Abstract
Salmonellosis is a major global foodborne infection, and strains that are resistant to a great variety of antibiotics have become a major public health concern. The aim of this study was to identify genes conferring resistance to fluoroquinolones and extended-spectrum β-lactams in nontyphoidal Salmonella (NTS) from patients and food-producing animals in China. In total, 133 and 21 NTS isolates from animals and humans, respectively, exhibiting concurrent resistance to ciprofloxacin and cefotaxime were cultured independently from 2009 to ∼2013. All of the isolates were identified, serotyped, and subjected to antimicrobial susceptibility testing. Importantly, the isolates with concurrent resistance to ciprofloxacin and cefotaxime all were confirmed as S. enterica serovar Indiana. The presence of fluoroquinolone resistance genes and extended-spectrum β-lactamases (ESBLs) was established by PCR and DNA sequencing. The occurrence and diversity of different genes conferring fluoroquinolone resistance [qepA, oqxAB, and aac(6′)-Ib-cr] with mutations in topoisomerase-encoding genes (gyrA and parC) and several ESBLs (including CTX-M-65, CTX-M-27, CTX-M-15, CTX-M-14, and CTX-M-14/CTX-M-15) were noteworthy. Genes located on mobile genetic elements were identified by conjugation and transformation. Pulsed-field gel electrophoresis, used to determine the genetic relationships between these isolates, generated 91 pulsotypes from 133 chicken isolates and 17 pulsotypes from the 21 clinical isolates that showed considerable diversity. Analysis of the pulsotypes obtained with the isolates showed some clones appeared to have existed for several years and had been disseminating between humans and food-producing animals. This study highlights the emergence of ciprofloxacin- and cefotaxime-resistant S. enterica serovar Indiana, posing a threat to public health.
INTRODUCTION
Salmonellosis, caused by Salmonella enterica, is a global foodborne disease of humans and livestock worldwide (1, 2). A total of 91,034 cases of human laboratory-confirmed Salmonella infections and 61 fatal cases were reported in 2012 in 27 European countries (3). In high-income regions of North America, there are an estimated 1.0 million Salmonella infections per year (4). In China, approximately 75% (30 million) of foodborne diseases every year are attributed to this bacterium (5). Specifically, children younger than 5 years were the group most at risk of infection by Salmonella, as noted in data obtained from several cities in China (2, 6, 7).
Salmonellosis is generally a self-limiting illness; however, antimicrobial agents may be required in severe cases, particularly in immunocompromised individuals, children, and the elderly (8). Fluoroquinolones and extended-spectrum β-lactams (ESBLs) are the front-line drugs of choice for treating salmonellosis (6, 9). However, in recent years, an increase in the occurrence of multidrug-resistant (MDR) Salmonella spp. expressing resistance to these compounds has been observed in several countries. Dissemination of antimicrobial resistance among nontyphoidal Salmonella isolates in humans is thought to be predominantly due to the use of antimicrobial agents in food animals (6, 10). Furthermore, infections with such drug-resistant Salmonella species are associated with increased morbidity and mortality.
Ciprofloxacin resistance is attributed mainly to mutations of quinolone resistance-determining regions (QRDRs) of the genes encoding the target bacterial topoisomerase enzymes (11). The presence of plasmid-mediated quinolone resistance (PMQR)-encoding genes also may contribute to the ciprofloxacin resistance phenotype (12). Although these PMQR genes confer only low-level resistance to fluoroquinolones, the presence of PMQR genes (particularly qnr genes) may provide a selective advantage for bacteria exposed to fluoroquinolones and facilitate the development of high-level chromosomal quinolone resistance.
On the other hand, it has been acknowledged that the horizontal transfer of plasmids carrying the ESBL genes is an important contributory factor in the epidemiology of this bacterial ecosystem. The predominant ESBL families of clinical importance include TEM, SHV, and CTX-M (13). In Salmonella, the most commonly found ESBL types in Asia are those of the CTX-M group, which are usually located on transmissible plasmids that have the capacity to disseminate among members of the Enterobacteriaceae (13). In other regions of the world, such as in the United States and Canada, the AmpC β-lactamase CMY-2 is the major contributor to ceftriaxone resistance in Salmonella (14).
Recently, concurrent resistance to ceftriaxone/cefotaxime and ciprofloxacin in S. enterica serovar Indiana has been reported for isolates cultured from food-producing animals (15, 16). Salmonella spp. have a variety of animal reservoirs and routes of transmission that can result in human infection (17). However, foods of animal origin, especially poultry and poultry products, often are involved in sporadic cases and outbreaks of human salmonellosis (15), and contaminated raw or undercooked chicken products are primary vehicles of Salmonella transmission to human beings (18). Consequently, concurrent resistance to ceftriaxone/cefotaxime and ciprofloxacin for S. enterica serovar Indiana poses a risk to humans, and this feature limits the treatment options available when the organisms are encountered. Currently, there is limited information describing the molecular epidemiology of ceftriaxone/cefotaxime- and ciprofloxacin-resistant S. enterica serovar Indiana isolates.
Therefore, the objectives of this study are to investigate and characterize those genes conferring resistance to cefotaxime and ciprofloxacin in coresistant S. enterica serovar Indiana isolates from both humans and food-producing animals in China.
MATERIALS AND METHODS
Ethics statement.
The fecal samples were acquired with written informed consent from the patients. This study was reviewed and approved by the ethics committee of the China National Center for Food Safety Risk Assessment (CFSA) according to the medical research regulations of the Ministry of Health, China. This research was conducted within China.
Bacteria and growth conditions.
