Abstract
Multidrug-resistant (MDR) Enterobacteriaceae infections are increasing in U.S. children; however, there is a paucity of multicentered analyses of antibiotic resistance genes responsible for MDR phenotypes among pediatric Enterobacteriaceae isolates. In this study, 225 isolates phenotypically identified as extended-spectrum β-lactamase (ESBL) or carbapenemase producers, recovered from children ages 0 to 18 years hospitalized between January 2011 and April 2015 at three Chicago area hospitals, were analyzed. We used DNA microarray platforms to detect ESBL, plasmid-mediated AmpC (pAmpC), and carbapenemase type β-lactamase (bla) genes. Repetitive-sequence-based PCR and multilocus sequence typing (MLST) were performed to assess isolate similarity. Plasmid replicon typing was conducted to classify plasmids. The median patient age was 4.2 years, 56% were female, and 44% presented in the outpatient setting. The majority (60.9%) of isolates were Escherichia coli and from urinary sources (69.8%). Of 225 isolates exhibiting ESBL- or carbapenemase-producing phenotypes, 90.7% contained a bla gene. The most common genotype was the blaCTX-M-1 group (49.8%); 1.8% were carbapenem-resistant Enterobacteriaceae (three blaKPC and one blaIMP). Overall, pAmpC (blaACT/MIR and blaCMY) were present in 14.2%. The predominant E. coli phylogenetic group was the virulent B2 group (67.6%) associated with ST43/ST131 (Pasteur/Achtman MLST scheme) containing the blaCTX-M-1 group (84%), and plasmid replicon types FIA, FII, and FIB. K. pneumoniae harboring blaKPC were non-ST258 with replicon types I1 and A/C. Enterobacter spp. carrying blaACT/MIR contained plasmid replicon FIIA. We found that β-lactam resistance in children is diverse and that certain resistance mechanisms differ from known circulating genotypes in adults in an endemic area. The potential impact of complex molecular types and the silent dissemination of MDR Enterobacteriaceae in a vulnerable population needs to be studied further.
INTRODUCTION
The dissemination of antibiotic-resistant Enterobacteriaceae during the last 2 decades has been rapid, resulting in a pandemic of infections associated with significant morbidity and mortality (1–3). The major driving force of antibiotic resistance within this family of Gram-negative bacteria is the β-lactamases (2, 4). Currently, more than 1,600 known β-lactamases are cataloged; a list that continues to expand (5).
Genes encoding β-lactamases (bla) may be chromosomal in origin; however, much of the global spread of β-lactam resistance is facilitated by mobile genetic elements (such as plasmids and transposons) harboring bla genes encoding extended-spectrum β-lactamases (ESBL), AmpC cephalosporinases (AmpC), and carbapenemases (e.g., Klebsiella pneumoniae carbapenemase [KPC] and New Delhi MBL [NDM]) conferring carbapenem resistance in Enterobacteriaceae (CRE) (2, 4, 6, 7). Many of these β-lactamase-producing organisms carry additional plasmid-borne genes against other classes of antibiotics rendering them multidrug resistant (MDR) (4, 7), i.e., resistant to three or more classes of antibiotics (8), leaving few, if any, antibiotics to treat these infections (9).
Recent studies describe the prevalence of MDR Enterobacteriaceae as increasing in the United States, including in children (10–12). However, few studies report the genetic determinants associated with MDR Enterobacteriaceae in pediatric populations, and there is a paucity of multicenter studies defining the molecular epidemiology of these organisms. Knowledge of the molecular epidemiology of MDR Enterobacteriaceae can have a profound effect on clinical practice, infection control measures, and public health policies for children. In this study, we sought to determine whether children cared for at three distinct institutions located in the same geographic area would be subject to similar antibiotic resistance threats. Understanding the composition and distribution of antibiotic resistance genotypes is a critical step in defining the impact of MDR Enterobacteriaceae infections in children and future treatment decisions.
MATERIALS AND METHODS
Study settings and population.
Hospital A is a tertiary care medical center that includes a children's hospital composed of 115 pediatric beds (level III neonatal, cardiac surgery, and pediatric intensive-care units and general pediatric and psychiatric wards) and a mother-newborn infant unit. Hospital B contains a 125-bed children's hospital (general pediatrics and newborn infant, neonatal, and pediatric intensive care units). Hospital C is a 288-bed, academic free-standing children's hospital providing complex quaternary pediatric care, including solid organ and stem cell transplantation services. All of the hospitals are located in the Chicago area.
This study included patients 0 to 18.99 years of age found to have a positive culture for an Enterobacteriaceae with an ESBL and/or carbapenem-resistant phenotype due to a carbapenemase. Isolates were collected between January 2011 and April 2015, and only one isolate per patient per admission was included. The study was approved by the institutional review boards of the three participating institutions. The institutions joined the study at various time points during the study period.
Bacterial isolates and antibiotic susceptibility testing.
The clinical microbiology laboratories of hospitals A, B, and C performed phenotypic identification and susceptibility testing of ESBL- and carbapenemase-producing isolates at the respective institutions using the MicroScan WalkAway system (Siemens Healthcare Diagnostics, Tarrytown, NY). Based on the guidelines of the Clinical and Laboratory Standards Institute (CLSI), screening β-lactam antibiotics for ESBL-producing bacteria included any one of the following: cefpodoxime, cefotaxime, ceftazidime, ceftriaxone, or aztreonam (13). ESBL production was confirmed by disk diffusion as necessary (BBL; Becton, Dickinson and Company, Sparks, MD) or on the Microscan instrument by comparing MICs of cefotaxime and ceftazidime with or without the addition of clavulanic acid. A 4-fold reduction in MIC or an increase in zone diameter of >5 mm associated with cefotaxime or ceftazidime in combination with clavulanic acid compared to the MIC of the antibiotic when tested alone confirmed an ESBL phenotype (13).
