Abstract
Polyhydroxybutyrate (PHB) granules, also designated as carbonosomes, are supra-molecular complexes in prokaryotes consisting of a PHB polymer core and a surface layer of structural and functional proteins. The presence of suspected phospholipids in the surface layer is based on in vitro data of isolated PHB granules and is often shown in cartoons of the PHB granule structure in reviews on PHB metabolism. However, the in vivo presence of a phospholipid layer has never been demonstrated. We addressed this topic by the expression of fusion proteins of DsRed2EC and other fluorescent proteins with the phospholipid-binding domain (LactC2) of lactadherin in three model organisms. The fusion proteins specifically localized at the cell membrane of Ralstonia eutropha but did not co-localize with PHB granules. The same result was obtained for Pseudomonas putida, a species that accumulates another type of polyhydroxyalkanoate (PHA) granules related to PHB. Notably, DsRed2EC-LactC2 expressed in Magnetospirillum gryphiswaldense was detected at the position of membrane-enclosed magnetosome chains and at the cytoplasmic membrane but not at PHB granules. In conclusion, the carbonosomes of representatives of α-proteobacteria, β-proteobacteria and γ-proteobacteria have no phospholipids in vivo and we postulate that the PHB/PHA granule surface layers in natural producers generally are free of phospholipids and consist of proteins only.
Polyhydroxybutyrate (PHB) and related polyhydroxyalkanoates (PHAs) are important storage compounds of carbon and energy in Eubacteria and Archaea. PHB and PHAs are accumulated when cells are living in an environment with a surplus of a suitable carbon source and/or when growth and cell division are impaired due to a limitation by a nutrient other than the carbon source. Research of the last decades in many laboratories has led to a detailed understanding of the biochemical pathways leading to the formation of PHB or PHA granules1,2,3,4,5,6,7,8. A large number of proteins/genes has been described that are necessary for and are involved in the formation of such storage PHB/PHA inclusion bodies. Detailed descriptions of key enzymes such as PHA synthases, PHA depolymerases and phasins (structural PHA granule associated proteins) are given in numerous publications9,10,11,12,13,14,15,16,17,18. Although only three genes are essential to provide the information for the biochemical pathway from the central metabolite acetyl-CoA to the PHB polymer, much more genes with putative functions in PHB metabolism have been identified in Ralstonia eutropha H16 (alternative designation Cupriavidus necator H16), the model organism of PHB research. The products of these genes comprise the key enzymes of polymerisation (PHB synthases)19,20,21, depolymerisation (PHB depolymerases)22,23,24,25,26, polymer surface displayed proteins, so-called phasins27,28,29,30,31,32,33, and other proteins that are necessary for the regulation and the subcellular localization of the granules34,35,36. Most of these proteins are specifically localized on the surface of PHB granules as was shown by immuno-gold-labelling and transmission electron microscopy27,35,37 or by fluorescence microscopy using fusions of green fluorescent protein variants and the target proteins26,32,33,38,39,40,41. The finding of so many proteins on the PHB granule surface with different functions has led to the classification of PHB granules as multifunctional units and the designation as carbonosomes42 has been proposed for these organelle-like structures43,44,45. A similar variety of surface-displayed proteins was detected in PHA granules of the model organism of PHA accumulating species, P. putida17,46,47,48,49,50 and in other well-studied PHA accumulating species; for a selection see51,52,53,54.
However, the exact molecular composition of the PHB/PHA granule surface layer is still not known. While it is generally accepted that several proteins are part of the PHB/PHA granule surface layer in vivo, the presence or absence of phospholipids on the PHB/PHA granule surface is controversially discussed4,5,8,50,55,56,57. The basis for the assumption that phospholipids are present in the surface layer of PHB granules is the identification of phosphatidic acid and at least one other lipid-like (acetone extractable) compound in purified PHB granules of Bacillus megaterium almost 50 years ago55 and the detection of phosphatidyl-ethanolamine, phosphatidyl-glycerol, diphosphatidyl-glycerol and a fourth not-identified compound (possibly phosphatidyl-serine) in isolated native PHB granules of R. eutropha (at that time designated as Alcaligenes eutrophus)58. However, it is possible that phospholipids bind artificially to the hydrophobic polymer after disrupture of the cells during the PHB granule isolation process. As a consequence, the identification of phospholipids in isolated PHB granules is not a proof for the in vivo presence of phospholipids in the PHB surface layer. Electron microscopy could be a suitable tool to image the surface layer of PHB granules. Indeed, electron micrographs of thin sections of PHB accumulating bacteria were frequently published in the past but the resolution of those images in most cases was of insufficient quality. One example of high technical quality in electron microscopy is the publication of Boatman in 1964 who investigated the inclusions of Rhodospirillum rubrum and showed that PHB granules were covered by a surface layer of 4 nm thickness59. This value corresponded to approximately half of the size of a cytoplasmic membrane and would be indicative for a (phospholipid) monolayer. If the PHB surface layer was composed of phospholipids, a single unit membrane (half of a double layer) with the polar groups facing to the cytoplasm and the hydrophobic tails pointing to the hydrophobic PHB polymer core would make sense. Similar determinations of the thickness of the PHB granule surface layer were made for R. eutropha (A. eutrophus) and other PHA accumulating bacteria by co-workers of Frank Mayer’s lab: they determined a value of about 2.9 to 3.8 nm60 and concluded that PHB granules in R. eutropha most likely are covered by a monolayer. However, the nature of the monolayer, proteins, phospholipids or a mixture of both, could not be resolved.
Recently, high resolution images of PHB accumulating R. eutropha cells were published but the nature of the PHB granule surface layer could not be determined61. Evidence for a discontinuous surface layer was provided that would not be in agreement with a continuous phospholipid layer. In another PHB accumulating species, Caryophanon latum, a relatively thick PHB granule boundary layer of 14 nm was detected in electron micrographs of thin sections. Globular shaped molecules were present in negatively stained samples of PHB granules liberated from C. latum and suggested that the surface layer could consist of proteins only62. In summary, none of the above-mentioned contributions was able to prove or disprove the in vivo presence of phospholipids in the surface layer of PHB granules.
In this study, we addressed this question by the expression of a set of fusion proteins of fluorescent proteins and the LactC2-domain of lactadherin in R. eutropha. Lactadherin is a protein of milk fat that specifically binds to phospholipids in vivo via its C2 domain63,64. Fluorescence microscopical analyses of the cells expressing the fusion proteins revealed that only the cytoplasmic membrane contained phospholipids but not the PHA granules of three model organisms. This finding excluded the presence of phospholipids in PHB/PHA granules in vivo.
Results
Construction of fluorescent fusion proteins and expression in E. coli
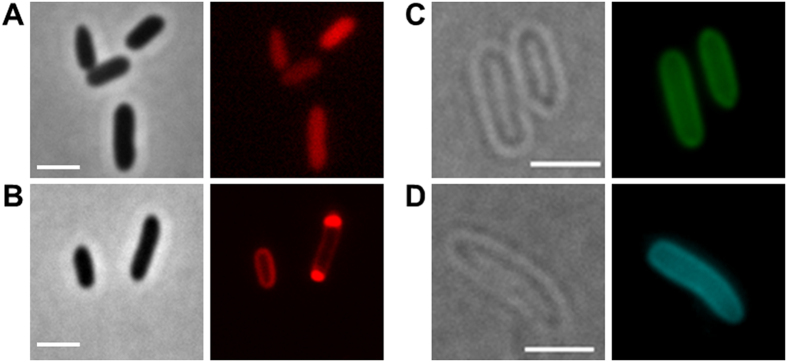
Fusions of the fluorescent protein DsRed2EC (or related fluorescent proteins as indicated in Table 1) with the phospholipid-binding domain of bovine lactadherin (LactC2) were constructed and ligated under control of the constitutively expressed promoter of the R. eutropha phaCAB operon into the broad host range plasmid pCM62. The expression of fluorescent proteins in E. coli (no producer of PHB granules) alone generally resulted in uniform fluorescence and confirmed that the proteins were soluble in the cytoplasm (Fig. 1A). In contrast to this, the cytoplasmic membrane and occasionally the cell periphery near the cell poles were fluorescent when the DsRed2EC-LactC2 fusion was expressed in E. coli and only a minor fluorescence signal was detected in the cytoplasm (Fig. 1B). Similar results (localization at the cell membrane) were obtained when fusions of super-folder GFP (sfGFP)65 or mTurquoise266 were fused to the LactC2 domain and expressed in E. coli (Fig. 1C,D). These results showed that the LactC2 fusion protein was functionally expressed and specifically bound to phospholipid molecules of the cytoplasmic membrane in E. coli.
