Abstract
The recent widespread emergence of carbapenem resistance in Enterobacteriaceae is a major public health concern, as carbapenems are a therapy of last resort against this family of common bacterial pathogens. Resistance genes can mobilize via various mechanisms, including conjugation and transposition; however, the importance of this mobility in short-term evolution, such as within nosocomial outbreaks, is unknown. Using a combination of short- and long-read whole-genome sequencing of 281 blaKPC-positive Enterobacteriaceae isolates from a single hospital over 5 years, we demonstrate rapid dissemination of this carbapenem resistance gene to multiple species, strains, and plasmids. Mobility of blaKPC occurs at multiple nested genetic levels, with transmission of blaKPC strains between individuals, frequent transfer of blaKPC plasmids between strains/species, and frequent transposition of blaKPC transposon Tn4401 between plasmids. We also identify a common insertion site for Tn4401 within various Tn2-like elements, suggesting that homologous recombination between Tn2-like elements has enhanced the spread of Tn4401 between different plasmid vectors. Furthermore, while short-read sequencing has known limitations for plasmid assembly, various studies have attempted to overcome this by the use of reference-based methods. We also demonstrate that, as a consequence of the genetic mobility observed in this study, plasmid structures can be extremely dynamic, and therefore these reference-based methods, as well as traditional partial typing methods, can produce very misleading conclusions. Overall, our findings demonstrate that nonclonal resistance gene dissemination can be extremely rapid, presenting significant challenges for public health surveillance and achieving effective control of antibiotic resistance.
INTRODUCTION
Although antibiotic resistance genes have been identified in ancient bacterial DNA (1), much of the recent, alarming increase in pathogen antimicrobial resistance is attributable to the dissemination of resistance genes via horizontal gene transfer (HGT) in response to selection imposed by widespread antibiotic use in medicine and agriculture (2, 3). Many resistance genes are located on plasmids that can be transferred between different bacterial strains or species, thus facilitating HGT (4). Furthermore, resistance gene mobility can be enhanced by integration into transposable elements, which are short stretches of DNA (several kilobases) that can autonomously mobilize between different genomic locations (5). However, the importance of HGT in short-term evolution is unclear, as capturing the processes in real time is challenging and outbreaks in health care settings are often thought to be dominated by clonal transmission (6–9).
Carbapenem resistance in Enterobacteriaceae has been recognized as a key threat to modern medicine (10, 11), as carbapenems often represent the therapy of last resort for serious infections (12, 13). One of the most prevalent carbapenem resistance genes is the Klebsiella pneumoniae carbapenemase (KPC) gene, blaKPC, first identified in 1996 and now endemic to many regions of the world (14). KPC is a beta-lactamase capable of hydrolyzing all beta-lactams, including penicillins, monobactams, cephalosporins, and carbapenems (15), leaving few treatment options for infected vulnerable hospitalized patients and resulting in worse treatment outcomes (16).
Most reports of blaKPC involve K. pneumoniae multilocus sequence type 258 (ST258) (9, 17), which has been found globally, indicating that clonal dissemination of this resistant lineage has been an important factor in the spread of blaKPC (9, 17–20). Nevertheless, blaKPC has also been observed in other K. pneumoniae lineages, as well as other species of Enterobacteriaceae, suggesting that blaKPC HGT has also played a role in resistance dissemination (21–25). As blaKPC is often found on conjugative plasmids, some of which have been identified in multiple strains or species, this provides a likely mechanism for HGT (21, 26, 27). In addition, blaKPC is usually present as part of the 10-kb Tn3-based mobile transposon Tn4401, which has been identified in various different plasmids, implicating Tn4401 transposition as another mechanism contributing to blaKPC spread (28, 29).
While Tn4401 transposition and plasmid conjugation have been measured in the laboratory (28, 30, 31), the frequencies with which these processes occur within real-world ecosystems are not fully understood. In clinical contexts, it is often assumed that short-term evolution is dominated by clonal propagation, such that transmission chains generally involve a single pathogenic strain. However, if HGT is frequent relative to transmission (e.g., a “plasmid outbreak”), then linked patients may show variation in strain composition. If transposition is also frequent, then both the host strain and the resistance plasmid may show high variability within a single outbreak. As current surveillance strategies tend to focus on the host strain, it is important to establish the relevance of blaKPC mobility within outbreak settings.
Traditional approaches to plasmid investigation, such as PCR-based replicon typing, are limited in resolution. Next-generation sequencing has been successfully applied to molecular epidemiological investigation of a number of pathogens at the host strain level; however, the application and limitations of this technology for transmission chains involving HGT are relatively unexplored. Whole-genome sequencing using short-read technologies (e.g., Illumina) has become cheap and accessible but is not ideal for plasmid analysis because of de novo assembly limitations, as it is often not possible to accurately reconstruct the genomic context surrounding repeated sequences (21, 32). Long-read sequencing (e.g., PacBio) can largely overcome this, often providing single-contig plasmid assemblies, but it is prohibitively expensive for many applications. Several studies have utilized reference-based methods for plasmid assembly or inference of plasmid structures using short-read data (33, 34); however, these approaches make the implicit assumption that plasmid structures are relatively stable. It will be important to understand the potential shortcomings of these assumptions in relation to mobile genetic elements, which may frequently be involved in plasmid rearrangements. Understanding when and how to successfully apply short- and/or long-read sequencing technologies to molecular epidemiology tracking will be important to the field as the incidence of HGT is increasingly recognized (35).
At our institution, blaKPC was first identified in 2007 in a patient simultaneously colonized with blaKPC-positive K. pneumoniae and Klebsiella oxytoca harboring blaKPC plasmids pKPC_UVA01 and pKPC_UVA02, respectively (36, 37). Since then, we have prospectively screened extended-spectrum cephalosporin-resistant/carbapenem-nonsusceptible isolates of all Enterobacteriaceae species for blaKPC, despite national guidelines that recommend that screening focus on carbapenem-nonsusceptible Klebsiella species and Escherichia coli (38–41). Here we describe the genetic basis of nonclonal blaKPC emergence in a single hospital setting by using a combination of short- and long-read whole-genome sequencing methods to provide genomic characterization of 281 Enterobacteriaceae isolates from the first 5 years of this multispecies blaKPC outbreak.
MATERIALS AND METHODS
Isolate collection and Illumina sequencing.
Isolates were prospectively collected from August 2007 to December 2012 through the Clinical Microbiology Laboratory of the University of Virginia Health System, which serves a 619-bed tertiary care hospital, outpatient clinics in central Virginia, and since August 2010, a 40-bed long-term acute care hospital. From April 2009, weekly surveillance by perirectal swab was performed in all inpatient units with historically high transmission or where there was a patient who was known to be colonized or infected with carbapenemase-producing Enterobacteriaceae (CPE) by previously described methods (40, 42, 43). Enterobacteriaceae isolates from nonsurveillance clinical samples that were flagged as possible extended-spectrum β-lactamase (ESBL) producing or had an ertapenem MIC of ≥1 μg/ml by automated susceptibility profiling (VITEK2; bioMérieux, Durham, NC) underwent carbapenemase phenotypic testing by the modified Hodge test (August 2007 to June 2008) or the indirect carbapenemase test (July 2008 to December 2012). Isolates with a positive carbapenemase phenotypic test and/or a meropenem or imipenem MIC of ≥1 μg/ml underwent blaKPC PCR analysis as previously described (39).
A subset of 37 K. pneumoniae isolates, with corresponding sequence data, have been previously described (37). For the rest of the study isolates, Illumina sequencing, de novo assembly, mapping, and variant calling were performed as previously described (37), with some exceptions (see the supplemental material), and including the use of additional, species-specific references for mapping (see Table S5 in the supplemental material). A total of 281 isolates from 182 patients were available for analysis; for the exclusion criteria used for additional isolates, see the supplemental material.
Classification to the species level.
Classification to the species level was performed by microbiological and sequenced-based methods (see the supplemental material for details).
Phylogenetic analysis and strain classification.