Isolates were considered to have a carbapenemase phenotype by Centers for Disease Control and Prevention criteria, if they were resistant to all expanded-spectrum cephalosporins (ceftriaxone, cefotaxime, or ceftazidime) and nonsusceptible to at least one carbapenem (ertapenem, meropenem, imipenem, or doripenem) (14). The presence of carbapenemases was phenotypically assessed using the Modified Hodge Test and/or the MBL Etest (bioMérieux, Athens, GA), as appropriate.
Analysis of bla genes.
Genomic DNA was purified from bacterial isolates by using a DNeasy blood and tissue kit (Qiagen, Inc., Valencia, CA). A DNA microarray-based assay was performed to evaluate for the presence of bla genes in isolates (Check-MDR CT101; Check-Points, Wageningen, The Netherlands). The assay detects blaCTX-M-1 group, blaCTX-M-2 group, blaCTX-M-8 plus -25 group, blaCTX-M-9 group, and blaTEM-WT (wild type) and blaTEM-type ESBL genes, blaSHV-WT (wild type) and blaSHV-type ESBL genes, plasmid-based AmpC cephalosporinases (pAmpC; blaACC, blaACT/MIR, blaCMY II, blaDHA, and blaFOX), and carbapenemases (blaKPC and blaNDM) (15). A more comprehensive DNA microarray (Check-MDR CT103XL; Check-Points) was performed on isolates identified as bla negative by the Check-MDR CT101 kit. The Check-MDR CT103XL kit includes additional ESBL (blaVEB, blaPER, blaBEL, and blaGES) and carbapenemase (blaVIM, blaIMP, blaGES, blaGIM, blaSPM, blaOXA-23, blaOXA-24, blaOXA-48, and blaOXA-58) targets (16). Experiments were performed according to the manufacturer's protocol.
Rep-PCR.
To assess for clonal relatedness among strains of E. coli, Klebsiella spp., and Enterobacter spp., repetitive-sequenced-based PCR (rep-PCR) was performed using DiversiLab (bioMérieux, Athens, GA) E. coli, Klebsiella, and Enterobacter fingerprinting kits. Genomic DNA was extracted using an UltraClean microbial DNA isolation kit (MO BIO Laboratories, Carlsbad, CA), followed by PCR amplification and separation of rep-PCR amplicons by electrophoresis on microfluidic chips using the DiversiLab manufacturer's protocol for detection (Agilent Bioanalyzer 2100; Agilent Technologies, Inc., Santa Clara, CA) and analysis (DiversiLab online software). Isolates in which band patterns demonstrated >95% similarity (Pearson's correlation) were considered to represent the same strain type (17). Among isolates analyzed by rep-PCR, PCR amplification and sequencing were also performed to further characterize bla genes as revealed by microarray testing.
MLST and hsp60 sequencing.
Gene amplification and sequencing of seven housekeeping genes (rpoB, gapA, mdh, pgi, phoE, tonB, and infB) for Klebsiella spp. and eight housekeeping genes (dinB, icdA, pabB, polB, putP, trpA, trpB, and uidA) for E. coli were performed as previously described (18, 19), and allele and sequence types (STs) were determined by using the multilocus sequence typing (MLST) Pasteur website (http://www.pasteur.fr/recherche/genopole/PF8/mlst/). We used hsp60 sequencing, which targets a single housekeeping gene, to further determine relatedness among Enterobacter spp. (20).
Phylogenetic analysis and plasmid replicon typing.
A previously described multiplex PCR-based method was used to assign E. coli to one of the four major phylogenetic groups (A, B1, B2, and D) (21). From rep-PCR strain types, plasmid replicon typing was performed on representative isolates according to the scheme described by Carattoli et al. (22).
RESULTS
Characteristics of pediatric patients in the study population.
Enterobacteriaceae strains phenotypically identified as ESBL- or carbapenemase-producing bacteria were recovered from 225 children during the study period. The median age was 4.2 years (range, 0.008 to 18.9 years), and 27% were younger than 1 year of age (Table 1). In this study, the majority (56%) were female, 36% were Hispanic, and 44% of the children presented in the outpatient setting.
TABLE 1.
Demographic and healthcare setting of Chicago children infected with MDR Enterobacteriaceaea
| Variable | No. (%) of children (n = 225) |
|---|---|
| Median age in yrs (range) | 4.2 (0.008–18.9) |
| Male | 98 (44) |
| Race | |
| Hispanic | 80 (36) |
| White | 71 (32) |
| Black | 34 (15) |
| Age <1 years | 60 (27) |
| Healthcare setting | |
| PICU | 59 (26) |
| NICU | 15 (7) |
| Pediatric ward | 52 (23) |
| Outpatient | 99 (44) |
MDR, multidrug resistant, i.e., resistant to three or more antibiotic classes. “Outpatient” includes ambulatory healthcare and the emergency department. PICU, pediatric intensive care unit; NICU, neonatal intensive care unit. All values represent “number (%)” unless indicated otherwise in column 1.
