Abstract
Cell fate and differentiation in the Arabidopsis root epidermis are genetically defined but remain plastic to environmental signals such as limited availability of inorganic phosphate (Pi). Root hairs of Pi-deficient plants are more frequent and longer than those of plants grown under Pi-replete conditions. To dissect genes involved in Pi deficiency-induced root hair morphogenesis, we constructed a co-expression network of Pi-responsive genes against a customized database that was assembled from experiments in which differentially expressed genes that encode proteins with validated functions in root hair development were over-represented. To further filter out less relevant genes, we combined this procedure with a search for common cis-regulatory elements in the promoters of the selected genes. In addition to well-described players and processes such as auxin signalling and modifications of primary cell walls, we discovered several novel aspects in the biology of root hairs induced by Pi deficiency, including cell cycle control, putative plastid-to-nucleus signalling, pathogen defence, reprogramming of cell wall-related carbohydrate metabolism, and chromatin remodelling. This approach allows the discovery of novel of aspects of a biological process from transcriptional profiles with high sensitivity and accuracy.
Root hairs are tip growing lateral extensions of specialized cells in the epidermis that are important for the uptake of water and mineral nutrients. In Arabidopsis, root epidermal cells are arranged in alternating, parallel files of root hairs (trichoblasts) and non-hair cells (atrichoblasts). The identity of epidermal cells is determined by their position relative to the underlying cortical cell layer; epidermal cells that are located over a cleft of two cortical cells (H position) develop into hair cells, whereas cells that lie over periclinal cell walls (N position) adopt the non-hair cell fate1,2,3. Root hair cell fate is conferred by a non-cell autonomous signal presumably generated in cortex cells by JACKDAW4 that is stronger in cells in the H position due to a larger contact area and higher relative abundance of the leucine-rich repeat receptor kinase SCRAMBLED (SCM), which is required for signal transduction5. In H cells, the cortical signal represses the expression of the MYB protein WEREWOLF (WER). Together with the WD40 repeat protein TRANSPARENT TESTA GLABRA 1 (TTG1) and the MYC transcription factors GLABRA 3 (GL3) and ENHANCER OF GLABRA 3 (EGL3), WER forms an activator complex that negatively regulates the root hair cell fate by supporting transcription of the homeodomain protein GLABRA 2 (GL2). GL2 prevents cells in the N position from entering the hair cell fate by repressing a suite of transcription factors that positively regulate root hair morphogenesis6. Cell identity is reinforced by cell-to-cell communication during which the R3 MYB transcription factor CAPRICE (CPC) is migrating to the H position where it competes with WER for binding to the activator complex. The CPC-GL3/EGL3-TTG complex does not support GL2 expression and the cell surrenders to the hair fate. GL3 and EGL3 are preferentially expressed in H cells and move to N-positioned cells to support their differentiation into non-hair cells7,8,9,10.
Although precisely patterned, the fate of epidermal cells is not irreversible and remains responsive to external and internal cues. Movement of epidermal cells from N to H positions after laser ablations or anticlinal cell divisions after which daughter cells occupy different positions relative to the underlying cortical cells caused epidermal cells to adopt the fate that is dictated by its position, indicating that positional information and not cell lineage controls cell specification11. Expression of GL2 is associated with an open chromatin structure, but perception of positional information can rapidly induce an alternative state of chromatin organization12.
Positional information is not only important for establishing the fate of root epidermal cells, it also determines their size13,14. Mutants that cannot perceive this information such as scm or wer form short, trichoblast-like epidermal cells in both the H and N position14. It appears that either the strength of the signal, its perception or the transduction of positional information to downstream targets can be modulated by environmental cues. Phosphate-deficient plants form shorter cells and more cortical cells, leading to an increase in root hair frequency per unit root length15,16. It has been suggested that Pi deficiency reduces the strength of the positional signal, leading to shorter cells and to a less stringent pattern of epidermal cell that allows the formation of hairs in ectopic positions14. In addition, Pi deficiency increases the growth rate and the duration of root hair elongation, resulting in significantly longer hairs16,17. Increased root hair length and density is part of a complex Pi starvation response (PSR) that comprises reprogramming of primary and secondary metabolic pathways, increased expression of genes involved in the acquisition, uptake, distribution and recycling of Pi as well as alterations in root architecture18,19. These disparate responses render dissection genes that are specifically involved in a particular aspect of the PSR difficult.