There were 52 patients with multiple isolates of the same species. One of these (patient FK) carried two strains of K. pneumoniae that were highly divergent from each other (>20,000 chromosomal single-nucleotide variants [SNVs]), clearly representing a separate acquisition of blaKPC by each strain. Excluding this divergent strain pair, the remaining cases had differences ranging from 0 to 60 SNVs (median, 2 SNVs). As these could plausibly represent clonal evolution within the patient, we conservatively chose to include only a single representative (the earliest isolate) for phylogenetic reconstruction, in order to avoid artificially inflating genetic clusters because of repeated patient sampling. Phylogenetic analysis was then performed separately for each species using PhyML (44) (see the supplemental material). Chromosomally distinct strains were defined by partitioning each phylogeny with a cutoff of ∼500 SNVs (see the supplemental material). On the basis of the molecular clock of Enterobacteriaceae (1 to 20 SNVs/chromosome/year) (6, 37, 45), we can be relatively confident that isolates belonging to distinct strains will not have a shared ancestor within the time frame of blaKPC dispersal, and the number of distinct strains thus provides a conservative estimate of the number of distinct blaKPC acquisition events.
Long-read PacBio sequencing.
For long-read sequencing, 17 isolates were randomly chosen from the entire set of sequenced isolates (i.e., including patient duplicates). Long-read sequencing and initial de novo assembly were performed as previously described (37). Refinement of assemblies and closure of plasmid/chromosomal sequences was performed as described in the supplemental material.
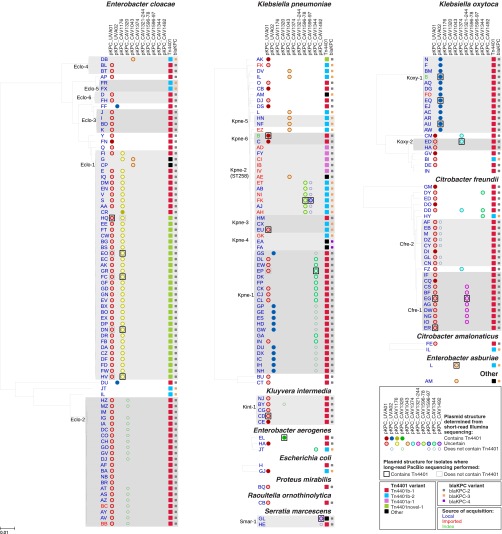
Since the isolates used for PacBio sequencing were randomly chosen from the set of all Illumina-sequenced isolates, some of them represented within-patient strain duplicates (see the previous section on phylogenetic analysis) and were therefore not included in the phylogenetic reconstruction. For display purposes (Fig. 1), the blaKPC structure(s) determined by long-read PacBio sequencing for each of these isolates is shown alongside the representative isolate of the same strain from the same patient. In all of the cases, the representative isolate has the same short-read plasmid profile and Tn4401 variant as the PacBio-sequenced isolate.
FIG 1.
Diversity of bacterial species, strains, plasmids, and Tn4401 variants. For each species, a phylogeny was generated from mapping to a species-specific chromosomal reference, after the deduplication of closely related isolates from the same patient (see Materials and Methods). Distinct strains are defined by a cutoff of ∼500 SNVs (see Materials and Methods); strains found in more than one patient are shaded gray. Circles show plasmid “presence” as determined from Illumina data, with the fill color indicating uncertainty about whether the plasmid contains blaKPC. Boxes show plasmid structures determined from long-read PacBio sequencing of 17 randomly chosen isolates, as well as the previously sequenced isolates from index patient B (37). Where the PacBio-sequenced isolate was excluded from the phylogeny as a patient duplicate, the plasmid structure of the corresponding closely related isolate from the same patient is shown. Tn4401 and blaKPC variants (Table 2) are indicated by large and small squares, respectively. The likely sources of blaKPC acquisition, as determined from epidemiological data, are indicated by text color.
Plasmid presence/absence classification.
The index blaKPC plasmids pKPC_UVA01 and pKPC_UVA02, together with the additional nine distinct blaKPC plasmids identified by long-read PacBio sequencing, were used as references to determine plasmid presence profiles for each isolate on the basis of the Illumina data. Plasmid presence was defined as ≥99% sequence identity over ≥80% of the length of the reference sequence, as determined by BLASTn comparison of each isolate's de novo assembly with the reference plasmid. The high identity cutoff was chosen to reduce false positives from sequences that are only distantly related (and therefore unlikely to have a common ancestor within the time frame of the outbreak), while the more permissive length cutoff allows for some rearrangement. It should be noted that the method does not take structural continuity into account.
Analysis of Tn4401 flanking sequences.
Where a plasmid was classified as being present in a particular isolate, it was not always certain to contain Tn4401. The plasmid presence classification was further refined as “containing Tn4401” if the isolate's de novo assembly supported Tn4401 being present within the expected sequence context of that plasmid, “not containing Tn4401” if the plasmid was assembled without Tn4401, or “uncertain” if structure could not be determined from the de novo assembly. The identification of novel Tn4401 insertion sites was also based on the de novo assemblies. These methods are described in detail in the supplemental material.
Variation in Tn4401.
Tn4401 isoform classification was performed by comparing each isolate's de novo assembly with the previously described isoform b reference sequence from EU176013.1 (29) using BLASTn to identify structural variation. SNVs were determined by mapping to a reference consisting of pKPC_UVA01 plus a species-specific chromosome as described above, followed by extraction of the Tn4401 region. Variation is reported for all sites where at least one isolate had a nonreference call, including any ambiguity at that site in other isolates. Ambiguity at nonvariable sites is not reported, which may result in an underestimate of true variation. However, any resulting underestimation is likely to be very minor, as the proportion of called sites, excluding deleted regions described above, was >96% for all isolates.
Epidemiological classification.
For epidemiologic analysis, patients were assigned a one- or two-letter code for deidentification. Routine perirectal surveillance cultures for silent colonization began in April 2009 (38, 40). Cases were classified as “imported” if they did not have any prior admission to the University of Virginia Medical Center/Long-term Acute Care Hospital (UVaMC) and either had a blaKPC-positive Enterobacteriaceae isolated within 48 h of admission or had a carbapenem-resistant Enterobacteriaceae culture before transfer to UVaMC with a subsequent isolate at UVaMC confirmed as blaKPC PCR positive. The index case was also classified as imported. In the remaining cases, the source of blaKPC acquisition was classified as “local.” The 48-h cutoff is arbitrary and may result in some misclassification if patients either acquire blaKPC within the first 48 h of admission or if blaKPC carriage/infection remains undetected for >48 h; however, this is expected to be minimal (see the supplemental material). Charts and patient contacts were reviewed by using bed tracing data and the electronic medical record. This study was approved by the University of Virginia Institutional Review Board (protocol 13558).
Transmission analysis.
Possible patient-to-patient transmission events were determined on the basis of having overlapping stays on the same ward, as well as genetically related blaKPC isolates. The analysis was performed separately for two different levels of genetic relatedness (strain or Tn4401 variant). This is described in detail in the supplemental material.
Nucleotide sequence accession number.
Sequence data obtained in this study have been deposited at the National Center for Biotechnology Information under BioProject no. PRJNA246471.
RESULTS
There were 204 patients infected/colonized with blaKPC-positive Enterobacteriaceae during the prospective sampling period, on the basis of clinical and surveillance sampling. We performed short-read Illumina sequencing of all 294 available isolates; 13 of them were excluded because of quality issues (see Materials and Methods), leaving 281 isolates, from 182/204 (89%) patients, for analysis (see Table S1 in the supplemental material). In all 281 isolates, blaKPC was carried within a complete or partial Tn4401 structure.
blaKPC is found in many different host strains, indicating frequent HGT.
There were 13 different species carrying blaKPC (Fig. 1). The four most prevalent species were Enterobacter cloacae (96 isolates from 80 patients), K. pneumoniae (94 isolates from 55 patients), Klebsiella oxytoca (35 isolates from 20 patients), and Citrobacter freundii (30 isolates from 25 patients), each of which showed substantial genetic diversity. Across all of the species, there were a total of 62 distinct strains (>500 chromosomal SNVs; see Materials and Methods). Of these, 18 strains were identified in multiple patients and 44 were seen in only a single patient (Fig. 1), with new strains continuing to appear throughout the study period. The very recent emergence of blaKPC on an evolutionary time scale (15) implies that each strain likely acquired blaKPC independently, demonstrating frequent HGT between different strains and species.
The blaKPC plasmids pKPC_UVA01 and pKPC_UVA02 are widely dispersed.