Characteristics of bacterial isolates in the study population.
Of the 225 isolates, the majority (60.9%) were E. coli, followed by 16.4% Klebsiella spp., 13.3% Enterobacter spp., 4.9% Proteus spp., 3.6% Serratia spp., and 0.9% other (including Morganella and Citrobacter spp.). The most common specimen source was urine (69.8%); 4.9% were from blood, 16.9% were from respiratory sources (sputum, tracheal aspirate and bronchoalveolar lavage), 2.7% were from wounds or abscesses, 2.2% were from peritoneal or abdominal sources, 0.4% were from the central nervous system, and 3.1% were from other sources.
Antimicrobial susceptibility testing.
The antibiotic susceptibility testing of the 222 available isolates are summarized in Table 2. Overall, carbapenems and amikacin retained the greatest activity, with 98.2% of isolates susceptible to meropenem or imipenem and 97.1% susceptible to amikacin; 82.2% of urinary isolates were susceptible to nitrofurantoin. Isolates had relatively high rates of resistance (∼40 to 60%) to gentamicin, tobramycin, trimethoprim-sulfamethoxazole, tetracycline, and fluoroquinolones (ciprofloxacin and levofloxacin, see Table 2).
TABLE 2.
Anatomical sites of isolation and antibiotic susceptibility patterns of ESBL- and carbapenemase-producing Enterobacteriaceae isolates from children
| Anatomical sitea | No. of isolatesb | % susceptiblec |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTX | CAZ | FEP | P/T | CPMd | GNT | TOB | AMK | FQ | TMP-SMX | TET | NIT | ||
| All | 222 | 2.5 | 3.7 | 4.5 | 79.1 | 98.2 | 56.2 | 45.0 | 97.1 | 50.8 | 60.4 | 42.6 | 81.8 |
| Urine | 155 | 2.8 | 4.5 | 3.8 | 86.7 | 98.1 | 54.3 | 44.8 | 97.4 | 43.1 | 37.5 | 43.5 | 82.2 |
| Respiratory | 38 | 2.6 | 2.6 | 2.8 | 64.3 | 97.3 | 44.7 | 45.9 | 97.4 | 73.7 | 56.8 | 60.0 | 0.0 |
| Blood | 10 | 0.0 | 0.0 | 0.0 | 33.3 | 100 | 55.6 | 55.6 | 100 | 55.6 | 33.3 | 0.0 | 0.0 |
| Wound | 7 | 0.0 | 0.0 | 28.6 | 100 | 100 | 57.1 | 28.6 | 100 | 28.6 | 57.1 | 0.0 | ND |
| Other | 7 | 0.0 | 0.0 | 0.0 | 69.0 | 100 | 71.4 | 57.1 | 100 | 57.1 | 28.6 | 50.0 | ND |
| Abd/Perit | 4 | 0.0 | 0.0 | 0.0 | 100 | 50.0 | 0.0 | 50.0 | 100 | 100 | 25.0 | ND | ND |
| CNS | 1 | 0.0 | 0.0 | 0.0 | ND | 100 | 100 | 100 | 100 | 0.0 | 100 | ND | ND |
“Respiratory” includes bronchoalveolar lavage fluid, trachea, and sputum cultures. “Wound” includes abscess and wound cultures. “Abd/Perit” includes intra-abdominal and peritoneal fluid cultures. CNS, central nervous system cultures.
That is, the number of isolates with antibiotic susceptibility data available.
CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; P/T, piperacillin/tazobactam; CPM, carbapenems (includes imipenem, meropenem, and ertapenem); GNT, gentamicin; TOB, tobramycin; AMK, amikacin; FQ, fluoroquinolones (includes ciprofloxacin and levofloxacin); TMP-SMX, trimethoprim-sulfamethoxazole; TET, tetracycline; NIT, nitrofurantoin. ND, no data.
Carbapenem resistance was due exclusively to the CPE isolates.
Composition of bla genes in Enterobacteriaceae isolates.
Table 3 summarizes the bla genes detected by DNA microarray testing. Molecular characterization revealed that 90.7% of isolates contained an ESBL, AmpC, or carbapenemase gene, and in some isolates more than one bla was found (238 bla genes in 225 isolates). CTX-M-type ESBLs were the most common bla genes detected; they were found in 152 of 225 Enterobacteriaceae isolates (67.6%). Approximately half (49.8%) belonged to the blaCTX-M-1 group, which contains blaCTX-M-15, the gene most frequently associated with pandemic CTX-M E. coli strains (1, 23). Additionally, blaTEM- and blaSHV-type ESBL genes were found in 5.3 and 16.4% of isolates, respectively, and only 0.4% isolates contained blaPER-type ESBL genes.