Gene regulatory networks involved in epidermal cell fate specification and morphogenesis have been inferred from transcriptional profiling approaches for standard growth conditions, reflecting genetically determined developmental programs20. Here, we report a co-expression-based approach to identify genes with root hair-related functions among the relatively large subset of genes that are transcriptionally regulated by Pi starvation. While most co-expression analyses are based on large, non-specific databases that comprise experiments conducted with various tissues and genotypes subjected to different experimental conditions, the current approach relies on a customized database, allowing the inference of genes that are tightly associated with trichoblast differentiation. Using this method, we identified functional modules that regulate or mediate processes critical for the phenotype typical of Pi-deficient plants by dissecting putatively orchestrated gene regulation directed by common cis-regulatory motifs on their promoters.
Results and Discussion
Identification of Pi-responsive genes related to root hair morphogenesis
To associate Pi-responsive genes with the induction of the root hair phenotype typical of Pi-deficient plants, we first normalized a set of 3,800 ATH1 microarray hybridizations collected from the NASCarray database. Microarray experiments that discriminate processes associated with root hair development were selected based on a positive gene list that comprised 56 genes with validated roles in root hair differentiation (Supplemental Table S1). We then selected microarrays in which more than 70% of the genes from this list were among the probe sets with the 25% highest or lowest signal strength. This procedure yielded 111 matches that fulfilled these criteria. Next, we selected genes that were defined as Pi-responsive at P < 0.05 from a previously conducted RNA-seq-based transcriptomic survey21. The pairwise correlation of the 1,701 Pi-responsive genes was then calculated based on this database using the in-house software package MACCU22, yielding five larger clusters with 120 (C0), 71 (C1), 50 (C2), 11 (C3) and 11 (C4) genes (Supplemental Table S2).
To further reduce the complexity of the network and to identify co-regulated modules that are particularly involved in altering the root hair phenotype in response to Pi deficiency, we searched for cis-regulatory elements (CREs) in the promoters that are significantly over-represented within these five clusters. To this end, we employed the MEME-LaB toolbox designed for ab initio motif finding in co-expressed gene clusters23. Subsequently, the motifs identified in the promoters of at least four genes were compared for similarity with known motifs of transcription factor binding sites (TFBS) using the motif database scanning algorithm Tomtom within the MEME Suite web server (http://meme-suite.org) and the regulatory sequence analysis tools (RSAT, http://rsat.ulb.ac.be/rsat/) to identify previously identified TFBS24,25. A total of 14 CREs were identified in the promoters of the genes in clusters 0 to 4 by this approach (Supplemental Table S3).
Genes involved in cell wall organization are up-regulated by Pi deficiency
The largest cluster (C0) contains genes that are mainly related to cell morphogenesis, with the GO categories ‘cell maturation’, ‘root hair development’ and ‘cell wall organization’ strongly over-represented (Figs 1 and 2). Most of the genes from this cluster encode proteins that are predicted to localize to the extracellular space or on the plasma membrane; almost all genes were up-regulated upon Pi deficiency21. A subset of 56 genes was previously defined as being preferentially expressed in root hairs26, some of which showed more than 1,000-fold enrichment (e.g. HRGP2). For mutants defective in the expression of the gene encoding the unknown protein At3g49960, the peroxidase superfamily protein At1g05240, the serine protease inhibitor family protein CCP3 and the ATPase AHA7, we previously reported a short root hair phenotype and reduced abundancy of root hairs, respectively under control conditions26. For aha7 also a reduced frequency of root hairs under iron-deficient conditions was observed27. Mutants defective in the expression of ROPGEF4 and At3g01730 were reported to form longer (ropgef4) or shorter (At3g01730) root hairs under Pi-deficient conditions when compared to wild-type plants22.