Table 1. Strains, plasmids used in this study.
| Strain/plasmid | Relevant characteristic | Source/reference |
|---|---|---|
| Escherichia coli JM109 | cloning strain | |
| E. coli HMS174 | 82 | |
| E. coli S17-1 | conjugation strain | 83 |
| E. coli WM3064 | conjugation strain | William Metcalf |
| E. coli BL21(DE3) | Heterologous expression of pET Duet vector | 84 |
| Ralstonia eutropha H16 | Wild type | DSMZ 428 |
| R. eutropha H16 ΔphaP1 | Chromosomal deletion of phaP1 | 31 |
| R. eutropha H16 Δ(phaP1-phaP4) | Chromosomal deletions of phaP1-phaP4 | 31 |
| R. eutropha H16 Δ(phaP1-phaP5) | Additional deletion of phaP5 in ΔphaP1-phaP4 background | 32 |
| Pseudomonas putida GPO1 | Pseudomonas putida wild type strain | 85 |
| M. gryphiswaldense MSR-1 R/S | Wild type | 86 |
| M. gryphiswaldense mamC-egfp | mamC-egfp chromosomal fusion | 67 |
| M. gryphiswaldense ΔmamAB | deletion of mamAB operon | 87 |
| M. gryphiswaldense ΔphbCAB | deletion of phbCAB operon | 67 |
| pJM9238 | expression of phaCAB | 82 |
| pBBR1MCS2 | broad host range vector, Kmr | 88 |
| pBBR1MCS2-PphaC-eyfp-c1 | universal vector for construction of fusions C-terminal to eYFP under the PphaC promoter | 32 |
| pBBR1MCS2-PphaC-eyfp-c1-psd | N-terminal fusion of Psd of R. eutropha to eYFP | this study |
| pBBR1MCS-2-PphaC-DsRed2EC-c1 | universal vector for construction of fusions C-terminal to DsRed2EC under control of the PphaC promoter of R. eutropha | this study |
| pCM62 | broad host range vector, Tcr | 89 |
| pCM62-PphaC-DsRed2EC-c1 | universal vector for construction of fusions C-terminal to DsRed2EC under control of the PphaC promoter of R. eutropha | this study |
| p416 | source of LactC2 | 76 |
| pETDuet-phaCerulean-phaAB-venus-lactC2 | Co-expression of phaC-Cerulean-phaAB and Venus-LactC2 | this study |
| pCM62-PphaC-DsRed2EC-c1-lactC2 | plasmid for expression of DsRed2EC-LactC2 | this study |
| pCM62-PphaC-mTurquoise2-c1-lactC2 | plasmid for expression of mTurquoise2-LactC2 | this study |
| pCM62-PphaC-sfgfp-c1-lactC2 | plasmid for expression of sfGFP-LactC2 | this study |
| pBBR1MCS2-PphaC-eyfp-c1-phaC1(C319A) | N-terminal fusion of inactive PhaC1(C319A) to eYFP | 33 |
| pBAM-PmamDC-dsRed2EC-c1-lactC2 | plasmid for expression of DsRed2EC-LactC2 | this study |
| pBAM-Ptet-dsRed2EC-c1-lactC2 | plasmid for expression of DsRed2EC-LactC2 | this study |
Resistance against kanamycin (Kmr), tetracycline (Tcr).
Figure 1. Expression of fluorescent proteins in E. coli.
(A) expression of DsRed2EC alone (phase contrast/red channel), fluorescence visible in the cytoplasm, (B) Expression of DsRed2EC-LactC2 fusion (phase contrast/red channel), note, uniform fluorescence of the cell membrane and additional fluorescent foci at the cell poles in some cells, (C) Expression of sfGFP-LactC2 fusion (bright field/green channel), (D) Expression of mTurquoise2-LactC2 (bright filed/blue channel). Scale bars correspond to 2 μm. Since E. coli is not able to synthesize PHB no granules are visible.
Expression of DsRed2EC and DsRed2EC-LactC2 in R. eutropha
Next, we transferred the LactC2 fusion constructs to R. eutropha. As shown in Fig. 2A, expression of DsRed2EC alone resulted in more or less uniform fluorescence of the cytoplasm and confirmed that DsRed2EC is a soluble protein in R. eutropha as in E. coli. R. eutropha accumulated PHB granules during growth on NB medium. PHB granules can be seen in the phase contrast image of Fig. 2A as dark stained globular structures. Remarkably, no fluorescence was visible in the red channel at the position of the PHB granule. This indicated that DsRed2EC alone is not able to bind to PHB granules.
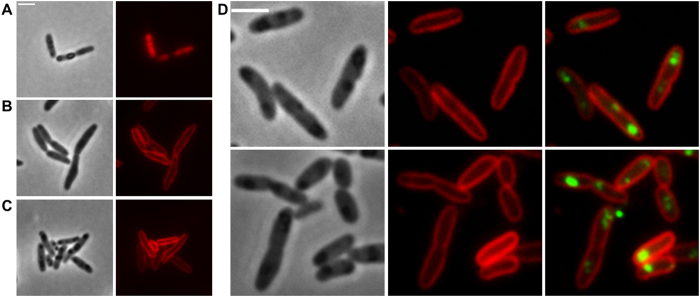
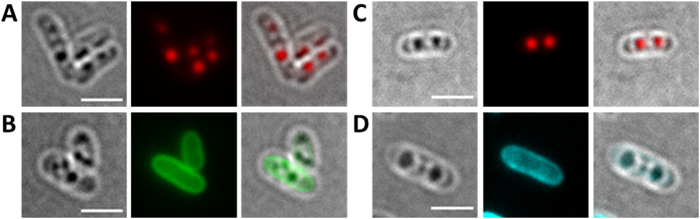
Figure 2. Expression of DsRed2EC and DsRed2EC-LactC2 in R. eutropha H16.
Expression of DsRed2EC alone in wild type (A). Expression of DsRed2EC-LactC2 in ∆phaC mutant (B) and in wild type (C). Phase contrast (left) and red channel (right) in (A–C). In (D), DsRed2EC-LactC2 was co-expressed with eYFP-PhaC (C319A) in R. eutropha wild type (from left to right: phase contrast, red channel, merge of red and green channels). Scale bars correspond to 2 μm.
When we expressed the DsRed2EC-LactC2 fusion first in a ∆phaC background (Fig. 2B), in which the cells cannot accumulate PHB granules because of the absence of the key enzyme of PHB biosynthesis (PHB synthase PhaC), a uniform fluorescence of the cytoplasmic membrane was achieved. We conclude that the DsRed2EC-LactC2 fusion specifically binds to the phospholipids of the cell membrane. Next, the DsRed2EC-LactC2 fusion was expressed in R. eutropha wild type cells (Fig. 2C). The same uniform fluorescence of the cytoplasmic membrane was observed as found for the ∆phaC R. eutropha mutant. However, PHB granules were present and could be detected in the corresponding phase contrast images. The PHB granules appeared as dark regions (in phase contrast) but DsRed2EC-LactC2-specific fluorescence was absent from the suspected positions of PHB granules. As an alternative to identification by phase contrast, PHB granules can be visualized by staining with Nile red but in this case, a differentiation between Nile red and DsRed2EC-LactC2 fluorescence is not possible. To provide a phase contrast-independent proof for the presence of PHB granules in the wild type, PHB granules were labelled by the expression of a fusion of the enhanced yellow fluorescent protein (eYFP) with an inactive PHB synthase (PhaC with C319A mutation) that specifically localizes at the surface of PHB granules. This construct was transferred to R. eutropha and expressed together with the DsRed2EC-LactC2 fusion (Fig. 2D). It has been shown previously that the inactive PHB synthase fusion (eYFP-PhaC) was specifically attached to PHB granules in R. eutropha without changing the PHB content of R. eutropha33. It is evident from the overlay images in Fig. 2D that the DsRed2EC-LactC2 fusion did not co-localize with the PhaC-eYFP-labelled PHB granules. The same results were obtained when the mTurquoise2-LactC2 or the sfGFP-LactC2 fusions were expressed in R. eutropha wild type in which PHB was stained with the lipophilic dye Nile red (Suppl. Fig. S1). mTurquoise2 and sfGFP have higher fluorescence intensities and shorter folding times, respectively, than DsRed2EC. These results are in line with the presence of phospholipids in the cytoplasmic membrane but contradict the presence of phospholipids on the surface of PHB granules.