In total, 133 S. enterica serovar Indiana isolates with concurrent resistance to ciprofloxacin and cefotaxime were recovered from poultry slaughterhouses (10.1%; n = 1,320; isolates from whole chicken carcasses after dehairing and precooling bath) in 3 cities (Hebi [n = 88], Kaifeng [n = 40], and Luohe [n = 5]) in Henan Province from February through November 2012. Twenty-one S. enterica serovar Indiana isolates with concurrent resistance to ciprofloxacin and cefotaxime were cultured from patients (2.6%; n = 802; isolates from fresh fecal swabs) in five different geographical regions (Dengfeng [n = 1], Jiyuan [n = 5], Shangqiu [n = 7], Zhenzhou [n = 3], and Zhoukou[n = 5]) in 28 sentinel hospitals and 6 regional Center for Disease Control and Prevention of Henan Province diagnostic laboratories, located in Henan Province, from 2009 to 2013. The ages of the patients varied from 1 month to 70 years, and more than half were less than 2 years old (Table 1).
TABLE 1.
Phenotypes and topoisomerase and PMQR genotypes of 21 human ciprofloxacin- and cefotaxime-resistant S. enterica serovar Indiana isolates
| Strain | Source | Age | Yr of isolation | MIC (mg/liter) |
QRDR amino acid substitutionsa |
Gene(s) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP | CTX | gyrA | gyrB | parC | parE | PMQR | β-Lactamase | ||||
| P1 | Zhoukou | 1 mo | 2009 | 128 | 128 | S83F/D87G | WT | T57S/S80R | WT | aac(6′)-Ib-cr, oqxAB | blaCTX-M-15 |
| P2 | Jiyuan | 7 yr | 2010 | 8 | 128 | S83F/D87G | WT | T57S/S80R | WT | oqxAB | blaCTX-M-14 |
| P3 | Zhoukou | 28 yr | 2010 | 128 | 128 | S83F/D87N | WT | T57S/S80R | WT | aac(6′)-Ib-cr, oqxAB | blaCTX-M-65 |
| P4 | Zhenzhou | 1 mo | 2011 | 32 | 128 | S83F/D87G | WT | T57S/S80R | WT | aac(6′)-Ib-cr | blaCTX-M-15 |
| P5 | Jiyuan | 28 yr | 2011 | 128 | 128 | S83F/D87N | WT | T57S/S80R | WT | aac(6′)-Ib-cr, oqxAB | blaCTX-M-65 |
| P6 | Jiyuan | 1 mo | 2011 | 128 | 128 | S83F/D87N | WT | T57S/S80R | WT | aac(6′)-Ib-cr, oqxAB | blaCTX-M-65 |
| P7 | Jiyuan | 36 yr | 2011 | 128 | 128 | S83F/D87N | WT | T57S/S80R | WT | aac(6′)-Ib-cr, oqxAB | blaCTX-M-65 |
| P8 | Jiyuan | 1 yr | 2011 | 128 | 128 | S83F/D87N | WT | T57S/S80R | WT | aac(6′)-Ib-cr, oqxAB | blaCTX-M-65 |
| P9 | Shangqiu | 22 yr | 2011 | 128 | 128 | S83F/D87G | WT | T57S/S80R | WT | aac(6′)-Ib-cr, oqxAB | blaCTX-M-15 |
| P10 | Shangqiu | 50 yr | 2011 | 128 | 128 | S83F/D87G | WT | T57S/S80R | WT | aac(6′)-Ib-cr, oqxAB | blaCTX-M-15 |
| P11 | Shangqiu | 49 yr | 2011 | 128 | 128 | S83F/D87G | WT | T57S/S80R | WT | aac(6′)-Ib-cr, oqxAB | blaCTX-M-14, blaCTX-M-15 |
| P13 | Shangqiu | 1 mo | 2011 | 128 | 128 | S83F/D87N | WT | T57S/S80R | WT | aac(6′)-Ib-cr, oqxAB | blaCTX-M-27 |
| P14 | Zhenzhou | 4 mo | 2012 | 128 | 128 | S83F/D87N | WT | T57S/S80R | WT | aac(6′)-Ib-cr, oqxAB | blaCTX-M-65 |
| P17 | Shangqiu | 15 mo | 2012 | 32 | 128 | S83F/D87G | WT | T57S/S80R | WT | oqxAB | blaCTX-M-14 |
| P18 | Zhoukou | 17 mo | 2012 | 128 | 128 | S83F/D87G | WT | T57S/S80R | WT | aac(6′)-Ib-cr, oqxAB | blaCTX-M-15 |
| P19 | Zhoukou | 9 mo | 2012 | 128 | 128 | S83F/D87G | WT | T57S/S80R | WT | aac(6′)-Ib-cr, oqxAB | blaCTX-M-65 |
| P21 | Shangqiu | 5 mo | 2012 | 128 | 128 | S83F/D87G | WT | T57S/S80R | WT | aac(6′)-Ib-cr, oqxAB | blaCTX-M-65 |
| P22 | Shangqiu | 74 yr | 2012 | 128 | 128 | S83F/D87G | WT | T57S/S80R | WT | aac(6′)-Ib-cr, oqxAB | blaCTX-M-65 |
| P23 | Dengfeng | 6 mo | 2012 | 128 | 128 | S83F/D87G | WT | T57S/S80R | WT | aac(6′)-Ib-cr, oqxAB | blaCTX-M-65 |
| P24 | Zhenzhou | 9 yr | 2013 | 128 | 128 | S83F/D87N | WT | T57S/S80R | WT | aac(6′)-Ib-cr, oqxAB | blaCTX-M-14 |
| P26 | Zhoukou | 9 mo | 2013 | 32 | 128 | S83F/D87G | WT | T57S/S80R | WT | oqxAB | blaCTX-M-14 |
WT, wild type.