We hypothesized that the spread of blaKPC could be due to conjugative transfer of the index blaKPC plasmids, pKPC_UVA01 and pKPC_UVA02. With plasmid presence defined as ≥99% sequence identity over ≥80% of the plasmid length, 121 (66%) and 32 (18%) patients had isolates carrying pKPC_UVA01 and pKPC_UVA02, respectively, corresponding to 39 and 5 distinct strains from 10 and 4 species, respectively (Fig. 1). Although the wide dispersal of these plasmids supports the plasmid-mediated outbreak hypothesis, short-read data can be limited in providing structural inferences when repetitive sequences are present, and for many isolates, it was not possible to confirm that blaKPC was actually colocated within pKPC_UVA01 or pKPC_UVA02 (Fig. 1).
blaKPC is found in many different plasmids, indicating frequent Tn4401 transposition.
To further investigate blaKPC plasmid structures, we performed long-read PacBio sequencing of 17 isolates that were chosen at random from the 281 available, yielding closed blaKPC structures in all of the cases. Fifteen isolates had a single blaKPC plasmid, and two isolates had two blaKPC plasmids, giving a total of 19 blaKPC plasmids from the 17 isolates (Table 1). One isolate additionally had a chromosomal insertion of Tn4401.
TABLE 1.
blaKPC-containing structures ascertained from long-read PacBio sequencing of 17 randomly chosen isolates
| Isolate | Species | Patienth | Date | blaKPC plasmid | Size (bp) | Groupa | Within-group genetic change(s)b | Tn4401 variant | Flanking sequencesc | Tn2-like elementd |
|---|---|---|---|---|---|---|---|---|---|---|
| CAV1344 | K. pneumoniae | EP | Dec 2010 | pKPC_CAV1344 | 176,497 | Singleton | NAi | Tn4401b-1 | GTTCT…GTTCT | Yes |
| CAV1392 | K. pneumoniae | EU | Mar 2011 | pKPC_CAV1392 | 43,621 | pKPC_UVA01 | 1 SNV | Tn4401b-2 | GTTCT…GTTCT | Yes |
| NA (chromosomal) | NA | NA | NA | Tn4401b-2 | AGATA…AGATA | No | ||||
| CAV1596 | K. pneumoniae | FK | Apr 2012 | pKPC_CAV1596-78 | 77,801 | Singleton | NA | Tn4401b-2 | GTTCT…GTTCT | Yes |
| pKPC_CAV1596-97 | 96,702 | Singleton | NA | Tn4401b-2 | TATCG…TATCG | No | ||||
| CAV1099 | K. oxytoca | AU | Apr 2009 | pKPC_CAV1099 | 113,105 | pKPC_UVA02 | 0 SNVs | Tn4401b-1 | ATGCA…GGCCAe | No |
| CAV1335 | K. oxytoca | EQ | Dec 2010 | pKPC_CAV1335 | 113,105 | pKPC_UVA02 | 0 SNVs | Tn4401b-1 | ATGCA…GGCCAe | No |
| CAV1374 | K. oxytoca | ED | Aug 2010 | pKPC_CAV1374 | 332,956 | Singleton | NA | Tn4401b-1 | GTTCT…GTTCT | Yes |
| CAV1043 | E. asburiae | L | Mar 2008 | pKPC_CAV1043 | 59,138 | Singleton | NA | Tn4401b-2 | GTTCT…GTTCT | Yes |
| CAV1176 | E. cloacae | DN | May 2010 | pKPC_CAV1176 | 90,452 | pKPC_CAV1176 | 0 SNVs | Tn4401novel-1 | GTTCT…GTTCT | Yes |
| CAV1311 | E. cloacae | EO | Jan 2011 | pKPC_CAV1311 | 90,452 | pKPC_CAV1176 | 0 SNVs | Tn4401novel-1 | GTTCT…GTTCT | Yes |
| CAV1411 | E. cloacae | FC | Jun 2011 | pKPC_CAV1411 | 90,452 | pKPC_CAV1176 | 1 SNV, 40-kb inversion | Tn4401novel-1 | GTTCT…GTTCT | Yes |
| CAV1669 | E. cloacae | HV | Aug 2012 | pKPC_CAV1669 | 90,452 | pKPC_CAV1176 | 40-kb inversion | Tn4401novel-1 | GTTCT…GTTCT | Yes |
| CAV1668 | E. cloacae | HQ | Aug 2012 | pKPC_CAV1668 | 43,433 | pKPC_UVA01 | 1 SNV, 188-bp deletion | Tn4401novel-1 | GTTCT…GTTCT | Yes |
| CAV1321 | C. freundii | EG | Nov 2010 | pKPC_CAV1321-45 | 44,846 | pKPC_UVA01 | 1,225-bp insertion | Tn4401b-1 | GTTCT…GTTCT | Yes |
| pKPC_CAV1321-244 | 243,709 | Singleton | NA | Tn4401b-1 | GTTCT…GTTCT | Yes | ||||
| CAV1741 | C. freundii | ER | Oct 2012 | pKPC_CAV1741 | 129,196 | pKPC_UVA01 | 14,960-bp duplication, 70,615-bp insertion | Tn4401b-1f | GTTCT…GTTCT | Yes |
| CAV1151 | K. intermedia | CD | Sep 2009 | pKPC_CAV1151 | 43,621 | pKPC_UVA01 | 0 SNVsg | Tn4401b-1 | GTTCT…GTTCT | Yes |
| CAV1320 | E. aerogenes | EL | Nov 2010 | pKPC_CAV1320 | 13,981 | Singleton | NA | Tn4401b-1 | TTGTT…TTGTT | No |
| CAV1492 | S. marcescens | GL | Dec 2011 | pKPC_CAV1492 | 69,158 | Singleton | NA | Tn4401b-8 | TTTTT…TTTTT | No |
Plasmids are defined as belonging to the same group if the sequences are largely identical, allowing for a small number of substitutions and/or rearrangements that may be expected to occur within the outbreak time frame. Different groups have very limited homology outside the Tn4401 region, indicative of independent integrations into distinct plasmid structures. “Singleton” indicates a plasmid backbone that is distinct from all of the others shown.
Difference(s) from the reference sequence of that plasmid group, as specified in the previous column.
Sequences immediately flanking Tn4401; generally expected to be identical because of 5-bp target site duplication during transposition (28).
Tn4401 integrated into the tnpA gene of a Tn2-like element.
No evidence of target site duplication.
Two copies.
It is noteworthy that this plasmid from K. intermedia CAV1151 is exactly identical to pKPC_UVA01 from K. pneumoniae CAV1016, with isolation dates 2 years apart.
Anonymized patient identifiers are used; they do not represent initials or any other personal information.
NA, not applicable.
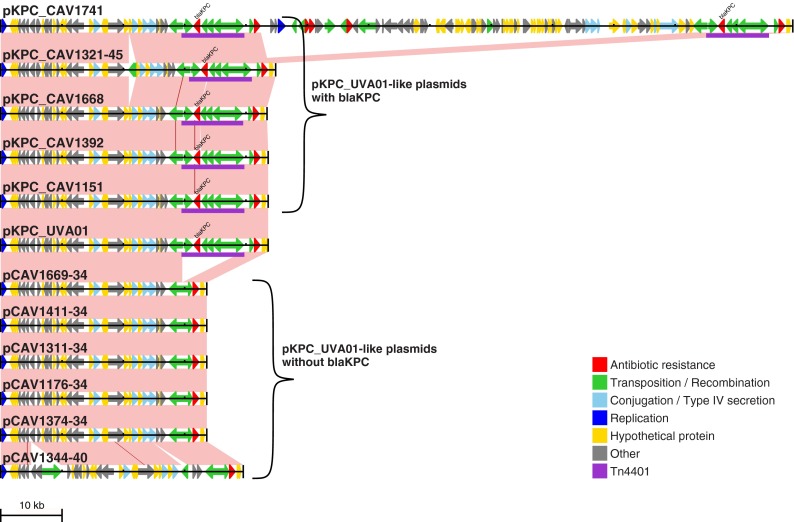
From the analysis of Illumina data described above, 11 of these 17 isolates contained pKPC_UVA01. As expected, the PacBio assemblies revealed a pKPC_UVA01-like plasmid in each of these isolates. However, only five of these pKPC_UVA01-like plasmids actually contained blaKPC (Fig. 2). The other six pKPC_UVA01-like plasmids lacked the entire Tn4401 element, which was present on a different plasmid in these isolates. Importantly, this demonstrates that plasmid presence (as defined by Illumina sequencing) is an unreliable indicator of the mobile unit carrying blaKPC, as the “presence” of pKPC_UVA01 was misleading in 55% (6/11) of the randomly selected PacBio isolates.