Expression of DsRed2EC-LactC2 in R. eutropha phasin mutants
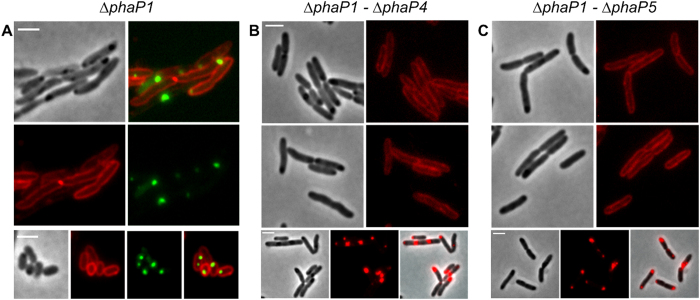
Phasin proteins, in particular PhaP1, constitute the major fraction of PHB granule associated proteins in R. eutropha26. To investigate whether the presence of phasin proteins protects the surface layer of PHB granules from interaction with the cytoplasmic membrane and from binding of phospholipids we expressed the DsRed2EC-LactC2 fusion in a ∆phaP1 background (Fig. 3A), in a ∆phaP1-∆phaP4 background (Fig. 3B) and in a ∆phaP1-∆phaP5 (Fig. 3C) background (in case of the ∆phaP1 strain with the additional presence of the inactive eYFP-PhaC (C319A) fusion (Fig. 3A)). Essentially, the same results as for the wild type were obtained. A DsRed2EC-LactC2-specific fluorescence could not be detected at the location of the PHB granules in any of the phasin mutant strains; only the cytoplasmic membrane was fluorescent in all cases. These results show that the absence of the major phasin PhaP1 or of any of the phasin proteins PhaP1 to PhaP5 did not enable DsRed2EC-LactC2 to bind to PHB granules. Our data suggest that phospholipids-even in the absence of phasin proteins-apparently do not bind to PHB granules.
Figure 3. Expression of DsRed2EC-LactC2 in R. eutropha phasin mutants.
Co-expression of DsRed2EC-LactC2 and eYFP-PhaC in ∆phaP1 mutant (A). Expression of DsRed2EC-LactC2 in ∆phaP1-∆phaP4 mutant (B) and in ∆phaP1-∆phaP5 mutant (C). Phase contrast and fluorescent images are shown. In the bottom rows of (B,C) cells were additionally stained with Nile red to indicate the position of PHB granules more clearly than in phase contrast images (phase contrast, red channel, merge). Scale bars correspond to 2 μm.
DsRed2EC-LactC2 binds to magnetosome chains but not to PHB granules in M. gryphiswaldense
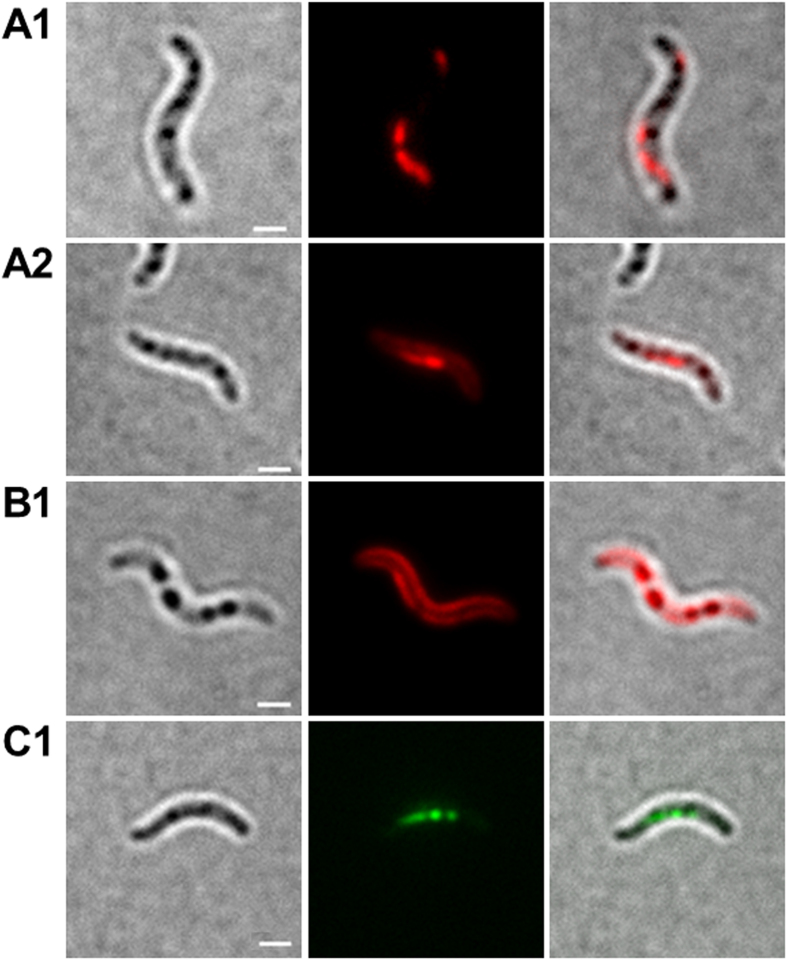
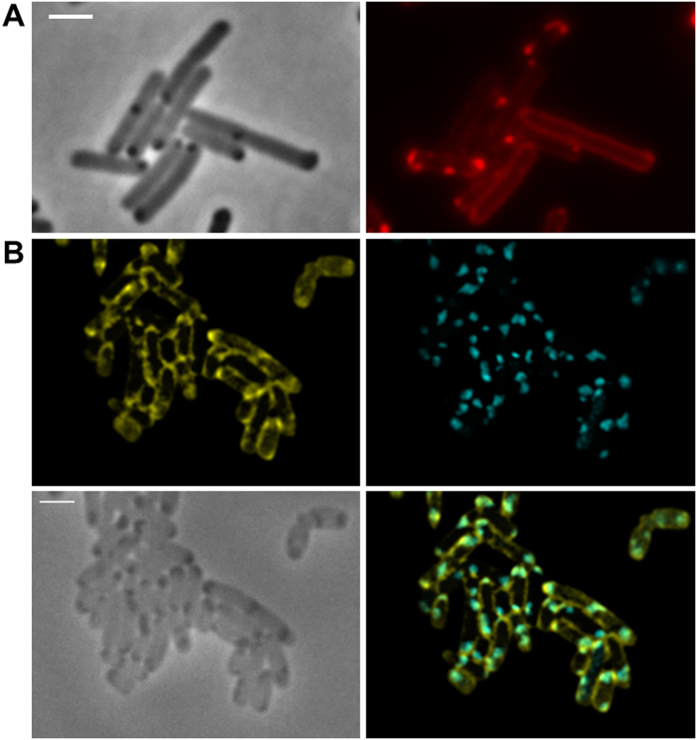
The attachment of phospholipid-binding proteins to membranes could depend on membrane curvature. Since PHB granules are much smaller than whole cells, the curvature of a potential phospholipid membrane of PHB granules would be substantially higher and potentially could prevent binding of LactC2. Moreover, the inner side of the cell membrane has a negative curvature while a potentially existing PHB granule membrane would have a positive curvature. To exclude that binding of the LactC2 domain of lactadherin is specific for a negative membrane curvature we looked for an appropriate positive control of LactC2-binding to membranes with positive curvature. Unfortunately, most inclusions of prokaryotes, as far as known, are not enclosed by phospholipid membranes. Magnetosomes of magnetotactic bacteria, however, are an exception and it is well known that magnetosomes are membrane-surrounded prokaryotic organelles with positive membrane curvature. Notably, magnetosome membranes have a similar composition as the cytoplasmic membrane43,45. Binding of dsRed-LactC2 to the magnetosome chains of magnetotactic bacteria would be therefore a suited positive control to test the ability of the dsRed-LactC2 fusion to detect membrane-enclosed organelles with a positive membrane curvature. Due to the small size of magnetosomes (≈35 nm in diameter in M. gryphiswaldense) in comparison to PHB granules (200–400 nm in diameter) individual magnetosomes cannot be resolved by light microscopy. However, magnetosomes of most magnetotactic bacteria form chains of many magnetosomes aligned on a cytoskeleton-like filament43. Therefore, the detection of a filament-like fluorescence signal of DsRed2EC-LactC2 expressed in a magnetotactic species would indicate the binding of the DsRed2EC-LactC2 fusion to the membranes of magnetosomes. We expressed the DsRed2EC-LactC2 fusion in M. gryphiswaldense and cultivated the recombinant bacteria under conditions at which they readily form magnetosome chains. The presence of magnetic particles in the recombinant strain was verified by the use of a magnet positioned near the microscope. As shown in Fig. 4A1,A2, chain-like fluorescence signals of recombinant DsRed2EC-LactC2 were visible. These signals resembled images of M. gryphiswaldense cells in which the magnetosome-specific MamC protein was fused to eGFP (Fig. 4C1). A non-magnetic M. gryphiswaldense mutant strain, in which the 16.4 kbp mamAB operon was deleted, did not form any chain-like fluorescent signals when DsRed2EC-LactC2 was expressed (Suppl. Fig. S2). However, DsRed2EC-LactC2 was bound to the cell membrane of M. gryphiswaldense cells (for image with focus to cell membrane, see Fig. 4B1). Occasionally, fluorescent foci were detected in the region of the cell membrane or within the cells (Suppl. Fig. S2B1,B2). Since M. gryphiswaldense is also able to synthesize PHB and since magnetosomes-containing M. gryphiswaldense cells usually harbour one to several PHB granules67 it could be that the observed fluorescent foci of DsRed2EC-LactC2 might represent PHB granules to which the fusion protein was attached. However, by comparing the cells shown in Suppl. Fig. S2B1,B2 in bright field and in fluorescence mode it is evident that the dark globular structures (representing PHB granules) did not co-localize with the foci of DsRed2EC-LactC2 that were found in some cells. Examination of other cells with fluorescent DsRed2EC foci never resulted in a co-localization with globular structures visible in bright field. This result corroborated the inability of the DsRed2EC-LactC2 fusion to bind to PHB granules in M. gryphiswaldense. Our finding was further verified by the expression