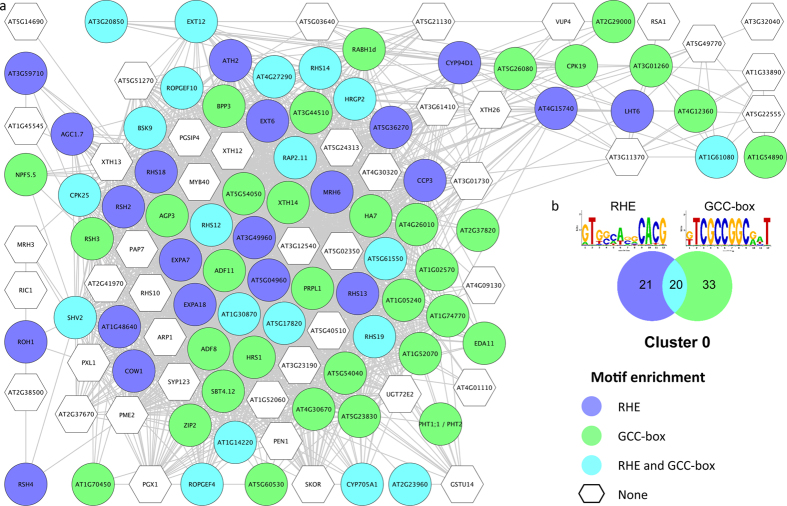
Figure 1. Cluster 0 of the co-expression network comprising genes that were differentially expressed in roots.
(a) Genes were clustered based on their co-expression relationships with a Pearson’s correlation coefficient of ≥0.90. The colour of the nodes indicates the presence of either the RHE, the GCC-box or both motifs as indicated in b. (b) Logo, colour code and number of genes that contain the motifs.
Figure 2. Visualization of the non-redundant biological gene ontology terms.
The size of the nodes corresponds to the number of the genes associated with a term. The significance is represented by the colour of the nodes. Networks were constructed by ClueGo and displayed in ‘significance view’ by Cytoscape (http://apps.cytoscape.org/apps/cluego).
Genes that define the length and shape of root hair cells of Pi-deficient plants are co-expressed with genes encoding cell wall-modifying proteins
The GARP transcription factor HRS1 HOMOLOGUE1 (HHO) is a Pi and nitrate signal integrator that represses primary root growth in the absence of Pi28. HHO is expressed in nuclei of elongating root cells and was up-regulated by Pi deficiency in our survey21,28. In contrast, POLYGALACTURONASE INVOLVED IN EXPANSION1 (PGX1) promotes cell elongation and is expressed in tissues undergoing cell expansion, including root tips29. PGX1 was down-regulated upon Pi starvation and may be involved in the regulation of the attenuation of root cell elongation upon Pi deficiency.
Expression of the proline-rich protein-like PRPL1 gene is restricted to trichoblasts26 and the protein was functionally associated with the elongation of root hairs30. Upon Pi starvation, the transcript level of PRPL1 was increased approximately 2-fold21. Thus, in conjunction with proteins involved in cell wall organization, induction of the proteins encoded by these genes might be involved in orchestrating the Pi deficiency root hair phenotype.
Root hair morphogenesis under Pi-deficient conditions is regulated by two major CREs
In cluster 0, the Root Hair Element (RHE31,32) was identified in the promoters of 42 genes. Two cysteine/histidine-rich C1 domain family proteins (At5g54050 and At5g54040), annotated as being involved in intracellular signal transduction, are up-regulated upon Pi deficiency with At5g54040 having substantially (more than 20-fold) higher expression levels21. The role of the two proteins in Pi deficiency-induced root hair formation is unclear at present. In eukaryotes, the C1 domain can bind diacylglycerol (DAG), which acts as a second messenger in animals. In plants, DAG is rapidly converted to phosphatidic acid (PA) by DAG kinase, which is more likely to act as a signalling molecule in plants33,34. Under Pi-deficient conditions, PA and DAG participate in membrane lipid remodelling, a pathway in which phospholipids are substituted by the galactolipid digalactosyldiacylglycerol (DGDG) and the sulfolipid sulfoquinovosyldiacylglycerol35,36. Lipid metabolism under Pi deficiency has been implicated in root hair development37,38, supporting the concept of tightly intertwined metabolic and developmental Pi starvation responses. Co-regulation of At5g54050 and At5g54040 with genes involved in root hair morphogenesis provides a further link of lipid metabolism and/or signalling with the root hair phenotype of Pi-deficient plants.