The protocols for isolating bacteria from animals used a modified method based on the United States Department of Agriculture-Food Safety and Inspection Service Microbiology Laboratory Guidebook (19), and protocols applied to patient samples were described previously (20). Finally, isolates from both animals and patients with typical Salmonella phenotypes were confirmed by the API 20E test (bioMérieux, Beijing, China) and amplification of the invA gene by PCR (21). For selecting concurrent resistance to ciprofloxacin and cefotaxime in Salmonella isolates, all confirmed isolates were incubated in LB supplemented with 1 mg/liter ciprofloxacin and 4 mg/liter cefotaxime. For all of the Salmonella isolates that grew, serotypes were determined by slide agglutination with commercial Salmonella antisera (Statens Serum Institute, Denmark) by following the Kauffmann-White scheme.
Antimicrobial susceptibility testing.
The antimicrobial susceptibility of all of the Salmonella isolates was determined by the agar dilution method and interpreted according to Clinical and Laboratory Standards Institute guidelines (22) and the European Committee on Antimicrobial Susceptibility Testing (35). The following antimicrobial compounds were assessed: ampicillin (AMP), cefotaxime (CTX), cefotaxime-clavulanic acid (CTX-CLA), ceftazidime (CAZ), ceftazidime-clavulanic acid (CAZ-CLA), chloramphenicol (CHL), ciprofloxacin (CIP), gentamicin (GEN), imipenem (IPM), tetracycline (TET), tigecycline (TGC), and trimethoprim-sulfamethoxazole (SXT). Multidrug resistance was defined as resistance to three or more different classes of agents. Isolates with MIC values of 1 mg/liter for either cefotaxime or ceftazidime were further screened for ESBL production by determination of synergy between 0.25 and 128 mg/liter ceftazidime or cefotaxime and 4 mg/liter clavulanate. Escherichia coli ATCC 25922 and Klebsiella pneumoniae ATCC 700603 were used as quality-control organisms in antimicrobial susceptibility tests.
PCR amplification and DNA sequence analysis.
The quinolone resistance determination regions (QRDRs) of gyrA, gyrB, parC, and parE in Salmonella isolates were amplified by PCR as described previously (11). All of the Salmonella isolates were screened by PCR for transferable PMQR-encoding genes, including qnrA, qnrB, qnrC, qnrD, qnrS, oqxAB, aac-(6′)-Ib, and qepA, as previously described (11, 12). A multiplex PCR was used to screen for blaCTX-M in all of the ESBL-producing isolates (23). Another multiplex PCR method was applied to screen for plasmid-mediated AmpC-encoding genes in clavulanic acid-resistant isolates (24). All of the PCR products were directly sequenced (TaKaRa Biotechnology Cooperation, Dalian, China) for sequence analysis and aligned using the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/).
Plasmid replicon typing and conjugation experiment.
Salmonella isolates were examined for the presence of 18 replicon (Inc) types by PCR-based replicon typing (PBRT) (25). Transfer of resistance was tested by filter-mating assays. Conjugation experiments were performed with Salmonella isolates as donors using a modified laboratory-based method and E. coli MG1655 as the recipient (26). Transconjugants were selected as pink colonies on MacConkey agar plates containing cefotaxime (4 mg/liter), and PCR and S1-pulsed-field gel electrophoresis (S1-PFGE) subsequently were performed to reconfirm the plasmid profiles in these transconjugants (6).
PFGE.
All Salmonella isolates were subtyped by PFGE to determine their DNA fingerprint profiles following digestion by the macrorestriction enzyme XbaI (New England BioLabs, Ltd., Beijing) according to the procedures developed by the U.S. Centers for Disease Control and Prevention PulseNet program (27). The interpretation of the PFGE patterns was performed with BioNumerics software (Applied Maths, St-Martens-Latem, Belgium) using the Dice similarity coefficient. Dendrograms were constructed on the basis of the unweighted pair group method using average linkages (UPGMA) with a position tolerance of 1%. Clusters were defined as DNA patterns sharing ≥85% similarity.
RESULTS
Antimicrobial susceptibility testing.
All 154 S. enterica serovar Indiana isolates were resistant to ampicillin, cefotaxime, ceftazidime, and ciprofloxacin and susceptible to imipenem and tigecycline. Resistance to chloramphenicol was common (98.7%; 152/154), followed by resistance to gentamicin (96.1%; 148/154), trimethoprim-sulfamethoxazole (92.2%; 142/154), and tetracycline (81.2%; 125/154) (Table 2). All Salmonella isolates expressed a multidrug-resistant phenotype (being resistant to at least three different classes of antimicrobial compounds). In total, 12 antimicrobial resistance profiles were identified among 154 S. enterica serovar Indiana isolates. The dominant resistance profiles identified were determined to be AMP-CAZ-CHL-CIP-CTX-GEN-SXT-TET (n = 109) and AMP-CAZ-CHL-CIP-CTX-GEN-SXT (n = 27) from both chicken (87.2%; 116/133) and human (95.2% 20/21) isolates. In total, 153 isolates (99.4%; 153/154) were confirmed as ESBL positive.
TABLE 2.
Resistance phenotypes of S. enterica serovar Indiana isolates for each antimicrobial from chicken and patients
| Antibiotic | Breakpoint(s)a (mg/liter) | No. (%) of resistant isolates from different sources |
||
|---|---|---|---|---|
| Chicken (n = 133) | Patients (n = 21) | Total | ||
| AMP | ≥32 | 133 (100) | 21 (100) | 154 (100) |
| CAZ | ≥16 | 133 (100) | 21 (100) | 154 (100) |
| CTX | ≥4 | 133 (100) | 21 (100) | 154 (100) |
| CHL | ≥32 | 131 (98.5) | 21 (100) | 152 (98.7) |
| CIP | ≥1 | 133 (100) | 21 (100) | 154 (100) |
| GEN | ≥16 | 128 (96.2) | 20 (95.2) | 148 (96.1) |
| IMP | ≥4 | 0 (0) | 0 (0) | 0 (0) |
| TET | ≥16 | 104 (78.2) | 21 (100) | 125 (81.2) |
| TGC | ≥2 | 0 (0) | 0 (0) | 0 (0) |
| SXT | ≥4/76 | 121 (91.0) | 21 (100) | 142 (92.2) |
Identification of quinolone resistance-encoding genes.