of DsRed2EC-LactC2 in a PHB-negative background of M. gryphiswaldense (∆phaCAB) (Suppl. Fig. S3). Again, chain-like fluorescent signals corresponding to a magnetosome chain were identified in the cells (see Suppl. Fig. S3A1–A4 for cells with focus to chain-like structures). Furthermore, occasionally some cells harboured fluorescent foci (Suppl. Fig. S3B1). Since the ∆phaCAB mutant does not produce PHB, the occasionally formed fluorescent foci cannot represent PHB granules. In summary, our data with the M. gryphiswaldense strains demonstrate that the DsRed2EC-LactC2 fusion protein was specifically bound to the cell membrane and the membrane-enclosed magnetosome chains but not to the surface layer of PHB granules. These finding are in agreement with the presence of phospholipids in the cell membrane and in the magnetosome membrane but with the absence of such phospholipids in the PHB granule surface layer.
Figure 4. Expression fusion proteins in M. gryphiswaldense.
Cells expressing DsRed2EC-LactC2 focussed to filament-like fluorescence representing magnetosome-filaments (A1 and A2). Cell expressing DsRed2EC-LactC2 focussed to cell membrane fluorescence (B1). Cell expressing MamC-GFP (C1). Note, presence of several globular inclusions in all images (PHB granules) that do not co-localize with DsRed2EC-LactC2 or with MamC-eGFP. Individual magnetosomes are too small (≈35 nm) to be visible in bright field. From left to right: bright field, fluorescence channel, merge. Scale bars correspond to 2 μm.
Expression of LactC2 in PHB accumulating E. coli
Previous fluorescence microscopical analysis of recombinant E. coli cells that harbour the PHB biosynthetic genes (phaCAB) of R. eutropha have shown that the first synthesized PHB granules were always located at the cell poles and in the area of the future septum (see Fig. 7 of68). Although the resolution of light microscopy did not allow an unambiguous detection it seemed possible that the PHB granules could come into physical contact with the cytoplasmic membrane in a recombinant PHB producing organism. To test if the proximity of the PHB granules to the cytoplasmic membrane in a recombinant E. coli strain resulted in the uptake and presence of phospholipids in PHB granules we expressed the DsRed2EC-LactC2 fusion in recombinant PHB accumulating E. coli cells. As shown in Fig. 5A, partial co-localization of PHB granules (as localized by phase contrast images) and DsRed2EC-LactC2 fluorescence was indeed observed. However, not all PHB granules revealed a DsRed2EC-LactC2-specific fluorescence. To study the recombinant system in more detail, an E. coli strain was constructed in which LactC2 fused to the yellow fluorescent Venus protein and the PHB synthase (PhaC) fused to Cerulean (a brighter CFP variant) were co-expressed from a pETDuet-1 vector. As shown in Fig. 5B, PHB granules-identified by phase contrast and by Cerulean-PHB synthase fluorescence-were indeed preferentially located at the cell poles. Remarkably, we detected an apparent co-localization of some but again not of all PHB granules (PhaC-Cerulean) and Venus-LactC2 fluorescence. These results imply that some recombinant PHB granules in E. coli could be covered by lipids. This finding will be discussed below.
Figure 5. Expression of DsRed2EC-LactC2 and Venus-LactC2 in recombinant PHB accumulating E. coli.
E. coli HMS174 cells co-expressing the phaCAB genes of R. eutropha (pJM9238) and DsRed2EC-LactC2 are shown in (A) phase contrast left, red channel right. Microscopic images of E. coli BL21(DE3) co-expressing the phaC-Cerulean-phaAB operon and Venus-LactC2 from the pETDuet-vector in (B) Venus channel middle left, Cerulean channel middle right, phase contrast bottom left, merge in bottom right. Scale bars correspond to 2 μm.
Figure 6. Expression of LactC2 fusion proteins in P. putida.
Cells of P. putida expressing sfGFP-LactC2 (A,B) or mTurquoise-LactC2 (C,D) were grown in mineral salts medium with sodium octanoate to promote PHA granule formation. Cells were stained with Nile red in (A,C) to visualize PHA granules. From left to right: bright field, red (top row) or green/turquoise (bottom row) channel and overlay images. Note, formation of globular inclusions visible in bright field that are stained by Nile-red (PHA granules) but that show no sfGFP-LactC2 or mTurquoise-LactC2 fluorescence. Scale bars correspond to 2 μm.
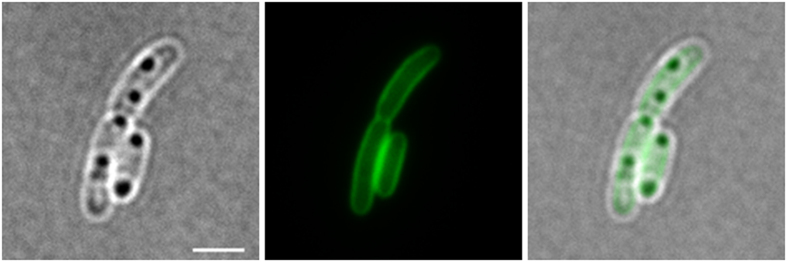
Figure 7. Expression of eYFP-Psd (phosphatidyl-serine decarboxylase) in R. eutropha H16.
From left to right: bright field, green channel, merge). eYFP-Psd co-localizes with the cell membrane but not with globular structures (PHB granules) that are visible in bright field. Scale bar corresponds to 2 μm.
Expression of LactC2-fusions in P. putida
To determine whether carbonosomes of medium-chain length PHA accumulating bacteria contained phospholipids in vivo we transferred the constructs carrying the mTurquoise2- and the sfGFP-LactC2 fusions to P. putida and determined the positions of PHA granules and of the fusion proteins (Fig. 6). When the cells were cultivated under PHA permissive conditions (mineral salts medium with sodium octanoate as precursor of PHA building blocks) only the cytoplasmic membrane was fluorescent. PHA granules, that became visible in bright field or after staining with Nile red, showed no LactC2-fusion protein fluorescence. These findings confirmed that medium chain length PHA granules in P. putida-similar to short-chain length PHA (PHB) granules in R. eutropha-do not contain phospholipids in vivo.