TABLE 3.
bla genes detected in the collection of Enterobacteriaceae isolates from Chicago children
| bla gene (no.) in Enterobacteriaceae samples (n = 225)a | % detection of bla genes in isolates (no. of isolates) by organismb |
||||||
|---|---|---|---|---|---|---|---|
| All (225) | E. coli (137) | Klebsiella spp. (37) | Enterobacter spp. (30) | Proteus spp. (11) | Serratia spp. (8) | Other (2) | |
| Genes encoding ESBL (202) | |||||||
| CTX-M-1 group (112) | 49.8 | 81.3 | 10.7 | 3.6 | 4.5 | 0.0 | 0.0 |
| CTX-M-9 group (36) | 16.0 | 83.3 | 11.1 | 2.8 | 2.8 | 0.0 | 0.0 |
| CTX-M-2 group (4) | 1.8 | 25.0 | 25.0 | 0.0 | 50.0 | 0.0 | 0.0 |
| TEM (12) | 5.3 | 66.7 | 8.3 | 8.3 | 16.7 | 0.0 | 0.0 |
| SHV (37) | 16.4 | 16.2 | 29.7 | 40.5 | 0.0 | 13.5 | 0.0 |
| PER (1) | 0.4 | 100 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| VEB (0) | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Genes encoding AmpC (32) | |||||||
| ACT/MIR (25) | 11.1 | 16.0 | 0.0 | 84.0 | 0.0 | 0.0 | 0.0 |
| CMY (7) | 3.1 | 85.7 | 0.0 | 0.0 | 0.0 | 0.0 | 14.3c |
| Genes encoding carbapenemases (4) | |||||||
| KPC (3) | 1.3 | 25.0 | 75.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| IMP (1) | 0.4 | 0.0 | 100 | 0.0 | 0.0 | 0.0 | 0.0 |
| Total bla genes (238) | |||||||
bla, β-lactamase gene, ESBL, extended-spectrum β-lactamases; AmpC, AmpC cephalosporinases; KPC, Klebsiella pneumoniae carbapenemase; IMP, active on imipenem.
Isolates may contain more than one bla gene; 9.3% (n = 21) of the isolates were bla gene negative. Wild-type, narrow-spectrum bla genes were not included in the totals.
This value represents an intrinsic chromosomal blaAmpC gene specific to Citrobacter spp., which was picked up by the DNA microarray (Check-Points) as a blaCMY-II-like gene.
blaAmpC cephalosporinase genes comprised the resistance determinants detected in 32/225 (14.2%) of isolates, of which 7 (3.1%) were blaCMY-type genes and 25 (11.1%) were blaACT/MIR-type AmpC genes. blaCMY genes were predominantly identified in E. coli (6 of 7), whereas the majority of 25 ACT/MIR genes were recovered from Enterobacter spp. (84%), with 16% in E. coli isolates. blaACT/MIR in Enterobacter spp. were often associated with blaSHV-type ESBL genes, 12/21 (57%), and coexistence of ESBL and AmpC genes were found in 21 of 32 (65.6%) Enterobacteriaceae isolates. blaAmpC genes were not detected in Proteus or Serratia spp.; however, a blaCMY-like gene was found in one Citrobacter freundii isolate, which represents an intrinsic chromosomal blaAmpC gene specific to Citrobacter spp., based on DNA sequence analysis. Carbapenemases were detected in four isolates (1.8%), three of which were identified as blaKPC and one K. pneumoniae contained a blaIMP metallo-β-lactamase gene. The blaKPC genes were carried by two K. pneumoniae and one E. coli strains, and all were identified by DNA sequencing to be blaKPC-2. Other ESBL, pAmpC, or carbapenemase bla genes were not detected in carbapenemase-containing isolates.
Rep-PCR, MLST, bla and hsp60 gene sequencing, and plasmid replicon typing.
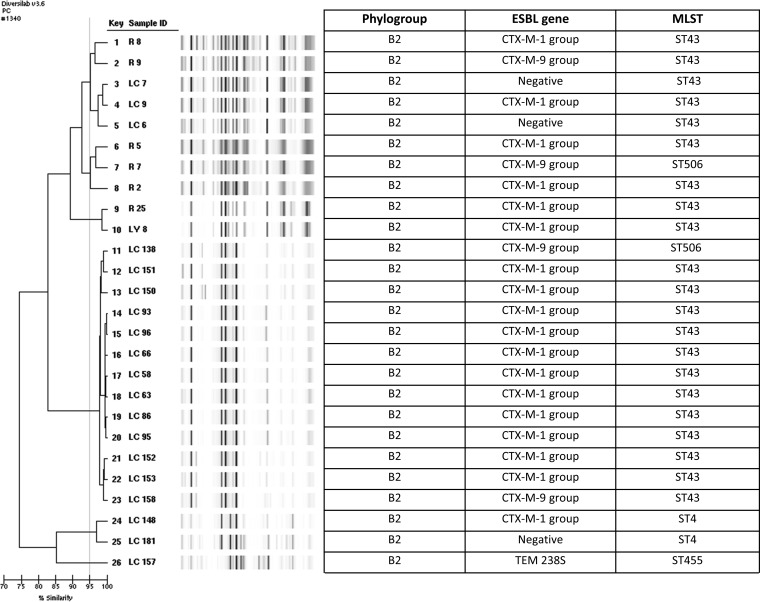
Rep-PCR was performed on all E. coli, Klebsiella spp. and Enterobacter isolates. We identified 40 different strain types of E. coli by rep-PCR. A dendrogram of the most highly represented rep-PCR profiles is shown in Fig. 1, representing 32.5% of the isolates. These corresponded to phylogroup B2. A subset of isolates with rep-PCR profiles related to one or more other strains were further studied by MLST, bla gene sequencing and replicon typing, and the majority were found to correspond to ST43 (ST131 in Achtman's MLST scheme), carry blaCTX-M-15, and contain plasmids of incompatibility types of replicon FIA, FII, and FIB. The E. coli strain harboring a blaKPC-2 gene belonged to phylogroup D, was identified as a novel sequence type (ST701), and had a distinct fingerprint pattern.