Of the 154 S. enterica serovar Indiana isolates we studied, four point mutations in QRDRs were found in 152 isolates, of which 80 possessed amino acid substitutions in GyrA (S83F and D87G) and ParC (T57S and S80R) as the dominant types (51.9%; 80/154), along with 72 isolates that contained GyrA (S83F and D87N) and ParC (T57S and S80R) alleles. Three point mutations were identified in 2 isolates from poultry slaughterhouses with amino acid substitutions in GyrA (S83F) and ParC (T57S and S80R).
PMQR-encoding genes were identified in 143 isolates (92.9%; 143/154) from both chicken (n = 122) and human (n = 21) isolates, including aac(6′)-Ib-cr (n = 117), oqxAB (n = 112), and qepA (n = 2). The PMQR genes qnrA, qnrB, qnrC, qnrD, and qnrS were not detected. Ninety-eight isolates (63.6%; 98/154) possessed more than one PMQR gene. Compared to the chicken isolates (60.9%; 81/133), human isolates (81.0%; 17/21) showed a higher prevalence with two PMQR genes. The ciprofloxacin MIC was measured and determined to be between 8 and 128 mg/liter in all of these isolates.
Among 132 isolates that were highly resistant to ciprofloxacin (MIC of ≥32 mg/liter), including 112 poultry and 20 human isolates, four topoisomerase point mutations were detected in each of 130 isolates, and a further three were detected in two isolates. PMQR genes were identified in 127 isolates (two PMQR genes in 96 isolates and one PMQR gene in 31 isolates), and only five isolates were negative for any of the PMQR genes. The 37 isolates that demonstrated a ciprofloxacin MIC of ≥128 mg/liter possessed four point mutations in QRDRs with two PMQR genes [aac(6′)-Ib-cr and oqxAB], of which 27 contained amino acid substitutions in GyrA (S83F and D87N) and ParC (T57S and S80R) and 10 had amino acid substitutions in GyrA (S83F and D87G) and ParC (T57S and S80R).
Identification of β-lactamase resistance-encoding genes.
Five blaCTX-M subtypes were identified among the 153 ESBL-positive Salmonella isolates, including blaCTX-M-65 (n = 131), blaCTX-M-14 (n = 15), blaCTX-M-15 (n = 5), blaCTX-M-27 (n = 1), and blaCTX-M-14/blaCTX-M-15 (n = 1). Salmonella isolates containing blaCTX-M-65 were recovered both from poultry slaughterhouses (n = 121) in all sampling cities and from humans (n = 10). Salmonella isolates containing blaCTX-M-14 were also recovered from poultry slaughterhouses (n = 11) in Hebi (n = 3) and Kaifeng (n = 8) and from humans (n = 4). The remaining three blaCTX-M subtypes were detected only in human isolates. Only three strains were AmpC-producing isolates, and these were cultured from poultry slaughterhouses. The blaCMY-2 gene was identified in two ESBL isolates with blaCTX-M-14 from Kaifeng and one clavulanic acid-resistant isolate from Luohe.
Association between ESBL and plasmid-mediated quinolone resistance genes.
Among all 154 S. enterica serovar Indiana isolates, the most frequent combination of aac(6′)-Ib-cr with oqxAB was in CTX-M-65-producing strains (n = 89). The combination of oqxAB with qepA was also found in only two CTX-M-65-producing isolates with amino acid substitutions in GyrA (S83F) and ParC (T57S and S80R). Isolates containing blaCTX-M-14 also possessed two PMQR genes, one PMQR gene, or no PMQR gene. Isolates with blaCTX-M-15 contained two PMQR genes or one PMQR gene.
Plasmid replicon typing and conjugation experiment.
Among 154 isolates, 106 isolates carried at least one replicon type and six different plasmid Inc types were detected, ranging from one to three replicon types in these isolates. The three most prevalent replicon types identified by PBRT included IncN (91.5%; 97/106), IncA/C (9.4%; 10/106), and IncI1-Ir (6.6%; 7/106). IncN was detected in all bacterial isolates that were positive for the blaCTX-M-65 gene. IncA/C, IncFIA, IncFIB, IncF, and IncI1-Ir were detected only in isolates positive for blaCTX-M-14 and blaCMY-2 genes. Forty randomly selected poultry isolates containing different plasmid Inc types, along with 21 clinical isolates, were selected as donors in the conjugation experiments. Five transconjugants were obtained. Conjugative transfer frequencies recorded ranged from 10−7 to 10−8 transconjugants per recipient. Analysis of the plasmids in these transconjugants identified IncA/C, IncI1-Ir, and IncN types along with three other isolates whose plasmid replicon types could not be confirmed with the primer sets used. Transconjugants obtained using S. enterica serovar Indiana isolates as donors harbored plasmids with sizes ranging from 130 to 285 kbp (Fig. 1 and Table 3). Genes present in the transconjugants included oqxA, oqxB, aac(6′)-Ib-cr, blaCTX-M-14, blaCTX-M-15, and blaCTX-M-65 (Fig. 1 and Table 3).
FIG 1.
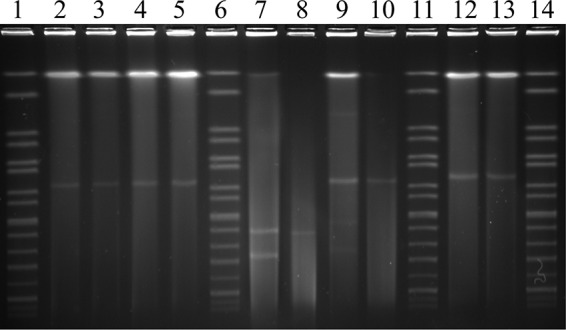
Plasmid profiles of representative S. enterica serovar Indiana isolates and transconjugants determined by S1-PFGE. Transmissible plasmids from S. enterica serovar Indiana and transconjugants (T) are highlighted. Lanes 1, 6, 11, and 14, marker H9812 with different bands labeled (used as a molecular size marker); lane 2, isolate P9; lane 3, isolate P9-(T); lane 4, isolate P10; lane 5, isolate P10-(T); lane 7, isolate D25; lane 8, isolate D25-(T); lane 9, isolate D26; lane 10, isolate D26-(T); lane 12, isolate D169; lane 13, isolate D169-(T).