Phosphatidyl-serine decarboxylase localizes at the cytoplasmic membrane and is absent from PHB granules in vivo
Phosphatidyl-ethanolamine is the main phospholipid in R. eutropha and is synthesized from phosphatidyl-serine by removal of the carboxylic acid group of the serine moiety. This decarboxylase step is catalysed by phosphatidyl-serine decarboxylase that is encoded by the psd (A1038) in R. eutropha. We constructed an eYFP-Psd fusion and expressed it in R. eutropha. Fluorescence microscopical analysis revealed that eYFP-Psd is associated to the cytoplasmic membrane. We assume that the decarboxylation of phosphatidyl-serine to phosphatidyl-ethanolamine is performed shortly before or after the insertion of phospholipids into the membrane. When the strain was cultivated under PHB permissive conditions only a cell membrane-localization of the eYFP-Psd protein was observed and no fluorescence was detected at the position of the PHB granules (Fig. 7). This result is in agreement with the absence of phospholipids in PHB granules.
Phospholipids bind to PHB granules in vitro
As shown above, in PHB accumulating R. eutropha cells, fluorescent proteins fused to the phospholipid-binding domain LactC2 of lactadherin were exclusively bound to the cytoplasmic membrane in vivo. No evidence for an in vivo attachment to the surface layer of PHB granules was obtained. To find an explanation for the in vitro detection of phospholipids in isolated PHB granules as described 50 years ago55 we purified native PHB granules via two glycerol gradients from R. eutropha strains that either expressed DsRed2EC alone or the DsRed2EC-LactC2 fusion and examined the granules by fluorescence microscopy. The PHB granule band of both glycerol density gradients had a white to beige colour and no evidence for the presence of large amounts of DsRed2EC was obtained. When PHB granules isolated from the DsRed2EC expressing strain were examined by fluorescence microscopy, no red-fluorescent granules were detected. This result is consistent with the data of Fig. 1 and confirmed that DsRed2EC has no affinity to PHB granules either in vivo or in vitro. When PHB granules that had been isolated from the DsRed2EC-LactC2-expressing strain were examined, most PHB granules were also non-fluorescent and showed that the DsRed2EC-LactC2 fusion also has no binding affinity to PHB granules. However, occasionally we detected PHB granules (less than 1% of isolated PHB granules) that showed red fluorescence (Suppl. Fig. S4). This indicated that in some cases the DsRed2EC-LactC2 fusion apparently can bind to PHB granules. We assume that fragments of the cytoplasmic membrane were artificially bound to PHB granules in a minor fraction of PHB granules and that the DsRed2EC-LactC2 fusion was able to bind to these phospholipid-contaminated PHB granules in vitro. This finding can explain the detection of trace amounts of phospholipids in isolated native PHB granules half a century ago by Griebel and co-workers55.
Discussion
A variety of phospholipid-binding proteins is known from literature (for overviews see69,70). Many of them depend on the presence of Ca2+ and/or are specific for phospholipids present in eukaryotes such as phosphatidyl-inositols and their mono- or multi-phosphorylated variants70,71. Blood clotting factor V and factor VIII as well as bovine lactadherin represent two Ca2+-independent phospholipid-binding proteins63,72. These proteins specifically bind to phospholipids via C-terminal located phospholipid-binding domains (C-domains). In lactadherin, two of such binding domains are present (C1 and C2 domain). In particular, the C2-domain (LactC2) is responsible for the high binding affinity of lactadherin to phospholipids64. The crystal structure of lactadherin was solved and the membrane binding part has been identified73. In an early publication using an in vitro binding assay, a low specificity of lactadherin for a variety of phospholipids such as phosphatidyl-serine (PS), phosphatidyl-ethanolamine (PE), phosphatidyl-inositol (PI), phosphatidyl-glycerol (PG), phosphatidic acid and cardiolipine (CL) but not to phosphatidyl-choline (PC) was shown63. Later, it was suggested that the LactC2 domain apparently conferred high membrane binding ability in particular to PS-containing membranes in vivo64,74. The extent to which the LactC2 domain binds also to phospholipids other than PS in vivo is not exactly known. However, lactadherin binds to a phospholipid mixture with an excess of PE in comparison to the concentration of PS even if the PS content is only 0.03%. Another factor that stimulates binding of lactadherin to PS-containing phospholipids is a high degree of curvature74 as it is present near the cell poles of rod-shaped bacteria and in small vesicles and PHB/PHA granules if such granules would contain phospholipids.
PE, PS and CL are the main components in eubacterial membranes. In R. eutropha, PG, PE, PS, and CL have been identified as membrane lipids75 with PE as the major component. Since lactadherin is able to bind to phospholipids with only trace amounts of PS, the DsRed2EC-LactC2 and related fusion constructs of our study should be suitable tools to detect PS-containing phospholipids in vivo. Indeed, a fusion of the LactC2 domain of bovine lactadherin with a green fluorescent protein has been previously used to detect PS-containing membranes in mammalian cells76. In this study, the specific fluorescence of the cell membranes of R. eutropha, E. coli, P. putida and of M. gryphiswaldense upon expression of the DsRed2EC-LactC2 construct is in full agreement with the presence of phospholipids in cell membranes of these species and confirms the specificity of the fluorescent LactC2 construct for the detection of phospholipids in prokaryotes. The finding that DsRed2EC-LactC2 was able to bind to the magnetosome chains of M. gryphiswaldense illustrates that also membranes of prokaryotic organelles with a strong positive curvature can be detected. The nature of the DsRed2EC-LactC2 foci that were observed in a minor fraction of the cell population remains unclear and might indicate the presence of a rare, yet to be identified membrane-embedded structure.
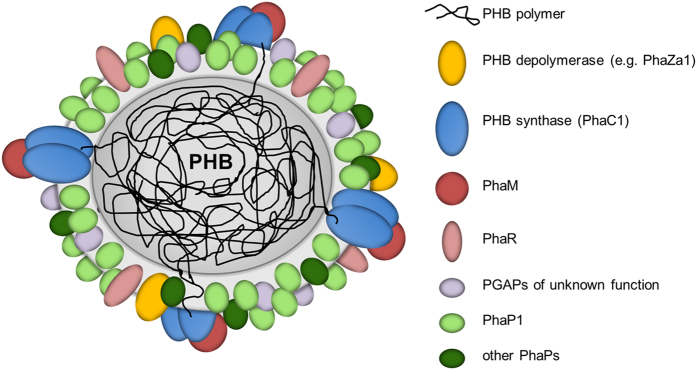
We never observed a co-localization of LactC2 fused to DsRed2EC or of one of the other fluorescent fusion constructs with carbonosomes in R. eutropha, P. putida. or M. gryphiswaldense. The simultaneous binding of DsRed2EC-LactC2 to the cell membrane and to the magnetosome chain but its absence at PHB granules in M. gryphiswaldense strongly indicates that PHB granules apparently do not contain phospholipids in the surface layer in vivo. A consequence of our findings is that the surface layer of carbonosomes is a protein layer. A current model of the structure of a PHB granule with all proteins identified in vivo for R. eutropha is given in Fig. 8.
Figure 8. Model of an in vivo PHB granule in R. eutropha H16.
The surface layer is free of phospholipids and consists of proteins only. The presently known PHB granule associated proteins (PGAPs) are symbolised by differentially coloured globules. All proteins in this model had been previously shown to be bound to PHB granules in vivo by expression of appropriate fusions with fluorescent proteins. For details and overview see references8,26. The dimension of the surface layer is enlarged relative to the polymer core for better visibility.