FIG 2.
pKPC_UVA01-like plasmids identified through long-read PacBio sequencing. The reference pKPC_UVA01 sequence is shown together with all 11 pKPC_UVA01-like plasmids identified through long-read PacBio sequencing, including the 6 that do not contain blaKPC. Arrows indicate predicted open reading frames; Tn4401 is purple. Pink shading indicates regions of identity between adjacent sequences, and SNVs are indicated by red lines.
After accounting for multiple variants of the same plasmid backbone (e.g., the pKPC_UVA01-like plasmids described above), the 19 blaKPC plasmids identified through long-read sequencing represented 11 distinct plasmid structures (Table 1; see Fig. S1 in the supplemental material). These consisted of five pKPC_UVA01-like plasmids, two pKPC_UVA02-like plasmids, four pKPC_CAV1176-like plasmids, and eight blaKPC plasmids that were each present in only a single PacBio-sequenced isolate. Using Illumina data to assess the presence of each of these 11 distinct blaKPC plasmids across the entire set of isolates as described above revealed varied patterns of plasmid presence (Fig. 1). However, in the majority of the cases, it was not possible to determine from Illumina data whether these plasmids contained blaKPC, so precise details regarding the distribution of blaKPC-containing plasmids across the 281 isolates remain elusive.
Taken together, these results demonstrate a great deal of blaKPC plasmid diversity, as 11 distinct blaKPC plasmids were identified through long-read sequencing of 17 isolates. Given that these isolates were randomly chosen, the total number of distinct blaKPC plasmids across the entire set of 281 isolates is likely to be much greater than this. Additional Tn4401 insertion sites were identified from the subset of isolates where flanking sequences could be adequately assembled using short-read data, further supporting this hypothesis (see Table S2 in the supplemental material). Therefore, HGT of the index blaKPC plasmids (pKPC_UVA01 and pKPC_UVA02) only partially explains blaKPC spread, and the large number of distinct blaKPC plasmids indicates high levels of Tn4401 mobility.
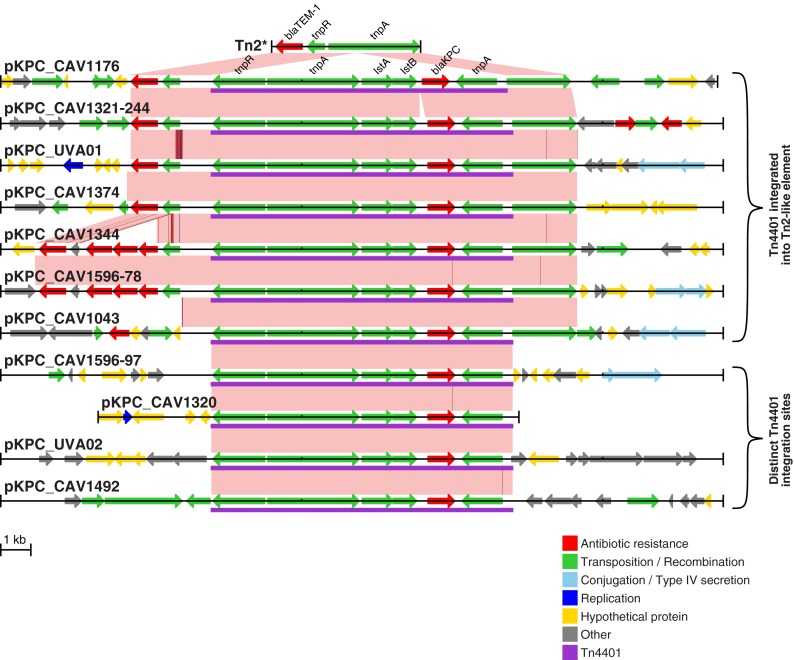
Tn4401 is present within a Tn2-like element in many different plasmids.
In 7 of the 11 distinct, fully characterized blaKPC plasmids, Tn4401 was surrounded by a sequence element related to the blaTEM-1-containing transposon Tn2 (Fig. 3). In all of the cases, the Tn4401 insertion site within the tnpA gene of Tn2 was the same, with approximately 1 kb of flanking sequence on either side of Tn4401 showing 100% identity, but the remainder of these Tn2-like elements showed substantial variation. For example, while the sequence surrounding Tn4401 in pKPC_CAV1176 was identical to the reference Tn2* sequence, the Tn2-like element in pKPC_CAV1043 was truncated. Additionally, pKPC_CAV1344 and pKPC_CAV1596-78 contained a Tn2 derivative, Tn1331, that contains the additional resistance genes blaOXA-9, aadA1, and aac(6′)-Ib and has been seen as a prior Tn4401 insertion site (46).
FIG 3.
Tn4401 is commonly integrated into a Tn2-like element. Tn4401 and the surrounding region (i.e., partial plasmid sequence, except for pKPC_CAV1320) are shown for each distinct blaKPC plasmid. Variants of the same plasmid backbone (Table 1) are not shown. Arrows indicate predicted open reading frames; Tn4401 is purple. Pink shading indicates regions of identity between adjacent sequences, SNVs are indicated by red lines, and short indels (1 or 2 bp) are indicated by blue lines. At the very top is the Tn2* reference sequence from AY123253 (49).
Tn4401 variation.
There were five different structural variants of Tn4401 (Table 2). The majority of the isolates, 230/281 (82%), had the Tn4401b isoform, with the remaining isolates containing Tn4401a (n = 8), a novel Tn4401 isoform with a 188-bp deletion upstream of blaKPC (n = 39) or one of two truncated Tn4401 structures (n = 4). At the nucleotide level, there were seven sites that were variable within Tn4401b. Three of these were located within blaKPC, giving rise to three different blaKPC alleles, blaKPC-2 (n = 179), blaKPC-3 (n = 44), and blaKPC-4 (n = 5). All non-Tn4401b isolates contained blaKPC-2. Taking all structural and nucleotide variations into account, there were a total of 12 different Tn4401 variants. However, most of these were very rare, with seven found only in single patients.
TABLE 2.
Tn4401 variations
| Tn4401 varianta | Structural isoform (29) | SNV(s)b | blaKPC variant | No. of: |
||
|---|---|---|---|---|---|---|
| Patients | Isolates | Strains | ||||
| Tn4401b-1c | b | blaKPC-2 | 121 | 176 | 42 | |
| Tn4401b-2 | b | 8015C→Td | blaKPC-3 | 22 | 40 | 19 |
| Tn4401b-3 | b | 8015C→T, 9621T→C | blaKPC-3 | 1 | 3 | 2 |
| Tn4401b-4 | b | 7199T→A, 8015C→T, 9621T→C | blaKPC-3 | 1 | 1 | 1 |
| Tn4401b-5 | b | 8015Ne | blaKPC-2/blaKPC-3 | 1 | 2 | 1 |
| Tn4401b-6 | b | 7509C→G, 7917T→Gf | blaKPC-4 | 1 | 1 | 1 |
| Tn4401b-7 | b | 6800T→C, 7509C→G, 7917T→G | blaKPC-4 | 1 | 4 | 1 |
| Tn4401b-8 | b | 9663T→C | blaKPC-2 | 1 | 3 | 1 |
| Tn4401a-1 | a (del 7020-7118) | blaKPC-2 | 5 | 8 | 1 | |
| Tn4401novel-1 | Novel (del 6919-7106) | blaKPC-2 | 28 | 39 | 2 | |
| Tn4401trunc-1 | Truncated (del 1-6654) | blaKPC-2 | 2 | 3 | 1 | |
| Tn4401trunc-2 | Truncated (del 1-6727) | 6800Ng | blaKPC-2 | 1 | 1 | 1 |
Variants are named such that letters indicate previously described structural isoforms and numbers indicate nucleotide level variations (SNVs) within an isoform (apart from the truncated Tn4401 structures, where numbers are used to indicate different truncation locations).
With respect to Tn4401b-1, which was considered the reference Tn4401 sequence in this study.
Tn4401b-1 differs from the reference isoform b sequence in EU176013.1 by the following 14 SNVs: 4939C→G, 4989C→T, 5099A→T, 5131A→G, 5154T→G, 5185G→C, 5255C→A, 5361G→C, 5375C→G, 5390A→C, 5996G→A, 5998G→C, 8112C→A, 8113A→C.
This substitution converts blaKPC-2 to blaKPC-3.