A second consensus motif, the GCC box (GCCGNM), was present in the promoters of 55 genes, among them the BRASSINOSTEROID-SIGNALING KINASE 9 (BSK9). BSKs have been shown to be part of the brassinosteroid (BR) signalling pathway down-stream of BRI139. BSK9 is co-expressed with several key genes in root hair development that are highly enriched in trichoblasts, but BSK9 itself is not preferentially expressed in root hairs26. Compromised BR signalling results in impaired expression of CPC and, consequently, in a decrease of hair formation in the H position40. Brassinosteroids have been further implicated in a mechanism that involves protein phosphorylation of the cell specification proteins EGL3 and TTG1 via the GSK3-like kinase BIN2, leading to inhibition of the WER-GL3/EGL3-TTG1 activator complex41. BSK9 was strongly up-regulated by Pi deficiency. Interestingly, the plasma membrane-bound Pi transporters PHT1;1 and PHT2 contain the GCC box in their promoters, suggesting a close co-regulation of genes involved in root hair elongation and Pi uptake.
Several AP/EREBP transcription factors have been identified as regulatory proteins that can interact with the GCC-box42,43,44,45. The AP2/ERF protein RAP2.11 is a putative trans-acting factor for genes harbouring the GCC-box in cluster 0. RAP2.11 is co-expressed with several genes with highly specific expression in root hairs such as EXP7 and EXP18, as well as some ROOT HAIR SPECIFIC (RHS) genes that contain the RHE consensus. Consistent with a role in nutrient signalling, RAP2.11 is preferentially expressed in root epidermal cells and in the root cap46. RAP2.11 over-expression lines form shorter primary roots and a more numerous root hairs, resembling Pi-deficient plants. Moreover, the H2O2 producing class III peroxidase PRX34 was down-regulated in rap2.11 plants46, indicative of a putative role of RAP2.11 in ROS homeostasis. PRX34 expression was also strongly decreased in mutants defective in the expression of the mediator subunit PFT1/MED2547. PFT1/MED25 is critical for root hair morphogenesis and pft1 mutants showed perturbed ROS distribution along roots, indicating a strong linkage of root hair differentiation and redox control via class III peroxidases.
Notably, 31 of the genes in cluster 0 are regulated by the bHLH transcription factor ROOT HAIR DEFECTIVE 6-LIKE 4 (RSL4), a direct target of ROOT HAIR DEFECTIVE 6 (RHD6) which positively regulates the root hair cell fate48,49. RSL4 is sufficient to initiate root hair growth and is up-regulated by Pi deficiency. It has been suggested that RSL4 integrates developmental and environmental signals to define the length of the hairs49.
A role for plastid-to-nucleus signalling in the regulation of PSR genes
Genes in cluster 1 are associated with the regulation of carbohydrate metabolic processes, in particular the biosynthesis of starch and glucan (Fig. 3). The majority of genes in this cluster (54 out of 72) encode proteins that are predicted to localize to plastids, indicating a possible connection between plastid metabolism and Pi deficiency-induced root hair formation. Pi starvation strongly compromises photosynthesis (PS) and regulatory (nucleic) nodes of the Pi starvation responses could be coupled to the down-regulation of PS-related genes. Such a regulatory circuit might be conserved among cells of different tissues independent on their photosynthetic activity. Cluster 1 contains several genes encoding proteins with RNA-binding and/or pentatricopeptide repeat (PPR) motifs (PGR3, SVR7, At2g17033, At1g70200, ERA-1, CP31B), transcriptional regulators (PTAC2, PTAC3, PTAC13, EMB1856) and translation initiation/elongation factors (SCO1, FUG1, IF3-2), which are mostly down-regulated in Pi-deficient plants. PPR-motif containing proteins such as PGR3 have been shown to stabilize RNA and to activate translation, thereby modulating plastid gene activity50. Also, several genes involved in carbohydrate metabolism are in this cluster.