Only when PHB granules were synthesized in a recombinant E. coli background, a potential co-localization of LactC2 was detected for some PHB granules. The artificial E. coli system lacks the native surface of carbonosomes as it can be found in R. eutropha or P. putida8,26,49,57. Hence, the hydrophobic surface of the polymer in E. coli is at least partially exposed to the cytoplasm where it might come in contact with the cytoplasmic membrane, in particular because PHB granules in E. coli tend to localize close to the cell poles. In R. eutropha and in P. putida, carbonosomes (and possibly also in M. gryphiswaldense) are attached to a subcellular scaffold (most likely the bacterial nucleoid) via interaction with PhaM36,77 and PhaF50 and do not localize at the cell poles even in the absence of PhaP1. As a consequence of over-production and close localization to the cell periphery, PHB granules in recombinant E. coli might abstract phospholipids to the PHB granule surface. The same artificial binding of phospholipids to the PHB granule surface layer happens in vitro during the PHB granule isolation process and was confirmed in this study by the occasional detection of DsRed2EC-LactC2 fluorescence in an isolated PHB granule fraction. This finding well explains the detection of traces of phospholipids in isolated PHB granules by Merrick and co-workers almost 50 years ago55. Since no LactC2-conferred fluorescence was detected in PHB or PHA granules in R. eutropha (PHB accumulating representative of β-proteobacteria), P. putida (PHA accumulating representative of γ-proteobacteria), or M. gryphiswaldense (PHB accumulating representative of α-proteobacteria), we conclude that membrane lipids are not present in the surface layer of native PHB/PHA granules in vivo. The fact that recombinant PHB granules in a foreign host can be covered by LactC2-reacting material demonstrates that the detection system is fairly sensitive and would detect phospholipids if present. In conclusion, we have ruled out the presence of phospholipids in vivo in naturally formed PHB and PHA granules and this is in line with cryo-tomography data61. We assume that the finding of phospholipids in isolated PHB granules most likely is an in vitro artefact and reflects the potential of carbonosomes to accommodate lipids on its surface under non-physiological conditions. As a further observation of this study, the enrichment of DsRed2EC-LactC2 near the cell poles in some cells might indicate that prokaryotic membranes can have a non-random distribution of phospholipids similar to the presence of so-called lipid rafts.
Methods
Bacterial strains, plasmids and culture conditions
Bacterial strains and plasmids used in this study are shown in Table 1. E. coli strains were grown in Lysogeny broth (LB) medium supplemented with the appropriate antibiotics at 37 °C. In some cases 0.5% (wt/vol) of glucose was added to promote accumulation of PHB of recombinant E. coli strains harbouring the PHB biosynthetic genes. R. eutropha H16 strains were grown on nutrient broth (NB, 0.8%, (wt/vol)) with or without addition of 0.2% (wt/vol) of sodium gluconate at 30 °C. Pseudomonas putida GPO1 (previously P. oleovorans) was grown in mineral salts medium with addition of sodium octanoate (0.3%, wt/vol) at 30 °C. M. gryphiswaldense was grown in modified flask standard medium (FSM) at 28 °C in 15 ml polypropylene tubes with sealed screw caps and a culture volume of 10 ml at micro-oxic conditions with moderate shaking (120 rpm)78.
Construction of fluorescent fusion proteins
Constructions of in frame fusion proteins generally were prepared via PCR. The DNA sequences of the used synthetic desoxyribo-oligonucleotides and the amino acid sequences of the resulting fusion proteins are shown in Suppl. Tables S5 and S6. All PCR constructs were ligated into appropriate vectors and transformed to E. coli. The DNA sequence of each construct was verified by commercial DNA sequencing of the entire length of the PCR-amplified region and only constructs with correct DNA sequence were used.
The plasmids were transformed to E. coli by standard transformation and were subsequently transferred via conjugation from recombinant E. coli S17-1 to R. eutropha H16. Selection was achieved by plating on mineral salts medium supplemented with 0.2% fructose and 15 μg ml−1 tetracycline or 350 μg ml−1 kanamycin, respectively. For P. putida GPO1, plasmids were transformed using electroporation as described elsewhere79. Selection was achieved by plating on NB medium and 15 μg ml−1 tetracycline. For M. gryphiswaldense, pBAM plasmids were transferred from E. coli WM3064 to M. gryphiswaldense MSR-1. Selection was achieved by addition of 5 μg/ml kanamycin.
Isolation of PHB granules
PHB granules were isolated from French press (twice) disrupted cells by two subsequent glycerol gradient centrifugations as described previously80.
Microscopical methods
Formation of PHB granules was followed by fluorescence microscopy using Nile red as dye (1–10 μg/ml DMSO, Nile red solution at 5–40% [vol/vol]). Fluorescence microscopy and detection of fluorescent proteins (eYFP, DsRed2EC, Venus, Cerulean, sfGFP, or mTurquoise2) was performed on a Zeiss Axioplan, Leica DM5500 B microscope or Nikon Ti-E microscope (MEA53100) by using F41-007 Cy3 and F41-54 Cy2 filters for analysis of the PHB granules (Nile red stained) and DsRed2EC and for eYFP analysis, respectively. Venus fluorescence was detected using an excitation filter ET500/20x and an emission filter ET535/30m. Cerulean fluorescence was detected with excitation filter ET436/20x and an emission filter ET480/40m. A specific filter set (excitation, 415/20 nm; emission, 520/60) was used to visualize mTurquoise2 and sfGFP was detected with the aid of a standard filter set (excitation, 500/24 nm; emission, 542/27nm).
Pictures were taken with a digital camera (Hamamatsu Orca Flash 4.0 sCMOS camera and processed with Nikon imaging software. To avoid a potential cross-talk between fluorescence channels images were recorded with and without Nile red. PHB granules could be also visualized by phase contrast or bright field microscopy. To image fluorescent protein fusions 5 μl portions of the sample were immobilized on agarose pads (1% (wt/vol) in phosphate buffered saline) and covered with a coverslip. Images were processed with ImageJ Fiji v1.50c81.
Additional Information
How to cite this article: Bresan, S. et al. Polyhydroxyalkanoate (PHA) Granules Have no Phospholipids. Sci. Rep. 6, 26612; doi: 10.1038/srep26612 (2016).
Supplementary Material
Acknowledgments
This work was supported by grants of the Deutsche Forschungsgemeinschaft to K.F. (GRK1708) and to D.J. (Je 152-17/1). We thank Anne-Claude Gavin for providing plasmid p416.
Footnotes
Author Contributions S.B. and A.S. performed most fluorescence microscopical experiments with natural carbonosome producers; W.H. constructed most fusion proteins and conducted the experiments with recombinant E. coli. D.P. performed construction of universal DsRed2EC fusion plasmids and M. gryphiswaldense strains. K.F. and D.J. designed the study. D.J. wrote the manuscript. All authors read and approved the manuscript.