TABLE 3.
Characteristics of blaCTX-M-positive S. enterica serovar Indiana isolates coharboring PMQR genes and their transconjugants
| Straina | Source | Resistance gene(s) | Antimicrobial resistance profile | Replicon typingb | Plasmid size(s) (kb) |
|---|---|---|---|---|---|
| D25 | Chicken | blaCTX-M-14, blaCMY2 | AMP-CAZ-CHL-CIP-CTX-SXT-TET | I1-Iγ,A/C | 130, 83 |
| D25-(T) | blaCTX-M-14 | AMP-CAZ-CHL-CTX-TET | A/C | 130 | |
| D26 | Chicken | oqxAB, blaCTX-M-14 | AMP-CAZ-CHL-CIP-CTX-GEN-SXT-TET | I1-Iγ, N, A/C | 260, 155, 94 |
| D26-(T) | oqxAB, blaCTX-M-14 | AMP-CAZ-CHL-CTX-SXT-TET | I1-Iγ, N, A/C | 260 | |
| D169 | Chicken | oqxAB, blaCTX-M-65 | AMP-CAZ-CHL-CIP-CTX-GEN-TET | NT | 285 |
| D169-(T) | blaCTX-M-65 | AMP-CAZ-CHL-CTX | NT | 285 | |
| P9 | Human | aac(6′)-Ib-cr, oqxAB, blaCTX-M-15 | AMP-CAZ-CHL-CIP-CTX-GEN-SXT-TET | NT | 256 |
| P9-(T) | aac(6′)-Ib-cr, oqxAB, blaCTX-M-15 | AMP-CAZ-CHL-CTX-GEN-SXT-TET | NT | 256 | |
| P10 | Human | aac(6′)-Ib-cr, oqxAB, blaCTX-M-15 | AMP-CAZ-CHL-CIP-CTX-GEN-SXT-TET | NT | 256 |
| P10-(T) | aac(6′)-Ib-cr, oqxAB, blaCTX-M-15 | AMP-CAZ-CHL-CTX-GEN-SXT-TET | NT | 256 |
(T), transconjugant.
NT, an isolate whose plasmid replicon was nontypeable.
PFGE analysis of Salmonella isolates.
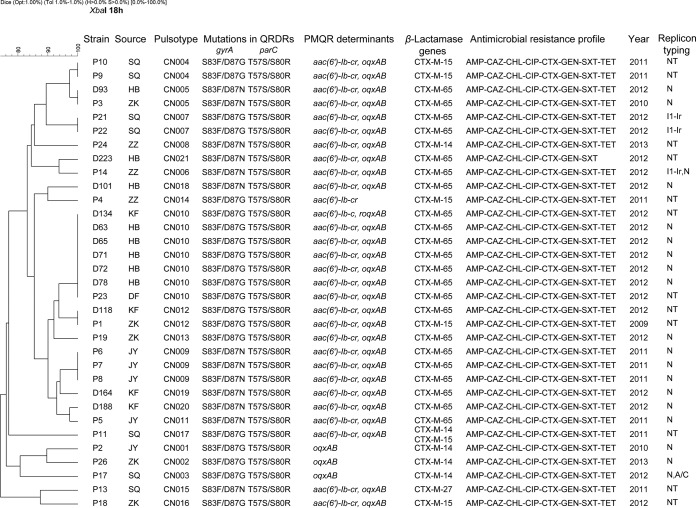
Ninety-one pulsotypes were identified among the 133 S. enterica serovar Indiana isolates from chicken by PFGE, and 14 different clusters were denoted as clusters CC1 to CC14 (see Fig. S1 in the supplemental material). There were 18 separate PFGE patterns containing more than one isolate. Isolates with the same PFGE patterns mostly were recovered from the same regions, but 4 indistinguishable pulsotypes contained isolates from different regions. Some isolates within the same pulsotype possessed the same type of blaCTX-M but not the same QRDR or PMQR profile. The blaCTX-M-65 gene was identified in isolates in 81 pulsotypes. Cluster CC1-1 had the greatest number of isolates with high-level resistance to ciprofloxacin (MIC, ≥128 mg/liter). In contrast, clusters CC13 and CC14 contained isolates with CTX-M-14 (see Fig. S1). The 21 clinical isolates were divided into 17 pulsotypes (Fig. 2). The predominant pulsotype was denoted INDX11.CN009, which included three isolates from the same city. Two pulsotypes (INDX11.CN004 and INDX11.CN007) were found to contain two each, and the remaining 14 pulsotypes contained a single isolate (Fig. 2).
FIG 2.
Dendrogram of 21 clinical and 12 poultry S. enterica serovar Indiana isolates that were resistant to both ciprofloxacin and cefotaxime, constructed based on PFGE with XbaI on patient samples. The isolate identity, source, pulsotype, year of isolation, antimicrobial resistance profile, ESBL genes, mutations in QRDRs, and PMQR determinants were identified. D in strain names indicates isolates from poultry, and P indicates isolates from patients. DF, Dengfeng; JY, Jiyuan; HB, Hebi; KF, Kaifeng; LH, Luohe; SQ, Shangqiu; ZK, Zhoukou; ZZ, Zhengzhou; NT, an isolate whose plasmid replicon typing was nontypeable.