FIG 1.
Genetic relatedness in representative ESBL E. coli isolates from Chicago children. Isolates in which band patterns demonstrated >95% similarity (Pearson's correlation) were considered to be clonal and of the same strain type. Phylogroup, phylogenetic group; ESBL, extended-spectrum β-lactamase; MLST, multilocus sequence type, Pasteur scheme. ST43 is equivalent to ST131 on the Achtman's scheme. MLST was performed on select isolates from rep-PCR strain types. The CTX-M-1 group of note contains CTX-M-15, associated with the pandemic CTX-M type ESBL in E. coli. The CTX-M-9 group of note contains CTX-M-9 and CTX-M-14, the second most common circulating CTX-M type ESBL. LC 157 is a novel ST type.
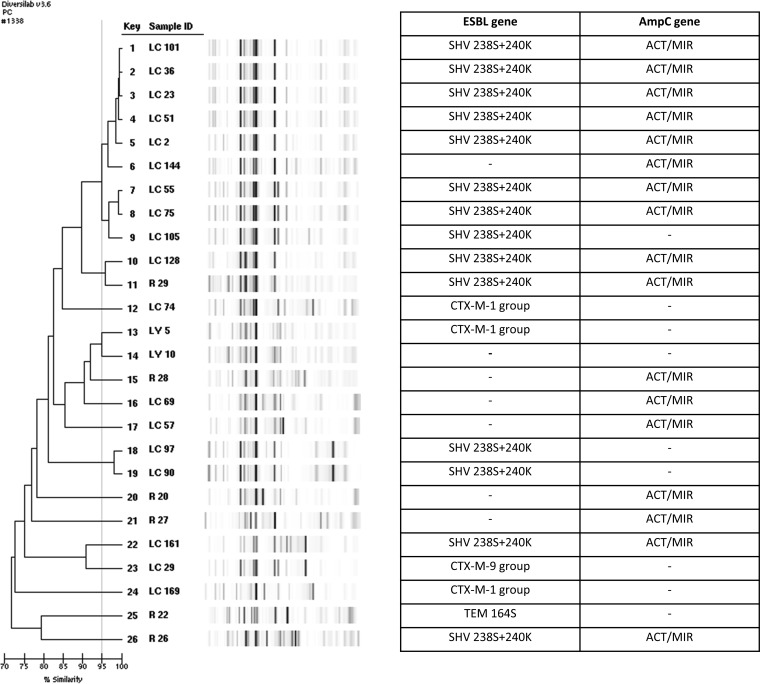
With respect to K. pneumoniae isolates, the rep-PCR profiles were mainly unrelated (see Fig. S1 in the supplemental material). Plasmid replicon typing revealed that the blaKPC-2-producing K. pneumoniae isolates contained plasmid replicon types I1 and A/C, and MLST showed that neither belonged to the ST258 lineage (ST22 and ST29). The K. pneumoniae containing the blaIMP gene was identified in ST253. Genetic relatedness in Enterobacter strains by rep-PCR (Fig. 2) correlated well with the hsp60 sequencing results and have been presented previously (24).
FIG 2.
Genetic relatedness of β-lactamase-carrying Enterobacter isolates from Chicago children. A dash indicates no β-lactamase gene was detected. Isolates in which band patterns demonstrated >95% similarity (Pearson's correlation) were considered clonal and of the same strain type. ESBL, extended-spectrum β-lactamase. AmpC, AmpC cephalosporinase; MLST, multilocus sequence type, Pasteur scheme. The term “ACT/MIR” means the gene may be an ACT- or MIR-type AmpC cephalosporinase gene but was not further differentiated by DNA microarray (Check-Points).
Phylogenetic grouping of E. coli.
Phylogenetic groups of E. coli include four main groups (A, B1, B2, and D), and groups B2 and D are most often associated with severe clinical disease attributed to increased virulence factors (21). Of 136 E. coli isolates tested, 119 (87.5%) were phylogenetic group B2 or D, with 67.6% and 19.1% belonging to B2 and D, respectively. One result was indeterminate (B2/D). Of all the E. coli strains, 78/136 (57.4%) were associated with blaCTX-M-1 group and 19.1% were associated with blaCTX-M-9 group. Most (65/78, 83.3%) belonged to the B2-E. coli blaCTX-M-1 group. Only 14 (10.3%) and 3 (2.2%) of the strains were of phylogroups A and B1, respectively.
DISCUSSION
In this unique survey, we linked resistance phenotypes with the genetic determinants of antibiotic resistance in Enterobacteriaceae isolates from children. The children from which these isolates were recovered were from three different centers located in the same city. As a result, we have an important “snapshot” on the molecular epidemiology of this emerging problem. A recent national study of trends of ESBL-producing Enterobacteriaceae in children using antimicrobial susceptibility data from 300 U.S. laboratories reported that the prevalence of the ESBL phenotype in Enterobacteriaceae isolated from children more than tripled during the study period, from 0.28% in 1999 to 0.92% in 2011, with the largest increases occurring in young children ages 1 to 5 years and in the intensive care unit setting (although the increase was seen in all age groups and health care settings) (11). This is consistent with our patient demographics. We also found that the most common circulating strain in children was phylogenetic group B2, multilocus sequence type 43 (ST43) E. coli harboring blaCTX-M-1 group ESBLs, which contain blaCTX-M-15, the predominant ESBL type disseminating globally. The pandemic, MDR, CTX-M-producing E. coli strains are of a large clonal lineage possessing the FimH30 allele (of the type 1 fimbriae fimH adhesin gene) and belong to the virulent phylogenetic group B2 (associated with extraintestinal pathogenic E. coli) and ST43 of the Pasteur MLST scheme, which is specific to the H30 subclone of the ST131 of the Achtman's MLST scheme (25).