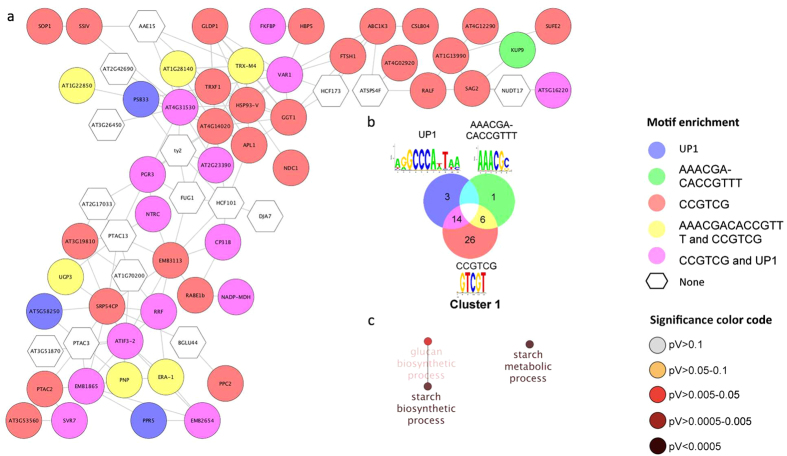
Figure 3. Cluster 1 of the co-expression network comprising genes that were differentially expressed in roots.
(a) Genes were clustered based on their co-expression relationships with a Pearson’s correlation coefficient of ≥0.90. The colour of the nodes indicates the presence of the various motifs indicated in b. (b) Logo, colour code and number of genes that contain the motifs. (c) Visualization the non-redundant biological gene ontology terms. The size of the nodes corresponds to the number of the genes associated to a term. The significance is represented by the colour of the nodes. Networks were constructed by ClueGo and displayed in ‘significance view’ by Cytoscape (http://apps.cytoscape.org/apps/cluego).
The chloroplast polynucleotide phosphorylase (PNPase) PNP/RIF10 has been associated with the Pi starvation response of roots. pnp plants form fewer lateral roots in response to Pi starvation and up-regulate a large subset of Pi starvation genes when grown on Pi-replete media, among them genes related to root hair formation specifically under Pi deficient conditions such as WRKY7551,52. Most interestingly, a Chlamydomonas reinhardtii mutant with compromised PNP expression was unable to induce Pi starvation responses, indicating conserved function of PNP in unicellular and multicellular organisms51. In our RNA-seq survey, PNP was down-regulated upon Pi deficiency21. As pointed out by Marchive et al.51, a plausible scenario could involve sugar signalling that is related to the nutritional state of plastids that, in interaction with Pi-responsive proteins, triggers the induction of systemic Pi starvation signals.
Promoter motif analysis supports a role of plastid-located proteins in cellular Pi homoeostasis
The motif AAACGACACCGTTT was found in the promoters of genes encoding PNP, the unknown protein At1g28140, the tetratricopeptide repeat (TPR)-like superfamily protein At3g53560, ERA-1 and a SNARE-associated Golgi protein family protein (At1g22850) which is tightly co-expressed with the chloroplast-located low affinity Pi transporter PHT2.1 (Fig. 3). At1g22850 is annotated as being involved in the biosynthesis of myo-inositol hexakisphosphate (InsP6), a storage form of Pi. Interestingly, inositol polyphosphate kinases have been implicated in root hair elongation and Pi sensing53. Regulation of genes in this module may affect root hair elongation either directly via signalling molecules or indirectly via alterations of the plant’s Pi status.
The promoters of PNP, At1g22850 and At1g28140 also contained the motif CCGTCG, shared with 49 genes in this cluster that are mainly related to carbohydrate metabolism, plastid organization and translation. PROTON GRADIENT REGULATION 3 (PGR3) has been associated with stabilization of photosynthetic electron transport L (petL) operon mRNA54, thereby supporting translation of the encoded proteins. Most genes in this sub-cluster were down-regulated under Pi-deficient conditions, suggesting decreased activity of genes encoding proteins that localize to plastids during Pi starvation. The consensus motif UP1, which was found to be enriched in the promoters of genes that were up-regulated after the decapitation of the main stem in Arabidopsis55, was present in 18 genes, which are enriched in the GO category ‘embryonic development’. A subset of six genes contained both CCGTCG and AAACGACACCGTTT, among them PNP, At1g28140, At1g22850 and ERA-1, indicative of tight co-regulation of these genes. The promoters of 15 genes, which include PGR3, contain both CCGTCG and UP1.