Quality filters failed at this position because of a mixture of reads supporting C and T (i.e., Tn4401b-5 actually represents a mixture of Tn4401b-1 and Tn4401b-2).
These two substitutions convert blaKPC-2 to blaKPC-4.
Quality filters failed at this position because of a lack of reads mapped in the reverse direction. All of the reads mapped in the forward direction supported a reference (T) call.
blaKPC mobility has occurred within the hospital.
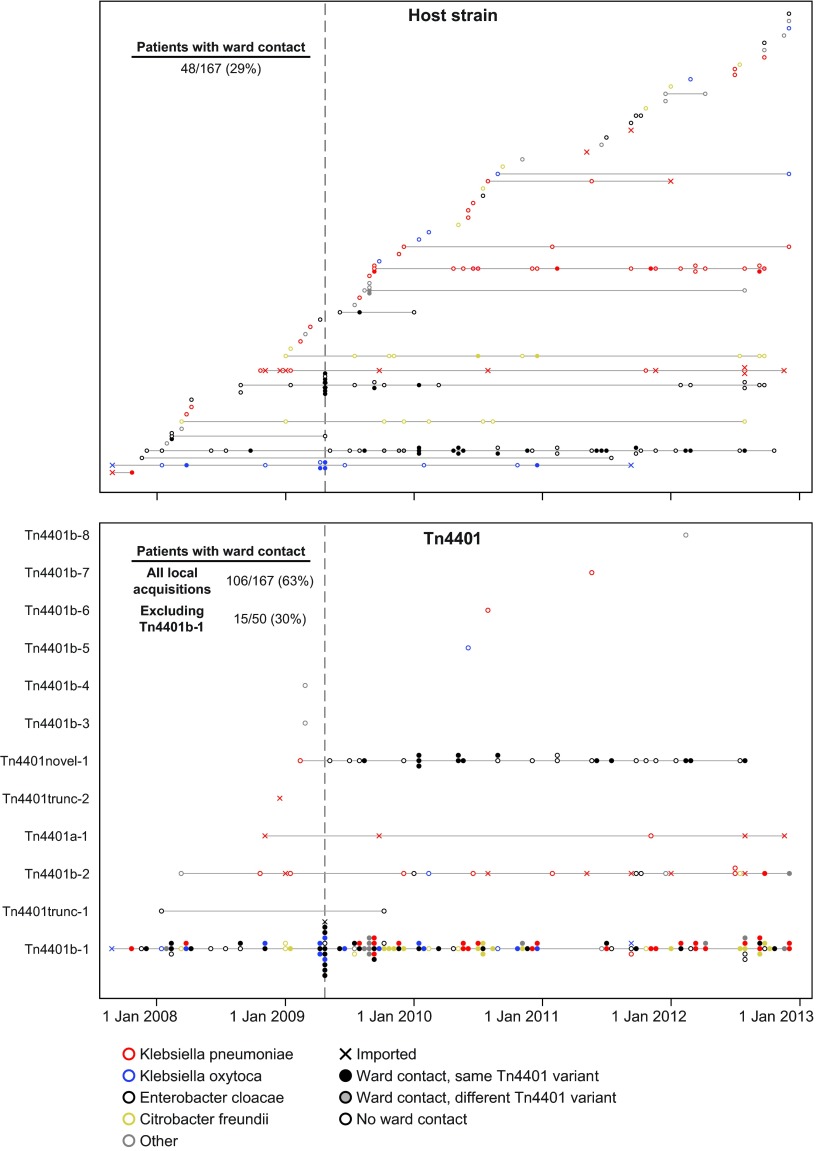
On the basis of prior health care exposure, the blaKPC acquisition source was classified as “imported” (likely acquisition prior to admission to our institution) for 15/182 (8%) patients and “local” (likely acquisition within our institution) for 167/182 (92%) patients (Fig. 1; see Materials and Methods). Imports were more likely to be infected/colonized with K. pneumoniae, particularly ST258 (see Table S3 in the supplemental material), consistent with previous reports of this strain being the dominant blaKPC carrier in the United States (9, 47). Thus, most host strain variation likely originated within the hospital via blaKPC HGT. In support of this, 15/16 (94%) patients infected/colonized with multiple strains/species had shared Tn4401 variants within the patient (see Table S4 in the supplemental material), suggesting recent blaKPC HGT. Notably, this included one patient with two different species carrying Tn4401b-3, which is not found in any other patient.
There was also some evidence of recent within-strain Tn4401 transposition. From the isolates that were randomly chosen for long-read sequencing, 4/17 (24%) had multiple Tn4401 copies (Table 1). If we assume that this randomly chosen subset is representative, this extrapolates to approximately 66/281 isolates across the whole data set. However, only 2/281 isolates had multiple Tn4401 variants (Tn4401b-5; Table 2), indicating that many isolates likely had multiple copies of the same Tn4401 variant, consistent with recent Tn4401 transposition.
Taken together, these results indicate that much of the genetic diversity observed is due to recent blaKPC mobility, likely within the hospital ecosystem over the described 5-year outbreak.
Direct patient-to-patient transmission does not explain blaKPC acquisition.
To further investigate the blaKPC acquisition source, we combined epidemiological and genetic data to trace possible transmission chains at two different genetic levels. We considered possible transmission events where the donor and recipient were on the same ward at the same time and carried the same host strain or Tn4401 variant. Considering only “local” acquisitions (see above), 48/167 (29%) patients had ward contact with another patient carrying the same blaKPC-positive strain (Fig. 4, top). A greater proportion of the patients, 106/167 (63%), had ward contact with another patient carrying the same Tn4401 variant. However, as Tn4401b-1 is very common (66% of the patients), these inferred transmissions may be spurious. With patients carrying this common variant excluded, only 15/50 (30%) had ward contact with another patient carrying the same Tn4401 variant (Fig. 4, bottom). Therefore, both genetic levels (strain or Tn4401 variant) demonstrated plausible transmissions for only a minority of the patients, indicating that direct patient-to-patient transmission is not the dominant mode of blaKPC acquisition or that there are many silently colonized patients below the limit of detection by our surveillance methods (40, 48).
FIG 4.
Ward contacts between patients with genetically related isolates. Each horizontal line represents a different strain (top) or Tn4401 variant (bottom). Filled circles indicate patients who had previous ward contact with another patient on the same horizontal line (i.e., possible patient-to-patient transmission). As Tn4401b-1 is present in two-thirds of the patients, many coincidental ward contacts may be expected to occur, resulting in a substantial overestimate of the number of transmission events. Therefore, the total number of Tn4401 acquisitions explainable by direct ward contact is indicated, as well as that with Tn4401b-1-carrying patients excluded. The vertical line indicates the onset of routine patient screening.
DISCUSSION
Here we have demonstrated high levels of genetic diversity in KPC-producing Enterobacteriaceae within a single institution over 5 years. This diversity occurs at multiple genetic levels, revealing a complex evolutionary history of the blaKPC gene involving many different host strains and plasmids.
In 7/11 distinct blaKPC plasmids identified through long-read sequencing, Tn4401 was located within a Tn2-like element. As these Tn2-like elements differed substantially from each other (Fig. 3), it is unlikely that this arose via the transposition of a composite Tn4401-Tn2-like structure. Instead, it suggests that Tn4401 has been repeatedly incorporated into pre-existing Tn2-like elements, which are known to be widespread, and genetically divergent, in Enterobacteriaceae (49, 50). However, the insertion site was identical in all of the cases, yet Tn4401 has been reported to have no insertion site specificity (28), suggesting that this was not facilitated by a standard transposition mechanism. Therefore, we suggest that this is most likely mediated by homologous recombination with other Tn2-like elements following an initial integration event, as recently suggested for another multidrug resistance gene, blaCTX-M-15 (51). This implies that Tn4401 mobility may have been enhanced via integration into a second, already widely dispersed, transposon. As the Tn4401-Tn2-like structure was present in the index case isolate (CAV1016, August 2007), we presume that the initial transposition of Tn4401 into a Tn2-like element occurred prior to entry into our hospital system. In support of this, one particular Tn2-like element, Tn1331, has been previously reported to contain Tn4401 (in exactly the same position within the tnpA gene as that described here) (21, 46, 52, 53), including one report describing a K. pneumoniae isolated in 2005, which predates blaKPC in our institution (46). We are not aware of any previous reports describing Tn4401 within a non-Tn1331 Tn2-like element.