DNA fingerprint comparisons between isolates cultured from patients and food-producing animals by PFGE exhibited diversity among the strains. Three pulsotypes (INDX11.CN005, INDX11.CN010, and INDX11.CN012) included isolates from both chicken and humans (Fig. 2). Some of the remaining pulsotypes from patients showed profiles highly similar to those from food-producing animals (Fig. 2). The results further confirmed that some predominant clones may have existed in Henan for several years, and chicken could be a source of concurrent resistance to ciprofloxacin and cefotaxime S. enterica serovar Indiana in humans.
DISCUSSION
In this study, we collected ciprofloxacin- and cefotaxime-resistant S. enterica serovar Indiana isolates from both processed chicken in poultry slaughterhouses and patients in Henan province, which is one of the largest production centers in China for broiler meat processing. Genes conferring resistance to fluoroquinolones and extended-spectrum β-lactams in these isolates were characterized, and the collection of bacteria was subtyped by PFGE.
In the present study, our aim was to characterize Salmonella isolates that were resistant to both fluoroquinolone and third-generation cephalosporins. All were identified as serovar Indiana, and interestingly this serovar has also been reported in livestock in Beijing and Guangdong (15, 16), suggesting that it has a broad geographical distribution in China. Our data showed that 10.1% of the isolates cultured from chicken were coresistant to cefotaxime and ciprofloxacin, a feature previously reported by Wang et al. (16), suggesting this strain has become established in some poultry flocks in China. Human clinical Salmonella isolates that were resistant to these two antimicrobial compounds recently were reported in S. enterica serovar Kentucky ST198-X1, a much less prevalent serotype from France (28), and in S. enterica serovar Typhimurium from China (29). In this study, only 2.6% of the isolates from humans were identified as cefotaxime- and ciprofloxacin-resistant S. enterica serovar Indiana, and this coresistance was first reported by our study. With the large-scale use of antimicrobial compounds over time, Salmonella isolates expressing resistance to critically important antimicrobial agents such as fluoroquinolones and/or extended-spectrum cephalosporins have been detected in cultures from animals and patients in numerous locations with various levels of prevalence in China (15, 16, 29).
The emergence of cefotaxime- and ciprofloxacin-resistant S. enterica serovar Indiana warrants continuous monitoring of the trend for development of antimicrobial resistance in China.
Double amino acid substitutions in GyrA (S83F and D87N)/GyrA (S83F and D87G) and ParC (T57S and S80R) were noted in most of the study isolates. No mutations were detected in either gyrB or parE. These data are consistent with a previous report (29) that showed only those mutations in gyrA and parC mediate quinolone and fluoroquinolone resistance.
PMQR-encoding genes were identified in 143 isolates, a number that was higher than the previous study reported (30) but similar to the report by Jiang et al. (15). Furthermore, two PMQR-encoding genes coexisting in a single isolate were frequently detected, a feature that was rarely observed in isolates from other countries (30). The frequent use of olaquindox as a growth promoter in animals has been suggested to exert a high selective pressure driving the acquisition of oqxAB and contributing to its subsequent dissemination (29). Uniquely, one of the isolates possessed three amino acid substitutions (denoted W102R, D179Y, and A160G) in aac(6′)-Ib-cr (31). Two others were detected with a qepA gene which was identified previously in E. coli. To our knowledge, this is the first description of qepA in S. enterica serovar Indiana in China, and this gene has been reported previously only in S. enterica serovar Typhimurium from Spain (32) and China (29). The qepA gene was found in CTX-M-65-producing isolates alone, along with substitutions in GyrA (S83F) and ParC (T57S and S80R). These data suggested that the acquisition of this transferable quinolone resistance gene is a rare event. Although PMQR confers only low-level quinolone resistance, it can facilitate the emergence of high-level resistance via mutation(s) in one or more of the topoisomerase genes (33). However, in most isolates, two PMQR-encoding genes combined with multiple mutations in topoisomerase genes resulted in high-level resistance to ciprofloxacin (MIC, ≥128 mg/liter).
ESBL screening and characterization for these S. enterica serovar Indiana isolates showed that different variants of the CTX-M family of β-lactamases were detectable in S. enterica serovar Indiana and that they most likely contributed to the cefotaxime resistance phenotypes of the host isolates. Subtypes of blaCTX-M genes identified in humans isolates were more variable than those identified in food-producing animals, which suggests that there were other sources in addition to chickens. blaCTX-M-65 was the predominant type existing both in food-producing animals and in humans, and these differed from the types reported earlier in S. enterica serovar Kentucky ST198-X1 (blaCTX-M-1 and blaCTX-M-15) (28) and in S. enterica serovar Typhimurium (blaCTX-M-14) (29). Genotypes blaCTX-M-14 and blaCTX-M-27 also have been found in S. enterica serovar Indiana isolates from chickens and pigs in Guangdong (15), and blaCTX-M-24 was identified in S. enterica serovar Indiana from chickens in Shandong (26). Why the predominant blaCTX-M in ciprofloxacin- and cefotaxime-resistant S. enterica serovar Indiana isolates was blaCTX-M-65 in Henan was not clear, but it suggests that the selection, dissemination, and maintenance of transmissible elements carrying these genes can occur during exposure to any of the agents to which resistance is conferred. Interestingly, blaCTX-M is often located on conjugative plasmids. It has been reported that ciprofloxacin-resistant S. enterica serovar Kentucky ST198-X1 acquired the genes conferring resistance to extended-spectrum cephalosporins only recently (28), and certain genotypes, such as blaCTX-M-55, are borne on plasmids that may have originated from E. coli possessing the ability to disseminate to Salmonella and other bacterial species (34). Conjugative systems in Gram-negative bacteria support transfer of mobile genetic elements between different genera, and in this study five transconjugants (8.25%; 5/61) were obtained. According to S1-PFGE, most of these harbored large plasmids of greater than 200 kbp (data not shown). Previously, several carbapenemase resistance mechanisms (such as KPC, OXA-48, NDM, and VIM) have been identified in S. enterica (28). None of the isolates in this study were found to be resistant to imipenem, although surveillance for the emergence of these types of S. enterica serovar Indiana isolates should be undertaken as a routine assay.