The E. coli ST43/ST131 CTX-M strains are diverse due to a broad range of plasmids carrying a variety of resistance determinants; however, these strains commonly carry genes associated with resistance to fluoroquinolones, concomitant resistance to aminoglycosides, and trimethoprim-sulfamethoxazole (TMP-SMX) (26). In the United States, E. coli ST43 and ST131 are discovered more commonly among health care-associated strains; however, there are increasing reports of community acquisition globally. These isolates are associated with serious infections, especially of the urinary tract and bloodstream, and have significant attributable morbidity and mortality (1, 23). In our pediatric population, we found that of 136 E. coli isolates, 68% were phylogenetic group B2, and 84% of those strains harbored blaCTX-M-1 group ESBL genes, an observation consistent with adult and Chicago area data (25–27). We also found significant coresistance with 51, 56, and 60% of isolates displaying fluoroquinolone, aminoglycoside, and TMP-SMX resistance, respectively.
The rise of ESBL-producing Enterobacteriaceae in the pediatric community is important for many reasons. There are few drugs available and approved to treat infections with these organisms in children (9, 23), and children who become colonized with MDR Enterobacteriaceae may be colonized for prolonged periods, even up to 4 years, thus potentially serving as ongoing, silent sources of spread (23, 28, 29). Although the predominance of blaCTX-M-type ESBL genes in E. coli in our population was consistent with data from adults, the genotypic information from other studied genera was different. Our study was performed in a region that is endemic for KPC-producing Enterobacteriaceae strains, but we did not find evidence of the ST258 K. pneumoniae which harbors the blaKPC gene (blaKPC-2 or blaKPC-3) in our patient population (30). The blaKPC-containing K. pneumoniae isolates that we identified were ST22 and ST29 and carried blaKPC-2. Our findings are consistent with single-center pediatric studies suggesting that U.S. children with carbapenemase-producing Enterobacteriaceae may be more commonly infected with endemic strain types circulating within their institution or region, which may differ from adult studies where long-term-care facilities have been shown to be a significant reservoir of KPC-containing K. pneumoniae affecting acute care hospitals via interfacility transfer (10, 29, 31). Furthermore, recent national phenotypic data suggest that carbapenem-resistant Enterobacteriaceae infections are increasing in U.S. children and that the genetic makeup of these carbapenem-resistant pathogens likely differs from adults (12).
Our finding of a blaIMP metallo-β-lactamase (MBL)-producing K. pneumoniae in our population likely represents a “sentinel event,” and cases of MBLs in U.S. children have only recently been described, including infections associated with New Delhi MBL (NDM) (32). Knowing the molecular mechanisms of β-lactam resistance in Enterobacteriaceae is extremely valuable since it may impact treatment decisions and infection control procedures such as patient isolation, cohorting, and environmental cleaning methodologies. The rapid global dissemination of carbapenemase genes, such as the blaKPC, blaNDM, blaVIM, and blaIMP MBL genes, as well as the blaCTX-M ESBL genes, is due to the successful integration and transfer of mobile genetic elements in Gram-negative bacteria. Local and regional surveillance identifying patients carrying these organisms can help prevent intra- and interfacility spread (33).
Our AmpC cephalosporinase data are unique in that the most common transmissible blaAmpC genes thought to be circulating in U.S. children and adults are the blaCMY-2 genes, which are most often found in E. coli (23, 34); however, in our isolates, the most common blaAmpC genes identified were blaACT/MIR-type genes in Enterobacter isolates (84%), which were associated with blaSHV-type ESBL genes in 40.5% of cases. Typing of a subset of Enterobacter isolates revealed blaACT-16 and blaACT-17 genes and plasmid replicon type FIIA. Our data suggest that for Enterobacter, the dissemination of ESBLs may be related to specific mobile genetic elements, i.e., “promiscuous plasmids” carrying bla genes, rather than predominant circulating clonal types (Fig. 2). Of note, 6% of isolates in this study were found to contain only blaAmpC genes; however, all were phenotypically identified as ESBL producers. We emphasize this as the treatment of ESBL and AmpC producers is different. For example, one might consider cefepime therapy in the treatment of infections due to AmpC-positive isolates, whereas for ESBL producers, cefepime is discouraged, and many institutions place patients with ESBL-producing Enterobacteriaceae infections under isolation precautions. Further detailed characterization of these strains is necessary to delineate whether “silent dissemination” of plasmid-mediated AmpC is occurring in the pediatric population at a greater frequency than previously recognized (28, 35, 36).
We recognize the limitations of our study. The design is a retrospective cohort study of resistance mechanisms in Enterobacteriaceae recovered from children located at three centers in a single geographic region, which may impact generalizability to other regions. Isolates were collected on the basis of phenotypic resistance suggestive of ESBL and carbapenemase production; therefore, no antibiotic-sensitive strains were collected. In addition, whereas a plasmid-based location of the bla genes by the majority of isolates is supported by DNA sequence analysis, some of these genes may represent chromosome-based mechanisms of resistance. Further studies are ongoing to assess plasmid localization of the determinants uncovered during the study. We did perform DNA sequence analysis for a subset of isolates and plasmid replicon typing in order to further strengthen our DNA microarray results.