The prevalence of Tn4401 insertions within Tn2-like elements also has important implications with regard to plasmid tracking. We previously published a method for arbitrary PCR to track the flanking regions around the Tn4401 element, as well as a PCR method to assay the presence of what we had wrongly assumed was a single plasmid, pKPC_UVA01. This PCR assay targeted the immediate Tn4401 insertion site within a Tn2-like element (54), which we have demonstrated here is present in many different plasmids, highlighting that PCR assay results, and indeed, those of any partial typing methods, need to be interpreted with a great deal of caution. We were further misled by the analysis of short-read whole-genome sequencing data that indicated the presence of pKPC_UVA01 in the majority of our isolates. Taking these findings together, it was tempting to conclude that horizontal transfer of pKPC_UVA01 was responsible for the great majority of the blaKPC carriage at our institution. However, long-read sequencing refuted this, revealing a far more complex picture.
More generally, this highlights certain limitations of plasmid reconstruction from short-read data. To illustrate by way of example, there were five isolates where long-read sequencing revealed pKPC_UVA01-like plasmids that were identical to the reference pKPC_UVA01 sequence apart from the absence of Tn4401 and the associated 5-bp target site duplication (Fig. 2). We presume that in these lineages, blaKPC may have been initially acquired via HGT of pKPC_UVA01, with subsequent homologous recombination transferring Tn4401 from pKPC_UVA01 to a different plasmid containing a Tn2-like element. In each of these five isolates, there are multiple Tn2-like elements that have 100% sequence identity over approximately 1 kb on either side of the Tn4401 insertion site. As this is longer than the fragment length used for paired-end sequencing, it is not possible to resolve the plasmid context of blaKPC by using short-read data. Importantly, any reference-based method for plasmid reconstruction (e.g., in this case, using the pKPC_UVA01 reference sequence to infer the presence of the plasmid in each isolate) is liable to produce misleading results. More generally, it is exactly the repetitive regions that cannot be resolved by using short-read data that could be expected to be involved in plasmid rearrangements, either through homologous recombination, as suggested here, or by virtue of the fact that transposable elements are often present in multiple copies. Therefore, having short-read data that are consistent with a known plasmid structure, even within the same outbreak, should not be sufficient to conclude that that structure is present, if the data are also consistent with an alternative structure. As several recent studies have utilized reference-based approaches for plasmid assembly/inference (33, 34), our results indicate that results obtained by any such methods should be interpreted with extreme caution.
Across the blaKPC-positive patients, there was large variation in both host strains and blaKPC plasmids, with Tn4401 being the largest genetic unit that was consistently present. Therefore, surveillance strategies aimed at tracking individual strains or plasmids could be misleading, and it may be more appropriate to focus on Tn4401. However, we found limited variation within the transposon, as Tn4401 sequences from 121/182 (66%) patients were identical to the index case (Table 2). This lack of variation implies that even the highest-resolution genetic methods may be insufficient for determining specific transmission routes. Even so, we have demonstrated that only a minority of blaKPC acquisition events can be explained by direct patient-to-patient transmission. Future studies should therefore contemporaneously investigate the possible involvement of unsampled reservoirs (e.g., environmental or silent colonization by additional carriers).
There are several limitations to this study. Because of the cost and effort involved in long-read sequencing, we were able to resolve only a minority of blaKPC plasmids. This means that although we have a compelling indicator of the diversity created by mobile genetic elements within a single hospital over a 5-year period, we are limited in the ability to genetically resolve pathways of blaKPC mobility between host strains and plasmid vectors, even within a single patient. We also speculate about the effect of Tn4401 insertion into Tn2-like elements, but future in vitro studies could be used to illuminate the effect of this composite structure on Tn4401 mobility. Another issue is the limit of detection of the culture-based screening methods and phenotypic tests used to identify blaKPC-positive clinical isolates. No single perirectal screening method to capture asymptomatically colonized patients is perfect (40, 48, 55), including the method used here, which has a sensitivity of ∼86% (40). In recognition of the fact that blaKPC expression and carbapenem susceptibility may be variable in different host species, and in the context of additional resistance mechanisms such as porin alterations, we lowered our surveillance thresholds to include all of the possible ESBL-producing organisms that subsequently tested positive in phenotypic carbapenemase tests. Even with this mitigation strategy, we anticipate that we have missed a proportion of the blaKPC-positive Enterobacteriaceae isolates that may be contributing to the evolution and transmission of blaKPC within our institution. Overall however, our broad screening approach across the members of the family Enterobacteriaceae has highlighted the importance of species other than E. coli and Klebsiella spp. in the transmission of blaKPC, with implications for the current CDC rectal surveillance protocol.
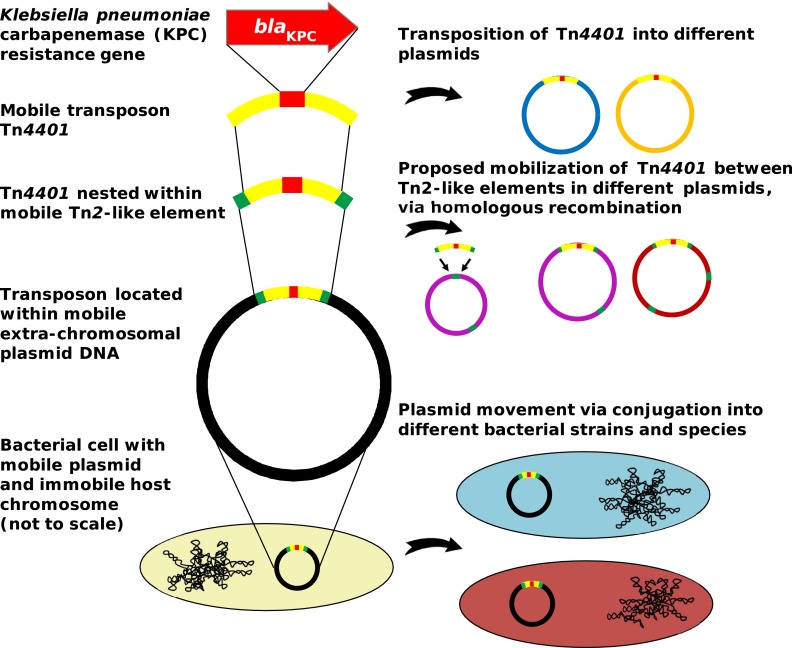
In conclusion, our detailed genetic analysis of the evolutionary events occurring in the early stages of antimicrobial resistance gene emergence in a single institution identifies several distinct processes occurring at high frequency (Fig. 5). First, the presence of shared blaKPC-containing strains in different patients reflects traditional (clonal) outbreak models. Second, blaKPC mobility between strains/species is facilitated by promiscuous blaKPC plasmids such as pKPC_UVA01. Third, blaKPC transfer between plasmids is likely enhanced by homologous recombination between Tn2-like elements, facilitating the movement of Tn4401 from one plasmid to another. Finally, blaKPC mobility is also enabled by standard Tn4401 transposition. Rather than a single process dominating, resistance dissemination is driven by a combination of these factors, with mobility occurring at multiple nested genetic levels, analogous to a Russian doll set. This has resulted in a high level of diversity in KPC-producing Enterobacteriaceae, at multiple genetic levels. As blaKPC prevalence continues to increase, so will this genetic diversity, inevitably resulting in a wider variety of more pathogenic strains carrying blaKPC.
FIG 5.
blaKPC spreads at multiple genetic levels, resulting in a high level of diversity in blaKPC-positive Enterobacteriaceae isolates.
Our results indicate that the current standard practice of screening only specific species for blaKPC carriage is likely to hamper surveillance efforts by grossly underestimating its true prevalence. Instead of the traditional view of an outbreak involving a single pathogenic strain, we propose that for KPC-producing Enterobacteriaceae, and possibly more generally, we should instead adopt the view of a “gene-based outbreak,” with surveillance strategies tracking the resistance gene itself rather than a specific host strain.
Supplementary Material
ACKNOWLEDGMENTS
We thank the UVaMC Clinical Microbiology staff for collection of study isolates and the UVaMC Infection Prevention and Control staff for assistance with patient tracking.
The Modernising Medical Microbiology (MMM) Informatics Group includes Jim Davies, Charles Crichton, Milind Acharya, and Carlos del Ojo Elias.
