system has already been approved by the FDA for clinical laboratory use for identifying cystic fibrosis-associated mutations with a turnaround time from sample to analyzed result of 24 to 32 h (16). Nonetheless, as an intermediate step, these technologies will first aid in the monitoring and follow-up of patients undergoing treatment for malaria rather than be used to guide immediate treatment at the presentation of the infection. In the future, not only would this technology permit the initial identification of the infecting malaria species, but it may also allow clinicians to rapidly select ideal antimalarial treatments at disease presentation and detect concerning mutations following the initiation of treatment.
ACKNOWLEDGMENTS
We thank Willard Dalton and Mary Eschete at the Terrebonne General Medical Center, Houma, Louisiana, who first identified and described the case.
The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Funding Statement
We acknowledge support from the Advanced Molecular Detection Initiative at the CDC. E.T. is supported by the Atlanta Research and Education Foundation.
REFERENCES
- 1.Wang P, Brooks DR, Sims PF, Hyde JE. 1995. A mutation-specific PCR system to detect sequence variation in the dihydropteroate synthetase gene of Plasmodium falciparum. Mol Biochem Parasitol 71:115–125. doi: 10.1016/0166-6851(95)00041-X. [DOI] [PubMed] [Google Scholar]
- 2.Alker AP, Mwapasa V, Meshnick SR. 2004. Rapid real-time PCR genotyping of mutations associated with sulfadoxine-pyrimethamine resistance in Plasmodium falciparum. Antimicrob Agents Chemother 48:2924–2929. doi: 10.1128/AAC.48.8.2924-2929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou Z, Poe AC, Limor J, Grady KK, Goldman I, McCollum AM, Escalante AA, Barnwell JW, Udhayakumar V. 2006. Pyrosequencing, a high-throughput method for detecting single nucleotide polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes of Plasmodium falciparum. J Clin Microbiol 44:3900–3910. doi: 10.1128/JCM.01209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alam MT, Vinayak S, Congpuong K, Wongsrichanalai C, Satimai W, Slutsker L, Escalante AA, Barnwell JW, Udhayakumar V. 2011. Tracking origins and spread of sulfadoxine-resistant Plasmodium falciparum dhps alleles in Thailand. Antimicrob Agents Chemother 55:155–164. doi: 10.1128/AAC.00691-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsiatis AC, Norris-Kirby A, Rich RG, Hafez MJ, Gocke CD, Eshleman JR, Murphy KM. 2010. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J Mol Diagn 12:425–432. doi: 10.2353/jmoldx.2010.090188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flaherty P, Natsoulis G, Muralidharan O, Winters M, Buenrostro J, Bell J, Brown S, Holodniy M, Zhang N, Ji HP. 2012. Ultrasensitive detection of rare mutations using next-generation targeted resequencing. Nucleic Acids Res 40:e2. doi: 10.1093/nar/gkr861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plucinski MM, Huber CS, Akinyi S, Dalton W, Eschete M, Grady K, Silva-Flannery L, Mathison BA, Udhayakumar V, Arguin PM, Barnwell JW. 2014. Novel mutation in cytochrome B of Plasmodium falciparum in one of two atovaquone-proguanil treatment failures in travelers returning from same site in Nigeria. Open Forum Infect Dis 1:ofu059. doi: 10.1093/ofid/ofu059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minoche AE, Dohm JC, Himmelbauer H. 2011. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems. Genome Biol 12:R112. doi: 10.1186/gb-2011-12-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Amore R, Ijaz UZ, Schirmer M, Kenny JG, Gregory R, Darby AC, Shakya M, Podar M, Quince C, Hall N. 2016. A comprehensive benchmarking study of protocols and sequencing platforms for 16S rRNA community profiling. BMC Genomics 17:55. doi: 10.1186/s12864-015-2194-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz E, Bujanover S, Kain KC. 2003. Genetic confirmation of atovaquone-proguanil-resistant Plasmodium falciparum malaria acquired by a nonimmune traveler to East Africa. Clin Infect Dis 37:450–451. doi: 10.1086/375599. [DOI] [PubMed] [Google Scholar]
- 11.White N, Pongtavornpinyo W. 2003. The de novo selection of drug-resistant malaria parasites. Proc Biol Sci 270:545–554. doi: 10.1098/rspb.2002.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musset L, Le Bras J, Clain J. 2007. Parallel evolution of adaptive mutations in Plasmodium falciparum mitochondrial DNA during atovaquone-proguanil treatment. Mol Biol Evol 24:1582–1585. doi: 10.1093/molbev/msm087. [DOI] [PubMed] [Google Scholar]
- 13.Cottrell G, Musset L, Hubert V, Le Bras J, Clain J, Atovaquone-Proguanil Treatment Failure Study Group. 2014. Emergence of resistance to atovaquone-proguanil in malaria parasites: insights from computational modeling and clinical case reports. Antimicrob Agents Chemother 58:4504–4514. doi: 10.1128/AAC.02550-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pak T, Kasarskis A. 2015. How next-generation sequencing and multiscale data analysis will transform infectious disease management. Clin Infect Dis 61:1695–1702. doi: 10.1093/cid/civ670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rusu LI, Wyres KL, Reumann M, Queiroz C, Bojovschi A, Conway T, Garg S, Edwards DJ, Hogg G, Holt KE. 2015. A platform for leveraging next generation sequencing for routine microbiology and public health use. Health Infect Sci Syst 3:S7. doi: 10.1186/2047-2501-3-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koser CU, Ellington MJ, Cartwright EJ, Gillespie SH, Brown NM, Farrington M, Holden MT, Dougan G, Bentley SD, Parkhill J, Peacock SJ. 2012. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog 8:e1002824. doi: 10.1371/journal.ppat.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]