The PFGE profile variation of the 154 isolates from poultry slaughterhouses and humans demonstrated extensive genetic heterogeneity. Several bacterial isolates from the same sampling place exhibited similar or indistinguishable PFGE pulsotypes, suggesting the potential for cross-contamination. Similar PFGE patterns of isolates from different regions highlighted the possibility for the clonal spreading of some coresistant strains. A comparison of PFGE patterns between bacteria cultured from food-producing animals and from humans further confirmed the existence of clones in Henan for several years. Most notably, the pulsotypes (INDX11.CN005, INDX11.CN010, and INDX11.CN012) from food-producing animals, such as chicken, suggested that chicken is a source of these coresistant S. enterica serovar Indiana isolates in humans.
Conclusions.
Characterization of the genetic basis for resistance to both third-generation cephalosporins and fluoroquinolones identified plasmids as a major contributing factor. Transmission of resistance genes, such as those encoding ESBLs, OqxAB, and AAC(6′)-Ib-c in Salmonella, will facilitate the selection of these MDR Salmonella isolates that pose a threat to public health. Ongoing surveillance by regulatory agencies is required to identify routes of transmission.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Beijing Natural Science Foundation (7154252).
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02849-15.
REFERENCES
- 1.Scallan E, Griffin PM, Angulo FJ, Tauxe RV, Hoekstra RM. 2011. Foodborne illness acquired in the United States-unspecified agents. Emerg Infect Dis 17:16–22. doi: 10.3201/eid1701.P21101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang Z, Ke B, Deng X, Liang J, Ran L, Lu L, He D, Huang Q, Ke C, Li Z, Yu H, Klena JD, Wu S. 2015. Serotypes, seasonal trends, and antibiotic resistance of non-typhoidal Salmonella from human patients in Guangdong Province, China, 2009-2012. BMC Infect Dis 15:53. doi: 10.1186/s12879-015-0784-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.European Food Safety Authority European Centre for Disease Prevention and Control. 2014. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2012. EFSA J 12:3547. doi: 10.2903/j.efsa.2014.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu H, Xia X, Cui Y, Hu Y, Xi M, Wang X, Shi X, Wang D, Meng J, Yang B. 2013. Prevalence of extended-spectrum β-lactamase–producing Salmonella on retail chicken in six provinces and two national cities in the People's Republic of China. J Food Prot 76:2040–2044. doi: 10.4315/0362-028X.JFP-13-224. [DOI] [PubMed] [Google Scholar]
- 6.Cui S, Li J, Sun Z, Hu C, Jin S, Li F, Guo Y, Ran L, Ma Y. 2009. Characterization of Salmonella enterica isolates from infants and toddlers in Wuhan, China. J Antimicrob Chemother 63:87–94. doi: 10.1093/jac/dkn452. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Xie X, Xu X, Wang X, Chang H, Wang C, Wang A, He Y, Yu H, Zeng M. 2014. Nontyphoidal Salmonella infection in children with acute gastroenteritis: prevalence, serotypes, and antimicrobial resistance in Shanghai, China. Foodborne Pathog Dis 11:200–206. doi: 10.1089/fpd.2013.1629. [DOI] [PubMed] [Google Scholar]
- 8.Van TT, Nguyen HN, Smooker PM, Coloe PJ. 2012. The antibiotic resistance characteristics of non-typhoidal Salmonella enterica isolated from food-producing animals, retail meat and humans in South East Asia. Int J Food Microbiol 154:98–106. doi: 10.1016/j.ijfoodmicro.2011.12.032. [DOI] [PubMed] [Google Scholar]
- 9.Liu HH. 2010. Safety profile of the fluoroquinolones. Drug Safety 33:353–369. doi: 10.2165/11536360-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 10.Glenn LM, Lindsey RL, Folster JP, Pecic G, Boerlin P, Gilmour MW, Harbottle H, Zhao S, Mcdermott PF, Fedorka-Cray PJ. 2013. Antimicrobial resistance genes in multidrug-resistant Salmonella enterica isolated from animals, retail meats, and humans in the United States and Canada. Microb Drug Resist 19:175–184. doi: 10.1089/mdr.2012.0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S, Cui S, McDermott PF, Zhao S, White DG, Paulsen I, Meng J. 2007. Contribution of target gene mutations and efflux to decreased susceptibility of Salmonella enterica serovar Typhimurium to fluoroquinolones and other antimicrobials. Antimicrob Agents Chemother 51:535–542. doi: 10.1128/AAC.00600-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J-H, Deng Y-T, Zeng Z-L, Gao J-H, Chen L, Arakawa Y, Chen Z-L. 2008. Coprevalence of plasmid-mediated quinolone resistance determinants QepA, Qnr, and AAC(6′)-Ib-cr among 16S rRNA methylase RmtB-producing Escherichia coli isolates from pigs. Antimicrob Agents Chemother 52:2992–2993. doi: 10.1128/AAC.01686-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutkind GO, Di Conza J, Power P, Radice M. 2013. β-Lactamase-mediated resistance: a biochemical, epidemiological and genetic overview. Curr Pharm Des 19:164–208. doi: 10.2174/138161213804070320. [DOI] [PubMed] [Google Scholar]
- 14.Hawkey P. 2008. Prevalence and clonality of extended-spectrum-β-lactamases in Asia. Clin Microbiol Infect 14:159–165. doi: 10.1111/j.1469-0691.2007.01855.x. [DOI] [PubMed] [Google Scholar]