Chromatin conformation dynamics modulates root hair formation in response to Pi deficiency
In cluster 2, genes that relate to the GO categories ‘cellular response to auxin stimulus’ and ‘auxin-activated signalling pathway’ are over-represented (Fig. 4). Most proteins encoded by genes in this cluster encode proteins that are predicted to localize to the nucleus. Genes related to cell division (i.e. TTN1, TTN3, TTN7, TTN8, MAD1, SCMC2, CYCB2;3, CYCB1;1, At1g20590) were all down-regulated by Pi deficiency. Also, several genes related to chromatin silencing (INO80, HEN1, TTN3, KAC2, At1g75150, and At2g27980) had decreased abundance in Pi-deficient plants, while the gene encoding the histone H2A variant HTA10 was up-regulated. These changes are indicative of dynamic restructuring of chromatin in response to Pi starvation. INO80 has been detected in root hair-specific proteomic analysis as being preferentially expressed in trichoblasts26, indicating robust presence of INO80 in root hairs both at the transcript and protein level. INO80 encodes a conserved chromatin-remodelling complex56,57 that interacts with H2A.Z. Proper deposition of H2A.Z is required for PSR gene induction58. The arp6 (actin-related protein 6) mutant is defective in a nuclear actin-related component of the chromatin remodelled SWR159, resulting in perturbed deposition of H2A.Z and constitutive formation of long and dense root hairs reminiscent of Pi-deficient plants58. It might thus be speculated that eviction of the canonical histones H2A/H2B is coupled to the deposition of H2A.Z and HDA10 in response to Pi starvation and may be required for proper regulation of a subset of PSR genes.
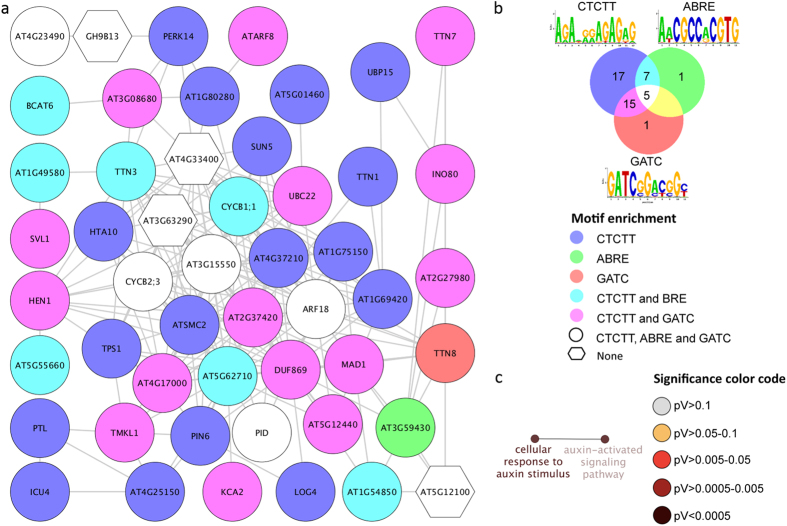
Figure 4. Cluster 2 of the co-expression network comprising genes that were differentially expressed in roots.
(a) Genes were clustered based on their co-expression relationships with a Pearson’s correlation coefficient of ≥0.90. The colour of the nodes indicates the presence of the various motifs indicated in b. (b) Logo, colour code and number of genes that contain the motifs. Network is visualized by Cytoscape 3.2.1 (http://www.cytoscape.org). (c) Visualization the non-redundant biological gene ontology terms. The size of the nodes corresponds to the number of the genes associated to a term. The significance is represented by the colour of the nodes. Networks were constructed by ClueGo and displayed in ‘significance view’ by Cytoscape (http://apps.cytoscape.org/apps/cluego).