Funding Statement
This publication presents independent research commissioned by the Health Innovation Challenge Fund (grants HICF-T5-358 and WT098615/Z/12/Z), a parallel funding partnership of the Department of Health and Wellcome Trust, the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre based at Oxford University Hospitals NHS Trust and the University of Oxford, and the NIHR Oxford Health Protection Research Unit on Healthcare Associated Infection and Antimicrobial Resistance (HPRU-2012-10041). The views expressed in this publication are those of the authors and not necessarily those of the funders. D.W.C. and T.E.P. are NIHR senior investigators. N.S. was supported by a Wellcome Trust University of Oxford research fellowship during this work. D.J.W. is a Sir Henry Dale Fellow jointly funded by the Wellcome Trust and the Royal Society (grant 101237/Z/13/Z).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00464-16.
REFERENCES
- 1.D'Costa VM, King CE, Kalan L, Morar M, Sung WW, Schwarz C, Froese D, Zazula G, Calmels F, Debruyne R, Golding GB, Poinar HN, Wright GD. 2011. Antibiotic resistance is ancient. Nature 477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 2.Datta N, Hughes VM. 1983. Plasmids of the same Inc groups in enterobacteria before and after the medical use of antibiotics. Nature 306:616–617. doi: 10.1038/306616a0. [DOI] [PubMed] [Google Scholar]
- 3.Hughes VM, Datta N. 1983. Conjugative plasmids in bacteria of the ‘pre-antibiotic’ era. Nature 302:725–726. doi: 10.1038/302725a0. [DOI] [PubMed] [Google Scholar]
- 4.Carattoli A. 2013. Plasmids and the spread of resistance. Int J Med Microbiol 303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Partridge SR. 2011. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol Rev 35:820–855. doi: 10.1111/j.1574-6976.2011.00277.x. [DOI] [PubMed] [Google Scholar]
- 6.Stoesser N, Giess A, Batty EM, Sheppard AE, Walker AS, Wilson DJ, Didelot X, Bashir A, Sebra R, Kasarskis A, Sthapit B, Shakya M, Kelly D, Pollard AJ, Peto TE, Crook DW, Donnelly P, Thorson S, Amatya P, Joshi S. 2014. Genome sequencing of an extended series of NDM-producing Klebsiella pneumoniae isolates from neonatal infections in a Nepali hospital characterizes the extent of community- versus hospital-associated transmission in an endemic setting. Antimicrob Agents Chemother 58:7347–7357. doi: 10.1128/AAC.03900-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poirel L, Savov E, Nazli A, Trifonova A, Todorova I, Gergova I, Nordmann P. 2014. Outbreak caused by NDM-1- and RmtB-producing Escherichia coli in Bulgaria. Antimicrob Agents Chemother 58:2472–2474. doi: 10.1128/AAC.02571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garza-Ramos U, Barrios H, Reyna-Flores F, Sánchez-Pérez A, Tamayo-Legorreta E, Ibarra-Pacheco A, Salazar-Salinas J, Núñez-Ceballos R, Silva-Sanchez J. 2014. Characteristics of KPC-2-producing Klebsiella pneumoniae (ST258) clinical isolates from outbreaks in 2 Mexican medical centers. Diagn Microbiol Infect Dis 79:483–485. doi: 10.1016/j.diagmicrobio.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Brolund A, Giske CG. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 53:3365–3370. doi: 10.1128/AAC.00126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance, 2014. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf. [Google Scholar]
- 11.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf. [Google Scholar]
- 12.Patel G, Huprikar S, Factor SH, Jenkins SG, Calfee DP. 2008. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol 29:1099–1106. doi: 10.1086/592412. [DOI] [PubMed] [Google Scholar]
- 13.Falagas ME, Tansarli GS, Karageorgopoulos DE, Vardakas KZ. 2014. Deaths attributable to carbapenem-resistant enterobacteriaceae infections. Emerg Infect Dis 20:1170–1175. doi: 10.3201/eid2007.121004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzouvelekis LS, Miriagou V, Kotsakis SD, Spyridopoulou K, Athanasiou E, Karagouni E, Tzelepi E, Daikos GL. 2013. KPC-producing, multidrug-resistant Klebsiella pneumoniae sequence type 258 as a typical opportunistic pathogen. Antimicrob Agents Chemother 57:5144–5146. doi: 10.1128/AAC.01052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leavitt A, Carmeli Y, Chmelnitsky I, Goren MG, Ofek I, Navon-Venezia S. 2010. Molecular epidemiology, sequence types, and plasmid analyses of KPC-producing Klebsiella pneumoniae strains in Israel. Antimicrob Agents Chemother 54:3002–3006. doi: 10.1128/AAC.01818-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adler A, Hussein O, Ben-David D, Masarwa S, Navon-Venezia S, Schwaber MJ, Carmeli Y, Post-Acute-Care Hospital Carbapenem-Resistant Enterobacteriaceae Working Group. 2015. Persistence of Klebsiella pneumoniae ST258 as the predominant clone of carbapenemase-producing Enterobacteriaceae in post-acute-care hospitals in Israel, 2008–13. J Antimicrob Chemother 70:89–92. doi: 10.1093/jac/dku333. [DOI] [PubMed] [Google Scholar]
- 19.Deleo FR, Chen L, Porcella SF, Martens CA, Kobayashi SD, Porter AR, Chavda KD, Jacobs MR, Mathema B, Olsen RJ, Bonomo RA, Musser JM, Kreiswirth BN. 2014. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci U S A 111:4988–4993. doi: 10.1073/pnas.1321364111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. 2014. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 22:686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conlan S, Thomas PJ, Deming C, Park M, Lau AF, Dekker JP, Snitkin ES, Clark TA, Luong K, Song Y, Tsai YC, Boitano M, Dayal J, Brooks SY, Schmidt B, Young AC, Thomas JW, Bouffard GG, Blakesley RW, NISC Comparative Sequencing Program, Mullikin JC, Korlach J, Henderson DK, Frank KM, Palmore TN, Segre JA. 2014. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med 6:254ra126. doi: 10.1126/scitranslmed.3009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruiz-Garbajosa P, Curiao T, Tato M, Gijon D, Pintado V, Valverde A, Baquero F, Morosini MI, Coque TM, Canton R. 2013. Multiclonal dispersal of KPC genes following the emergence of non-ST258 KPC-producing Klebsiella pneumoniae clones in Madrid, Spain. J Antimicrob Chemother 68:2487–2492. doi: 10.1093/jac/dkt237. [DOI] [PubMed] [Google Scholar]
- 23.Ocampo AM, Chen L, Cienfuegos AV, Roncancio G, Chavda KD, Kreiswirth BN, Jiménez JN. 2015. A two-year surveillance in five Colombian tertiary care hospitals reveals high frequency of non-CG258 clones of carbapenem-resistant Klebsiella pneumoniae with distinct clinical characteristics. Antimicrob Agents Chemother 60:332–342. doi: 10.1128/AAC.01775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tavares CP, Pereira PS, Marques Ee Faria AC, de Souza Ma de Almeida PR, Alves Ce Asensi FMD, Carvalho-Assef AP. 2015. Molecular epidemiology of KPC-2-producing Enterobacteriaceae (non-Klebsiella pneumoniae) isolated from Brazil. Diagn Microbiol Infect Dis 82:326–330. doi: 10.1016/j.diagmicrobio.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Bonura C, Giuffrè M, Aleo A, Fasciana T, Di Bernardo F, Stampone T, Giammanco A, MDR-GN Working Group, Palma DM, Mammina C. 2015. An update of the evolving epidemic of blaKPC carrying Klebsiella pneumoniae in Sicily, Italy, 2014: emergence of multiple non-ST258 clones. PLoS One 10:e0132936. doi: 10.1371/journal.pone.0132936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L, Chavda KD, Melano RG, Jacobs MR, Koll B, Hong T, Rojtman AD, Levi MH, Bonomo RA, Kreiswirth BN. 2014. Comparative genomic analysis of KPC-encoding pKpQIL-like plasmids and their distribution in New Jersey and New York hospitals. Antimicrob Agents Chemother 58:2871–2877. doi: 10.1128/AAC.00120-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tijet N, Muller MP, Matukas LM, Khan A, Patel SN, Melano RG. 2016. Lateral dissemination and inter-patient transmission of blaKPC-3: role of a conjugative plasmid in spreading carbapenem resistance. J Antimicrob Chemother 71:344–347. doi: 10.1093/jac/dkv356. [DOI] [PubMed] [Google Scholar]