In conclusion, our study represents the first multicentered U.S. study of the molecular epidemiology of ESBL-, AmpC-, and carbapenemase-producing Enterobacteriaceae isolates from children cared for at pediatric acute care facilities within a single metropolitan area. We found that the characterization of plasmid-mediated β-lactam resistance in children is complex and diverse and that the molecular characteristics in pediatric isolates exhibit differences compared to strain types circulating in adults in an area where such infections are endemic. This diversity and complexity must be further studied to assess the potential impact of various molecular types in pediatric infections and the imminent threat of silent dissemination of these dangerous bacteria within a vulnerable population. Our study also highlights the unique challenges that will be faced when developing strategies to control the spread of MDR organisms in children: having novel strain types as carriers of carbapenemase genes portends an even more complex molecular epidemiology.
Supplementary Material
ACKNOWLEDGMENTS
We thank Paul Schreckenberger and the microbiology laboratories of the participating institutions for providing isolates for this study. We thank Pamela Hagen, Jane Stevens, Joyce Houlihan, Kathleen McKinley, Violeta Rekasiu, and Donna Carter for collection, shipping, and cultivation of organisms. We thank Robert A. Weinstein, James B. McAuley, and Kenneth M. Boyer for thoughtful comments and guidance. We thank the team of curators of the Institut Pasteur MLST and whole-genome MLST databases for curating the data and making them publicly available at http://bigsdb.web.pasteur.fr/. We report no conflicts of interest relevant to this study.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the Department of Veterans Affairs or the National Institutes of Health.
Funding Statement
R.A.B. is also supported by the Department of Veterans Affairs Research and Development under award number I01BX001974, VISN 10 Geriatrics Research, Education and Clinical Center. L.K.L. is also supported by the Rush-Stroger Collaborative Award and The Children's Foundation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00098-16.
REFERENCES
- 1.Cantón R, Coque TM. 2006. The CTX-M β-lactamase pandemic. Curr Opin Microbiol 9:466–475. doi: 10.1016/j.mib.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 2.Bush K, Jacoby GA. 2010. Updated functional classification of β-lactamases. Antimicrob Agents Chemother 54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paterson DL. 2006. Resistance in gram-negative bacteria: Enterobacteriaceae. Am J Infect Control 34:S20–S28. doi: 10.1016/j.ajic.2006.05.238. [DOI] [PubMed] [Google Scholar]
- 4.Bush K, Fisher J. 2011. Epidemiological expansion, structural studies, and clinical challenges of new β-lactamases from gram-negative bacteria. Annu Rev Microbiol 65:455–478. doi: 10.1146/annurev-micro-090110-102911. [DOI] [PubMed] [Google Scholar]
- 5.Bush K. 2014. ICAAC Award Lecture: β-lactamases: ubiquitous and formidable. 54th Intersci Conf Antimicrob Agents Chemother (ICAAC) American Society for Microbiology, Washington, DC. [Google Scholar]
- 6.Paterson DL, Bonomo RA. 2005. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev 18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munoz-Price LS, Quinn JP. 2009. The Spread of Klebsiella pneumoniae carbapenemases: a tale of strains, plasmids, and transposons. Clin Infect Dis 49:1739–1741. doi: 10.1086/648078. [DOI] [PubMed] [Google Scholar]
- 8.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 9.Hsu AJ, Tamma PD. 2014. Treatment of multidrug-resistant Gram-negative infections in children. Clin Infect Dis 58:1439–1448. doi: 10.1093/cid/ciu069. [DOI] [PubMed] [Google Scholar]
- 10.Logan LK. 2012. Carbapenem-resistant enterobacteriaceae: an emerging problem in children. Clin Infect Dis 55:852–859. doi: 10.1093/cid/cis543. [DOI] [PubMed] [Google Scholar]
- 11.Logan LK, Braykov NP, Weinstein RA, Laxminarayan R. 2014. Extended-spectrum β-lactamase-producing and third-generation cephalosporin-resistant Enterobacteriaceae in children: trends in the United States, 1999-2011. J Pediatr Infect Dis Soc 3:320–328. doi: 10.1093/jpids/piu010. [DOI] [PubMed] [Google Scholar]
- 12.Logan L, Renschler J, Gandra S, Weinstein R, Laxminarayan R. 2015. Carbapenem-resistant Enterobacteriaceae in children, United States, 1999-2012. Emerg Infect Dis 21:2014–2021. doi: 10.3201/eid2111.150548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial susceptibility testing: 20th informational supplement (June 2010 update). Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 14.Centers for Disease Control and Prevention. 2012. CDC-CRE toolkit: guidance for control of carbapenem-resistant Enterobacteriaceae (CRE). Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 15.Bogaerts P, Hujer AM, Naas T, de Castro RR, Endimiani A, Nordmann P, Glupczynski Y, Bonomo RA. 2011. Multicenter evaluation of a new DNA microarray for rapid detection of clinically relevant bla genes from β-lactam-resistant gram-negative bacteria. Antimicrob Agents Chemother 55:4457–4460. doi: 10.1128/AAC.00353-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cunningham SA, Vasoo S, Patel R. 2016. Evaluation of the Check-Points Check MDR CT103 and CT103 XL microarray kits by use of preparatory rapid cell lysis. J Clin Microbiol 54:1368–1371. doi: 10.1128/JCM.03302-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pitout JDD, Campbell L, Church DL, Wang PW, Guttman DS, Gregson DB. 2009. Using a commercial DiversiLab semiautomated repetitive sequence-based PCR typing technique for identification of Escherichia coli clone ST131 producing CTX-M-15. J Clin Microbiol 47:1212–1215. doi: 10.1128/JCM.02265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaureguy F, Landraud L, Passet V, Diancourt L, Frapy E, Guigon G, Carbonnelle E, Lortholary O, Clermont O, Denamur E, Picard B, Nassif X, Brisse S. 2008. Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genomics 9:560–2164-9–560. doi: 10.1186/1471-2164-9-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann H, Roggenkamp A. 2003. Population genetics of the nomenspecies Enterobacter cloacae. Appl Environ Microbiol 69:5306–5318. doi: 10.1128/AEM.69.9.5306-5318.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bingen-Bidois M, Clermont O, Bonacorsi S, Terki M, Brahimi N, Loukil C, Barraud D, Bingen E. 2002. Phylogenetic analysis and prevalence of urosepsis strains of Escherichia coli bearing pathogenicity island-like domains. Infect Immun 70:3216–3226. doi: 10.1128/IAI.70.6.3216-3226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 23.Lukac PJ, Bonomo RA, Logan LK. 2015. Extended-spectrum β-lactamase-producing Enterobacteriaceae in children: old foe, emerging threat. Clin Infect Dis doi: 10.1093/cid/civ020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viau R, Kiedrowski L, Perez F, Marchaim D, Guerrero D, Kaye K. 2014. K-1676: outbreak analysis of Enterobacter cloacae: hsp60 compares favorably to rep-PCR. Intersci Conf Antimicrob Agents Chemother (ICAAC) American Society for Microbiology, Washington, DC. [Google Scholar]
- 25.Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother 66:1–14. doi: 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 26.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan M, Gomes Moriel D, Peters KM, Davies M, Rogers BA, Dougan G, Rodriguez-Baño J, Pascual A, Pitout JDD, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A 111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peirano G, Costello M, Pitout JD. 2010. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli from the Chicago area: high prevalence of ST131 producing CTX-M-15 in community hospitals. Int J Antimicrob Agents 36:19–23. doi: 10.1016/j.ijantimicag.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 28.Zerr DM, Qin X, Oron AP, Adler AL, Wolter DJ, Berry JE, Hoffman L, Weissman SJ. 2014. Pediatric infection and intestinal carriage due to extended-spectrum cephalosporin-resistant Enterobacteriaceae. Antimicrob Agents Chemother 58:3997–4004. doi: 10.1128/AAC.02558-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viau RA, Hujer AM, Marshall SH, Perez F, Hujer KM, Briceno DF, Dul M, Jacobs MR, Grossberg R, Toltzis P, Bonomo RA. 2012. “Silent” dissemination of Klebsiella pneumoniae isolates bearing K. pneumoniae carbapenemase in a long-term care facility for children and young adults in Northeast Ohio. Clin Infect Dis 54:1314–1321. doi: 10.1093/cid/cis036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin MY, Lyles-Banks RD, Lolans K, Hines DW, Spear JB, Petrak R, Trick WE, Weinstein RA, Hayden MK, for the Centers for Disease Control and Prevention Epicenters Program . 2013. The importance of long-term acute care hospitals in the regional epidemiology of Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae. Clin Infect Dis 57:1246–1252. doi: 10.1093/cid/cit500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stillwell T, Green M, Barbadora K, Ferrelli JG, Roberts TL, Weissman SJ, Nowalk A. 2014. Outbreak of KPC-3-producing carbapenem-resistant Klebsiella pneumoniae in a US pediatric hospital. J Pediatr Infect Dis doi: 10.1093/jpids/piu080. [DOI] [PubMed] [Google Scholar]
- 32.Pannaraj P, Bard J, Cerini C, Weissman S. 2015. Pediatric carbapenem-resistant Enterobacteriaceae in Los Angeles, California, a high-prevalence region in the United States. Pediatr Infect Dis J 34:11–16. doi: 10.1097/INF.0000000000000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trick W, Lin M, Cheng-Leidig R, Driscoll M, Tang A, Wei G, Runningdeer E, Arwady M, Weinstein R. 2015. Electronic public health registry of extensively drug-resistant organisms, Illinois, USA. Emerg Infect Dis 21:1725–1732. doi: 10.3201/eid2110.150538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacoby GA. 2009. AmpC β-lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin X, Zerr DM, Weissman SJ, Englund JA, Denno DM, Klein EJ, Tarr PI, Kwong J, Stapp JR, Tulloch LG, Galanakis E. 2008. Prevalence and mechanisms of broad-spectrum β-lactam resistance in Enterobacteriaceae: a children's hospital experience. Antimicrob Agents Chemother 52:3909–3914. doi: 10.1128/AAC.00622-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weissman SJ, Adler A, Qin X, Zerr DM. 2013. Emergence of extended-spectrum β-lactam resistance among Escherichia coli at a US academic children's hospital is clonal at the sequence type level for CTX-M-15, but not for CMY-2. Int J Antimicrob Agents 41:414–420. doi: 10.1016/j.ijantimicag.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.