References
- Anderson A. J. & Dawes E. A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54, 450–472 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison L. L. & Huisman G. W. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol. Mol. Biol. Rev. 63, 21–53 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbe J. et al. Nontemplate-dependent polymerization processes: polyhydroxyalkanoate synthases as a paradigm. Annu. Rev. Biochem. 74, 433–480 (2005). [DOI] [PubMed] [Google Scholar]
- Pötter M. & Steinbüchel A. Biogenesis and Structure of polyhydroxyalkanoate granules. Microbiol. Monogr. 1, 110–136 (2006). [Google Scholar]
- Grage K. et al. Bacterial polyhydroxyalkanoate granules: biogenesis, structure, and potential use as nano-/micro-beads in biotechnological and biomedical applications. Biomacromolecules 10, 660–669 (2009). [DOI] [PubMed] [Google Scholar]
- Rehm B. H. A. Bacterial polymers: biosynthesis, modifications and applications. Nat. Rev. Microbiol. 8, 578–592 (2010). [DOI] [PubMed] [Google Scholar]
- Chen G.-Q. A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chem Soc Rev 38, 2434–2446 (2009). [DOI] [PubMed] [Google Scholar]
- Jendrossek D. & Pfeiffer D. New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate). Environ Microbiol 16, 2357–2373 (2014). [DOI] [PubMed] [Google Scholar]
- Rehm B. H. A. Polyester synthases: natural catalysts for plastics. Biochem J 376, 15–33 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbe J. & Tian J. Polyhydroxyalkanoate (PHA) hemeostasis: the role of PHA synthase. Nat. Prod. Rep. 20, 445–457 (2003). [DOI] [PubMed] [Google Scholar]
- Wodzinska J. et al. Polyhydroxybutyrate synthase: Evidence for covalent catalysis. J. Am. Chem. Soc. 118, 6319–6320 (1996). [Google Scholar]
- Cho M., Brigham C. J., Sinskey A. J. & Stubbe J. Purification of a polyhydroxybutyrate synthase from its native organism, Ralstonia eutropha: implications in the initiation and elongation of polymer formation in vivo. Biochemistry 2276–2288 (2012). 10.1021/bi2013596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek R., Steinbüchel A. & Schmidt B. Occurrence of polyhydroxyalkanoic acid granule-associated proteins related to the Alcaligenes eutrophus H16 GA24 protein in other bacteria. FEMS Microbiol Lett 135, 23–30 (1996). [DOI] [PubMed] [Google Scholar]
- Jendrossek D. & Handrick R. Microbial degradation of polyhydroxyalkanoates. Annu. Rev. Microbiol. 56, 403–432 (2002). [DOI] [PubMed] [Google Scholar]
- Martinez V. et al. Identification and biochemical evidence of a medium-chain-length polyhydroxyalkanoate Depolymerase in the Bdellovibrio bacteriovorus predatory hydrolytic arsenal. Applied and environmental microbiology 78, 6017–6026 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers J. & Steinbüchel A. Poly (3-hydroxybutyrate) degradation in Ralstonia eutropha H16 is mediated stereoselectively to (S)-3-hydroxybutyryl-CoA via crotonyl-CoA. J Bacteriol 195, 3213–3223 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestro B. et al. A new family of intrinsically disordered proteins: structural characterization of the major phasin PhaF from Pseudomonas putida KT2440. PLoS ONE 8, e56904 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzina M. P. et al. A phasin with many faces: Structural insights on PhaP from Azotobacter sp. FA8. PLoS ONE 9, e103012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples O. P. & Sinskey A. J. Poly-beta-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbC). J Biol Chem 264, 15298–15303 (1989). [PubMed] [Google Scholar]
- Schubert P., Steinbüchel A. & Schlegel H. G. Cloning of the Alcaligenes eutrophus genes for synthesis of poly-beta-hydroxybutyric acid (PHB) and synthesis of PHB in Escherichia coli. J Bacteriol 170, 5837–5847 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater S. C., Voige W. H. & Dennis D. E. Cloning and expression in Escherichia coli of the Alcaligenes eutrophus H16 poly-beta-hydroxybutyrate biosynthetic pathway. J Bacteriol 170, 4431–4436 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handrick R., Reinhardt S. & Jendrossek D. Mobilization of poly(3-hydroxybutyrate) in Ralstonia eutropha. J Bacteriol 182, 5916–5918 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saegusa H., Shiraki M., Kanai C. & Saito T. Cloning of an intracellular Poly[D(−)-3-Hydroxybutyrate] depolymerase gene from Ralstonia eutropha H16 and characterization of the gene product. J Bacteriol 183, 94–100 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- York G. M. et al. Ralstonia eutropha H16 encodes two and possibly three intracellular poly[D-(−)-3-hydroxybutyrate] depolymerase genes. J Bacteriol 185, 3788–3794 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sznajder A. & Jendrossek D. To be or not to be a PHB depolymerase: PhaZd1 (PhaZ6) and PhaZd2 (PhaZ7) of Ralstonia eutropha are highly active PHB depolymerases but have no detectable role in mobilization of accumulated PHB. Applied and environmental microbiology 16, 4936–4946 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sznajder A., Pfeiffer D. & Jendrossek D. Comparative proteome analysis reveals four novel polyhydroxybutyrate (PHB) granule-associated proteins in Ralstonia eutropha H16. Applied and environmental microbiology 81, 1854–1858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek R., Pries A., Steinbüchel A. & Mayer F. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J Bacteriol 177, 2425–2435 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbüchel A. et al. Considerations on the structure and biochemistry of bacterial polyhydroxyalkanoic acid inclusions. Can. J. Microbiol. 41 Suppl 1, 94–105 (1995). [DOI] [PubMed] [Google Scholar]
- York G. M., Junker B. H., Stubbe J. A. & Sinskey A. J. Accumulation of the PhaP phasin of Ralstonia eutropha is dependent on production of polyhydroxybutyrate in cells. J Bacteriol 183, 4217–4226 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- York G. M., Stubbe J. & Sinskey A. J. New insight into the role of the PhaP phasin of Ralstonia eutropha in promoting synthesis of polyhydroxybutyrate. J Bacteriol 183, 2394–2397 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pötter M. et al. The complex structure of polyhydroxybutyrate (PHB) granules: four orthologous and paralogous phasins occur in Ralstonia eutropha. Microbiology (Reading, Engl) 150, 2301–2311 (2004). [DOI] [PubMed] [Google Scholar]
- Pfeiffer D. & Jendrossek D. Interaction between poly(3-hydroxybutyrate) granule-associated proteins as revealed by two-hybrid analysis and identification of a new phasin in Ralstonia eutropha H16. Microbiology 157, 2795–2807 (2011). [DOI] [PubMed] [Google Scholar]
- Pfeiffer D. & Jendrossek D. Localization of poly(3-hydroxybutyrate) (PHB) granule-associated proteins during PHB granule formation and identification of two new phasins, PhaP6 and PhaP7, in Ralstonia eutropha H16. J Bacteriol 194, 5909–5921 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- York G. M., Stubbe J. & Sinskey A. J. The Ralstonia eutropha PhaR protein couples synthesis of the PhaP phasin to the presence of polyhydroxybutyrate in cells and promotes polyhydroxybutyrate production. J Bacteriol 184, 59–66 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pötter M., Madkour M. H., Mayer F. & Steinbüchel A. Regulation of phasin expression and polyhydroxyalkanoate (PHA) granule formation in Ralstonia eutropha H16. Microbiology (Reading, Engl) 148, 2413–2426 (2002). [DOI] [PubMed] [Google Scholar]
- Pfeiffer D., Wahl A. & Jendrossek D. Identification of a multifunctional protein, PhaM, that determines number, surface to volume ratio, subcellular localization and distribution to daughter cells of poly(3-hydroxybutyrate), PHB, granules in Ralstonia eutropha H16. Mol Microbiol 82, 936–951 (2011). [DOI] [PubMed] [Google Scholar]
- Gerngross T. U., Reilly P., Stubbe J., Sinskey A. J. & Peoples O. P. Immunocytochemical analysis of poly-beta-hydroxybutyrate (PHB) synthase in Alcaligenes eutrophus H16: localization of the synthase enzyme at the surface of PHB granules. J Bacteriol 175, 5289–5293 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard G. C., McCool J. D., Wood D. W. & Gerngross T. U. Integrated recombinant protein expression and purification platform based on Ralstonia eutropha. Applied and environmental microbiology 71, 5735–5742 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchino K., Saito T., Gebauer B. & Jendrossek D. Isolated poly(3-hydroxybutyrate) (PHB) granules are complex bacterial organelles catalyzing formation of PHB from acetyl coenzyme A (CoA) and degradation of PHB to acetyl-CoA. J Bacteriol 189, 8250–8256 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann L. et al. Binding of the major phasin, PhaP1, from Ralstonia eutropha H16 to poly(3-hydroxybutyrate) granules. J Bacteriol 190, 2911–2919 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf W. et al. Metabolic Changes in Synechocystis PCC6803 upon Nitrogen-Starvation: Excess NADPH Sustains Polyhydroxybutyrate Accumulation. Metabolites 3, 101–118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendrossek D. Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J Bacteriol 191, 3195–3202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffel A. et al. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 440, 110–114 (2006). [DOI] [PubMed] [Google Scholar]