- 28.Cuzon G, Naas T, Nordmann P. 2011. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob Agents Chemother 55:5370–5373. doi: 10.1128/AAC.05202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P. 2008. Genetic structures at the origin of acquisition of the beta-lactamase blaKPC gene. Antimicrob Agents Chemother 52:1257–1263. doi: 10.1128/AAC.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siu LK, Lin JC, Gomez E, Eng Chiang RT. 2012. Virulence and plasmid transferability of KPC Klebsiella pneumoniae at the Veterans Affairs Healthcare System of New Jersey. Microb Drug Resist 18:380–384. doi: 10.1089/mdr.2011.0241. [DOI] [PubMed] [Google Scholar]
- 31.Cai JC, Zhou HW, Zhang R, Chen GX. 2008. Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli isolates possessing the plasmid-mediated carbapenem-hydrolyzing beta-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob Agents Chemother 52:2014–2018. doi: 10.1128/AAC.01539-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brouwer MS, Tagg KA, Mevius DJ, Iredell JR, Bossers A, Smith HE, Partridge SR. 2015. IncI shufflons: assembly issues in the next-generation sequencing era. Plasmid 80:111–117. doi: 10.1016/j.plasmid.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 33.Lanza VF, de Toro M, Garcillán-Barcia MP, Mora A, Blanco J, Coque TM, de la Cruz F. 2014. Plasmid flux in Escherichia coli ST131 sublineages, analyzed by plasmid constellation network (PLACNET), a new method for plasmid reconstruction from whole genome sequences. PLoS Genet 10:e1004766. doi: 10.1371/journal.pgen.1004766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pecora ND, Li N, Allard M, Li C, Albano E, Delaney M, Dubois A, Onderdonk AB, Bry L. 2015. Genomically informed surveillance for carbapenem-resistant Enterobacteriaceae in a health care system. mBio 6:e01030. doi: 10.1128/mBio.01030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Partridge SR. 2015. Resistance mechanisms in Enterobacteriaceae. Pathology 47:276–284. doi: 10.1097/PAT.0000000000000237. [DOI] [PubMed] [Google Scholar]
- 36.Mathers AJ, Cox HL, Bonatti H, Kitchel B, Brassinga AK, Wispelwey B, Sawyer RG, Pruett TL, Hazen KC, Patel JB, Sifri CD. 2009. Fatal cross infection by carbapenem-resistant Klebsiella in two liver transplant recipients. Transpl Infect Dis 11:257–265. doi: 10.1111/j.1399-3062.2009.00374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathers AJ, Stoesser N, Sheppard AE, Pankhurst L, Giess A, Yeh AJ, Didelot X, Turner SD, Sebra R, Kasarskis A, Peto T, Crook D, Sifri CD. 2015. Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae at a single institution: insights into endemicity from whole-genome sequencing. Antimicrob Agents Chemother 59:1656–1663. doi: 10.1128/AAC.04292-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Enfield KB, Huq NN, Gosseling MF, Low DJ, Hazen KC, Toney DM, Slitt G, Zapata HJ, Cox HL, Lewis JD, Kundzins JR, Mathers AJ, Sifri CD. 2014. Control of simultaneous outbreaks of carbapenemase-producing enterobacteriaceae and extensively drug-resistant Acinetobacter baumannii infection in an intensive care unit using interventions promoted in the Centers for Disease Control and Prevention 2012 carbapenemase-resistant Enterobacteriaceae Toolkit. Infect Control Hosp Epidemiol 35:810–817. doi: 10.1086/676857. [DOI] [PubMed] [Google Scholar]
- 39.Mathers AJ, Carroll J, Sifri CD, Hazen KC. 2013. Modified Hodge test versus indirect carbapenemase test: prospective evaluation of a phenotypic assay for detection of Klebsiella pneumoniae carbapenemase (KPC) in Enterobacteriaceae. J Clin Microbiol 51:1291–1293. doi: 10.1128/JCM.03240-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathers AJ, Poulter M, Dirks D, Carroll J, Sifri CD, Hazen KC. 2014. Clinical microbiology costs for methods of active surveillance for Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae. Infect Control Hosp Epidemiol 35:350–355. doi: 10.1086/675603. [DOI] [PubMed] [Google Scholar]
- 41.Centers for Disease Control and Prevention (CDC). 2009. Guidance for control of infections with carbapenem-resistant or carbapenemase-producing Enterobacteriaceae in acute care facilities. MMWR Morb Mortal Wkly Rep 58:256–260. http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5810a4.htm. [PubMed] [Google Scholar]
- 42.Lewis JD, Bishop M, Heon B, Mathers AJ, Enfield KB, Sifri CD. 2013. Admission surveillance for carbapenemase-producing Enterobacteriaceae at a long-term acute care hospital. Infect Control Hosp Epidemiol 34:832–834. doi: 10.1086/671263. [DOI] [PubMed] [Google Scholar]
- 43.Mathers AJ, Hazen KC, Carroll J, Yeh AJ, Cox HL, Bonomo RA, Sifri CD. 2013. First clinical cases of OXA-48-producing carbapenem-resistant Klebsiella pneumoniae in the United States: the “menace” arrives in the New World. J Clin Microbiol 51:680–683. doi: 10.1128/JCM.02580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 45.Reeves PR, Liu B, Zhou Z, Li D, Guo D, Ren Y, Clabots C, Lan R, Johnson JR, Wang L. 2011. Rates of mutation and host transmission for an Escherichia coli clone over 3 years. PLoS One 6:e26907. doi: 10.1371/journal.pone.0026907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen L, Chavda KD, Al Laham N, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. 2013. Complete nucleotide sequence of a blaKPC-harboring IncI2 plasmid and its dissemination in New Jersey and New York hospitals. Antimicrob Agents Chemother 57:5019–5025. doi: 10.1128/AAC.01397-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kitchel B, Sundin DR, Patel JB. 2009. Regional dissemination of KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 53:4511–4513. doi: 10.1128/AAC.00784-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis JD, Enfield KB, Mathers AJ, Giannetta ET, Sifri CD. 2015. The limits of serial surveillance cultures in predicting clearance of colonization with carbapenemase-producing Enterobacteriaceae. Infect Control Hosp Epidemiol 36:835–837. doi: 10.1017/ice.2015.57. [DOI] [PubMed] [Google Scholar]
- 49.Partridge SR, Hall RM. 2005. Evolution of transposons containing blaTEM genes. Antimicrob Agents Chemother 49:1267–1268. doi: 10.1128/AAC.49.3.1267-1268.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bailey JK, Pinyon JL, Anantham S, Hall RM. 2011. Distribution of the blaTEM gene and blaTEM-containing transposons in commensal Escherichia coli. J Antimicrob Chemother 66:745–751. doi: 10.1093/jac/dkq529. [DOI] [PubMed] [Google Scholar]
- 51.Zong Z, Ginn AN, Dobiasova H, Iredell JR, Partridge SR. 2015. Different IncI1 plasmids from Escherichia coli carry ISEcp1-blaCTX-M-15 associated with different Tn2-derived elements. Plasmid 80:118–126. doi: 10.1016/j.plasmid.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 52.Rice LB, Carias LL, Hutton RA, Rudin SD, Endimiani A, Bonomo RA. 2008. The KQ element, a complex genetic region conferring transferable resistance to carbapenems, aminoglycosides, and fluoroquinolones in Klebsiella pneumoniae. Antimicrob Agents Chemother 52:3427–3429. doi: 10.1128/AAC.00493-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martínez T, Vázquez GJ, Aquino EE, Martínez I, Robledo IE. 2014. ISEcp1-mediated transposition of blaKPC into the chromosome of a clinical isolate of Acinetobacter baumannii from Puerto Rico. J Med Microbiol 63:1644–1648. doi: 10.1099/jmm.0.080721-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mathers AJ, Cox HL, Kitchel B, Bonatti H, Brassinga AK, Carroll J, Scheld WM, Hazen KC, Sifri CD. 2011. Molecular dissection of an outbreak of carbapenem-resistant Enterobacteriaceae reveals intergenus KPC carbapenemase transmission through a promiscuous plasmid. mBio 2:e00204-11. doi: 10.1128/mBio.00204-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Viau R, Frank KM, Jacobs MR, Wilson B, Kaye K, Donskey CJ, Perez F, Endimiani A, Bonomo RA. 2016. Intestinal carriage of carbapenemase-producing organisms: current status of surveillance methods. Clin Microbiol Rev 29:1–27. doi: 10.1128/CMR.00108-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.