An auxin-induced regulatory module links chromatin dynamics to root hair development
The kinase PINOID (PID), a positive key regulator of polar auxin transport and a negative regulator of root hair growth60, was down-regulated upon Pi deficiency21. Proper PID expression was shown to be dependent on the expression of a neighbouring transcript, the long intergenic non-coding RNA (lincRNA) APOLO, which is transcribed by both Pol II and Pol V61. This dual expression induces a chromatin loop that encompasses the promoter of PID and is controlled by DNA methylation. The dynamics of loop formation results in an oscillatory gene expression pattern, which resembles similar oscillatory patterns that determines the formation of lateral roots62. Both root hair formation and lateral root development are modulated by auxin dynamics and are responsive to the Pi supply; altered PID expression result in changes in root development63. It is thus tempting to speculate that such auxin-based oscillatory gene expression patterns represent important adaptive mechanisms that alter developmental programs in response to environmental cues. Interestingly, RNAi APOLO plants with compromised PID expression form longer roots, a phenotype that is opposite to that of Pi-deficient plants61.
All but two genes in this cluster carry the CRE CCGTCG in their promoters, suggesting tight co-regulation of the genes in this module. A second CRE (TGATCR) was found in 22 genes, and a subset of 20 genes, including INO80, HEN1, the auxin response factors ARF8 and ARF18, as well as PID, carry both DNA motifs in their promoters. This module also contains the glycerophosphoryl diester phosphodiesterase-like protein GDPDL4/SVL3. SVL3 is an allele of the shaven mutant in which root hairs are ruptured during tip growth due to impaired primary cell wall organization64,65. GDPDL4 links this module to root hair development.
A third CRE (CACGTGGC) is present in the promoters of 13 genes. A small subset of genes contains all three motifs (PID, ARF18, CYCB2;3 and two genes encoding unknown proteins, At4g23490 and At3g15550). At4g23490 is co-expressed with genes involved in cell wall-related processes. For At3g15550, BLAST results suggest similarity with chromatin remodelling proteins.
Genes involved in secondary cell wall formation are down-regulated upon Pi deficiency
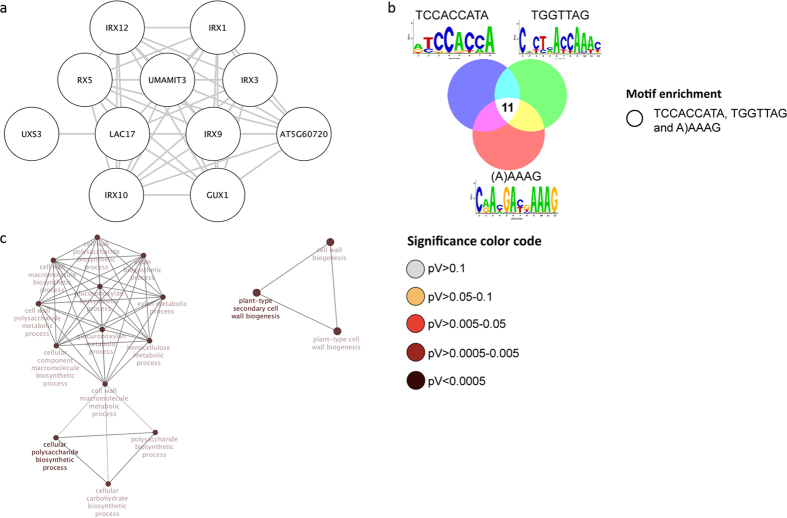
Cluster 3 contains genes that are related to the biosynthesis of polysaccharides and secondary cell walls (Fig. 5). Several IRREGULAR XYLEM (IRX) genes and the xylan glucuronyl transferase GUX1 are part of this cluster. GUX1 is involved in the addition of glucuronic acid and methylglucuronic acid residues onto xylan and for secondary wall deposition66,67. Another gene in this cluster, UXS3, produces UDP-xylose, a sugar donor for many cell wall carbohydrates such as hemicellulose and pectin. LAC17 encodes a protein that controls lipid deposition during protoxylem tracheary element development68, an unknown gene, At5g60720, has been associated with secondary wall formation in transcriptomic profiling experiments69,70. All genes were down-regulated upon Pi starvation and all genes contain three different CREs, ACCACCAAA, TGATTAG and AAAAG.