- Cornejo E., Abreu N. & Komeili A. Compartmentalization and organelle formation in bacteria. Curr. Opin. Cell Biol. 26, 132–138 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A. Molecular mechanisms of compartmentalization and biomineralization in magnetotactic bacteria. FEMS Microbiol. Rev. 36, 232–255 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto M. A., Bühler B., Jung K., Witholt B. & Kessler B. PhaF, a polyhydroxyalkanoate-granule-associated protein of Pseudomonas oleovorans GPo1 involved in the regulatory expression system for pha genes. J Bacteriol 181, 858–868 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Eugenio L. I. et al. Biochemical evidence that phaZ gene encodes a specific intracellular medium chain length polyhydroxyalkanoate depolymerase in Pseudomonas putida KT2442: characterization of a paradigmatic enzyme. J Biol Chem 282, 4951–4962 (2007). [DOI] [PubMed] [Google Scholar]
- de Eugenio L. I. et al. The PhaD regulator controls the simultaneous expression of the pha genes involved in polyhydroxyalkanoate metabolism and turnover in Pseudomonas putida KT2442. Environ Microbiol 12, 1591–1603 (2010). [DOI] [PubMed] [Google Scholar]
- Dinjaski N. & Prieto M. A. Swapping of phasin modules to optimize the in vivo immobilization of proteins to medium-chain-length polyhydroxyalkanoate granules in Pseudomonas putida. Biomacromolecules 14, 3285–3293 (2013). [DOI] [PubMed] [Google Scholar]
- Galán B. et al. Nucleoid-associated PhaF phasin drives intracellular location and segregation of polyhydroxyalkanoate granules in Pseudomonas putida KT2442. Mol Microbiol 79, 402–418 (2011). [DOI] [PubMed] [Google Scholar]
- McCool G. J. & Cannon M. C. Polyhydroxyalkanoate inclusion body-associated proteins and coding region in Bacillus megaterium. J Bacteriol 181, 585–592 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCool G. J. & Cannon M. C. PhaC and PhaR are required for polyhydroxyalkanoic acid synthase activity in Bacillus megaterium. J Bacteriol 183, 4235–4243 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handrick R. et al. Unraveling the function of the Rhodospirillum rubrum activator of polyhydroxybutyrate (PHB) degradation: the activator is a PHB-granule-bound protein (phasin). J Bacteriol 186, 2466–2475 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S. et al. Identification of the haloarchaeal phasin (PhaP) that functions in polyhydroxyalkanoate accumulation and granule formation in Haloferax mediterranei. Applied and environmental microbiology 78, 1946–1952 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griebel R., Smith Z. & Merrick J. M. Metabolism of poly-beta-hydroxybutyrate. I. Purification, composition, and properties of native poly-beta-hydroxybutyrate granules from Bacillus megaterium. Biochemistry 7, 3676–3681 (1968). [DOI] [PubMed] [Google Scholar]
- Ruth K., de Roo G., Egli T. & Ren Q. Identification of two acyl-CoA synthetases from Pseudomonas putida GPo1: one is located at the surface of polyhydroxyalkanoates granules. Biomacromolecules 9, 1652–1659 (2008). [DOI] [PubMed] [Google Scholar]
- Dinjaski N. & Prieto M. A. Smart polyhydroxyalkanoate nanobeads by protein based functionalization. Nanomedicine 11, 885–899 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz D. M. & Sanders K. M. Amorphous, biomimetic granules of polyhydroxybutyrate: preparation, characterization, and biological implications. J. Am. Chem. Soc. 116, 2695–2702 (1994). [Google Scholar]
- Boatman E. S. Observations on the fine structure of spheroplasts of Rhodospirillum rubrum. J. Cell Biol. 20, 297–311 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer F. & Hoppert M. Determination of the thickness of the boundary layer surrounding bacterial PHA inclusion bodies, and implications for models describing the molecular architecture of this layer. J Basic Microbiol 37, 45–52 (1997). [Google Scholar]
- Beeby M., Cho M., Stubbe J. & Jensen G. J. Growth and localization of polyhydroxybutyrate granules in Ralstonia eutropha. J Bacteriol 194, 1092–1099 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendrossek D., Selchow O. & Hoppert M. Poly (3-hydroxybutyrate) granules at the early stages of formation are localized close to the cytoplasmic membrane in Caryophanon latum. Applied and environmental microbiology 73, 586–593 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen M. H., Berglund L., Rasmussen J. T. & Petersen T. E. Bovine PAS-6/7 binds alpha v beta 5 integrins and anionic phospholipids through two domains. Biochemistry 36, 5441–5446 (1997). [DOI] [PubMed] [Google Scholar]
- Andersen M. H., Graversen H., Fedosov S. N., Petersen T. E. & Rasmussen J. T. Functional analyses of two cellular binding domains of bovine lactadherin. Biochemistry 39, 6200–6206 (2000). [DOI] [PubMed] [Google Scholar]
- Pédelacq J.-D., Cabantous S., Tran T., Terwilliger T. C. & Waldo G. S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006). [DOI] [PubMed] [Google Scholar]
- Goedhart J. et al. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat Commun 3, 751 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschdorf O., Plitzko J. M., Schüler D. & Müller F. D. A tailored galK counterselection system for efficient markerless gene deletion and chromosomal tagging in Magnetospirillum gryphiswaldense. Applied and environmental microbiology 80, 4323–4330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendrossek D. Fluorescence microscopical investigation of poly(3-hydroxybutyrate) granule formation in bacteria. Biomacromolecules 6, 598–603 (2005). [DOI] [PubMed] [Google Scholar]
- Stace C. L. & Ktistakis N. T. Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim. Biophys. Acta 1761, 913–926 (2006). [DOI] [PubMed] [Google Scholar]
- Lemmon M. A. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111 (2008). [DOI] [PubMed] [Google Scholar]
- Várnai P. & Balla T. Live cell imaging of phosphoinositide dynamics with fluorescent protein domains. Biochim. Biophys. Acta 1761, 957–967 (2006). [DOI] [PubMed] [Google Scholar]
- Gilbert G. E. & Drinkwater D. Specific membrane binding of factor VIII is mediated by O-phospho-L-serine, a moiety of phosphatidylserine. Biochemistry 32, 9577–9585 (1993). [DOI] [PubMed] [Google Scholar]
- Shao C., Novakovic V. A., Head J. F., Seaton B. A. & Gilbert G. E. Crystal structure of lactadherin C2 domain at 1.7A resolution with mutational and computational analyses of its membrane-binding motif. J Biol Chem 283, 7230–7241 (2008). [DOI] [PubMed] [Google Scholar]
- Otzen D. E., Blans K., Wang H., Gilbert G. E. & Rasmussen J. T. Lactadherin binds to phosphatidylserine-containing vesicles in a two-step mechanism sensitive to vesicle size and composition. Biochim. Biophys. Acta 1818, 1019–1027 (2012). [DOI] [PubMed] [Google Scholar]
- Thiele O. W., Dreysel J. & Hermann D. The ‘free’ lipids of two different strains of hydrogen-oxidizing bacteria in relation to their growth phases. Eur J Biochem 29, 224–236 (1972). [DOI] [PubMed] [Google Scholar]
- Yeung T. et al. Membrane phosphatidylserine regulates surface charge and protein localization. Science 319, 210–213 (2008). [DOI] [PubMed] [Google Scholar]
- Wahl A., Schuth N., Pfeiffer D., Nussberger S. & Jendrossek D. PHB granules are attached to the nucleoid via PhaM in Ralstonia eutropha. BMC Microbiol. 12, 262 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyen U. & Schüler D. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Applied microbiology and biotechnology 61, 536–544 (2003). [DOI] [PubMed] [Google Scholar]
- Wang Q. et al. Quick and efficient method for genetic transformation of biopolymer-producing bacteria. J. Chem. Technol. Biotechnol. 85, 775–778 (2009). [Google Scholar]
- Handrick R. et al. A new type of thermoalkalophilic hydrolase of Paucimonas lemoignei with high specificity for amorphous polyesters of short chain-length hydroxyalkanoic acids. J Biol Chem 276, 36215–36224 (2001). [DOI] [PubMed] [Google Scholar]
- Schindelin J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell J., Valentin H. E. & Dennis D. Regulated expression of the Alcaligenes eutrophus pha biosynthesis genes in Escherichia coli. Applied and environmental microbiology 61, 1391–1398 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R., Priefer U. & Pühler A. A broad host- range mobilization system for in vivo genetic engineering: trans- poson mutagenesis in Gram-negative bacteria. Nat. Biotechnol. 1, 784–791 (1983). [Google Scholar]
- Studier F. W. & Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189, 113–130 (1986). [DOI] [PubMed] [Google Scholar]
- de Smet M. J., Eggink G., Witholt B., Kingma J. & Wynberg H. Characterization of intracellular inclusions formed by Pseudomonas oleovorans during growth on octane. J Bacteriol 154, 870–878 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss D., Kube M. & Schüler D. Inactivation of the flagellin gene flaA in Magnetospirillum gryphiswaldense results in nonmagnetotactic mutants lacking flagellar filaments. Applied and environmental microbiology 70, 3624–3631 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohße A. et al. Functional analysis of the magnetosome island in Magnetospirillum gryphiswaldense: the mamAB operon is sufficient for magnetite biomineralization. PLoS ONE 6, e25561 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach M. E. et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176 (1995). [DOI] [PubMed] [Google Scholar]
- Marx C. J. & Lidstrom M. E. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147, 2065–2075 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.