- 15.Jiang HX, Song L, Liu J, Zhang XH, Ren YN, Zhang WH, Zhang JY, Liu YH, Webber MA, Ogbolu DO, Zeng ZL, Piddock LJ. 2014. Multiple transmissible genes encoding fluoroquinolone and third-generation cephalosporin resistance co-located in non-typhoidal Salmonella isolated from food-producing animals in China. Int J Antimicrob Agents 43:242–247. doi: 10.1016/j.ijantimicag.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Chen Q, Cui S, Xu X, Zhu J, Luo H, Wang D, Li F. 2013. Enumeration and characterization of Salmonella isolates from retail chicken carcasses in Beijing, China. Foodborne Pathog Dis 11:126–132. doi: 10.1089/fpd.2013.1586. [DOI] [PubMed] [Google Scholar]
- 17.Li R, Lai J, Wang Y, Liu S, Li Y, Liu K, Shen J, Wu C. 2013. Prevalence and characterization of Salmonella species isolated from pigs, ducks and chickens in Sichuan Province, China. Int J Food Microbiol 163:14–18. doi: 10.1016/j.ijfoodmicro.2013.01.020. [DOI] [PubMed] [Google Scholar]
- 18.M'ikanatha NM, Sandt CH, Localio AR, Tewari D, Rankin SC, Whichard JM, Altekruse SF, Lautenbach E, Folster JP, Russo A. 2010. Multidrug-resistant Salmonella isolates from retail chicken meat compared with human clinical isolates. Foodborne Pathog Dis 7:929–934. doi: 10.1089/fpd.2009.0499. [DOI] [PubMed] [Google Scholar]
- 19.USDA-FSIS. 2011. Laboratory guidebook: isolation and identification of Salmonella from meat, poultry, pasteurized egg and catfish products. MLG 4.05. U.S. Department of Agriculture-Food Safety and Inspection Service, Washington, DC. [Google Scholar]
- 20.Xia S, Hendriksen RS, Xie Z, Huang L, Zhang J, Guo W, Xu B, Ran L, Aarestrup FM. 2009. Molecular characterization and antimicrobial susceptibility of Salmonella isolates from infections in humans in Henan Province, China. J Clin Microbiol 47:401–409. doi: 10.1128/JCM.01099-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malorny B, Hoorfar J, Hugas M, Heuvelink A, Fach P, Ellerbroek L, Bunge C, Dorn C, Helmuth R. 2003. Interlaboratory diagnostic accuracy of a Salmonella specific PCR-based method. Int J Food Microbiol 89:241–249. doi: 10.1016/S0168-1605(03)00154-5. [DOI] [PubMed] [Google Scholar]
- 22.CLSI. 2012. Performance standards for antimicrobial susceptibility testing: seventeenth informational supplement M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 23.Xu L, Ensor V, Gossain S, Nye K, Hawkey P. 2005. Rapid and simple detection of blaCTX-M genes by multiplex PCR assay. J Med Microbiol 54:1183–1187. doi: 10.1099/jmm.0.46160-0. [DOI] [PubMed] [Google Scholar]
- 24.Perez-Perez FJ, Hanson ND. 2002. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol 40:2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 26.Lai J, Wang Y, Shen J, Li R, Han J, Foley SL, Wu C. 2013. Unique class 1 integron and multiple resistance genes co-located on IncHI2 plasmid is associated with the emerging multidrug resistance of Salmonella Indiana isolated from chicken in China. Foodborne Pathog Dis 10:581–588. doi: 10.1089/fpd.2012.1455. [DOI] [PubMed] [Google Scholar]
- 27.Ribot EM, Fair M, Gautom R, Cameron D, Hunter S, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157: H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 28.Le Hello S, Harrois D, Bouchrif B, Sontag L, Elhani D, Guibert V, Zerouali K, Weill FX. 2013. Highly drug-resistant Salmonella enterica serotype Kentucky ST198-X1: a microbiological study. Lancet Infect Dis 13:672–679. doi: 10.1016/S1473-3099(13)70124-5. [DOI] [PubMed] [Google Scholar]
- 29.Wong MH, Yan M, Chan EW, Biao K, Chen S. 2014. Emergence of clinical Salmonella enterica serovar Typhimurium isolates with concurrent resistance to ciprofloxacin, ceftriaxone, and azithromycin. Antimicrob Agents Chemother 58:3752–3756. doi: 10.1128/AAC.02770-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veldman K, Cavaco LM, Mevius D, Battisti A, Franco A, Botteldoorn N, Bruneau M, Perrin-Guyomard A, Cerny T, Escobar CDF. 2011. International collaborative study on the occurrence of plasmid-mediated quinolone resistance in Salmonella enterica and Escherichia coli isolated from animals, humans, food and the environment in 13 European countries. J Antimicrob Chemother 66:1278–1286. doi: 10.1093/jac/dkr084. [DOI] [PubMed] [Google Scholar]
- 31.Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. 2006. Prevalence in the United States of aac (6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother 50:3953–3955. doi: 10.1128/AAC.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lunn AD, Fàbrega A, Sánchez-Céspedes J, Vila J. 2010. Prevalence of mechanisms decreasing quinolone-susceptibility among Salmonella spp. clinical isolates. Int Microbiol 13:15–20. doi: 10.1128/AAC.00915-06. [DOI] [PubMed] [Google Scholar]
- 33.Hernández A, Sánchez MB, Martínez JL. 2011. Quinolone resistance: much more than predicted. Front Microbiol 2:22. doi: 10.3389/fmicb.2011.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong MH, Liu L, Yan M, Chan EW, Chen S. 2015. Dissemination of IncI2 plasmids that harbor blaCTX-M element among clinical Salmonella isolates. Antimicrob Agents Chemother 59:5026–5028. doi: 10.1128/AAC.00775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.EUCAST. 2012. Clinical breakpoints—bacteria, version 2.0. http://www.eucast.org/ast_of_bacteria/previous_versions_of_documents.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.