Figure 5. Cluster 3 of the co-expression network comprising genes that were differentially expressed in roots.
(a) Genes were clustered based on their co-expression relationships with a Pearson’s correlation coefficient of ≥0.90. Nodes containing all three motifs indicated in b. (b) Logo, colour code and number of genes that contain the motifs. (c) Visualization the non-redundant biological gene ontology terms. The size of the nodes corresponds to the number of the genes associated to a term. The significance is represented by the color of the nodes. Networks were constructed by ClueGo and displayed in ‘significance view’ by Cytoscape (http://apps.cytoscape.org/apps/cluego).
Glucosinolate biosynthesis genes are up-regulated upon Pi deficiency
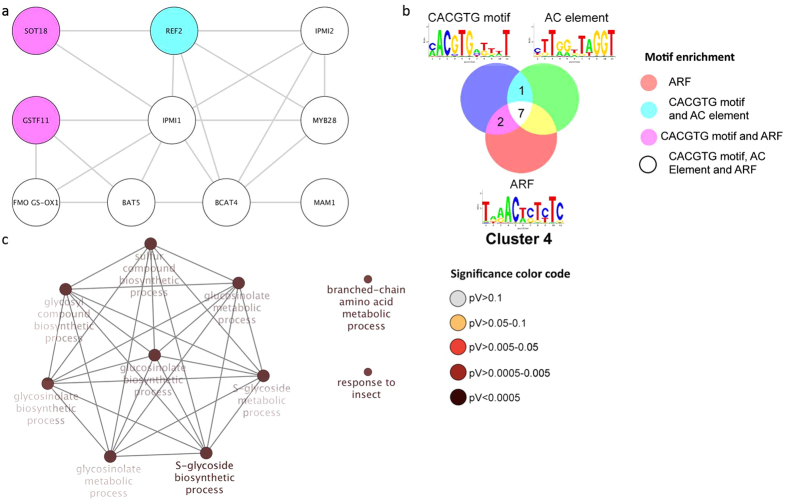
Cluster 4 contains genes related to methionine-derived glucosinolate biosynthesis: MAM1, IPMI1, IPMI2, IMD1, BAT5, GSTF11, BCAT4, REF2, SOT18, FMO GS-OX1 and MYB28 (Fig. 6). The genes in this cluster appear to be tightly co-regulated, indicated by the presence of three different motifs in the promoters of most of the genes. Ten genes carry the CACGTG motif and the AC element, all three CREs together are found in the promoters of seven genes. Unsurprisingly, the GO categories ‘S-glycoside biosynthetic process’, ‘branched-chain amino acid metabolic process’ and ‘response to insect’ are strongly over-represented in this cluster (Fig. 6).
Figure 6. Cluster 4 of the co-expression network comprising genes that were differentially expressed in roots.
(a) Genes were clustered based on their co-expression relationships with a Pearson’s correlation coefficient of ≥0.90. Nodes contain all three motifs indicated in b. (b) Logo, colour code and number of genes that contain the motifs. (c) Visualization the non-redundant biological gene ontology terms. The size of the nodes corresponds to the number of the genes associated to a term. The significance is represented by the colour of the nodes. Networks were constructed by ClueGo and displayed in ‘significance view’ by Cytoscape (http://apps.cytoscape.org/apps/cluego).
Conclusions
The present study describes a feasible approach to filter gene expression profiles with the aim of discovering novel aspects of a biological process. We reckon that with this approach relatively subtle but nevertheless critical changes in transcript abundance can be separated from stochastic noise that is inherent with transcription. It can be further stated that the identification of such aspects is strongly facilitated by the RNA-seq technology that allows the detection of robust changes in gene expression independent of a set threshold that is often used for microarray analyses because of high fluctuations among replicate experiments caused by the relatively low sensitivity of the probe sets. Our approach captured a large subset of cell wall-modulating genes (cluster 0) that were previously shown to be critical for proper root hair development. These data suggest that many genes that mediate critical functions in root hair differentiation under control (Pi-replete) conditions are also important for the induction of the elongated root hairs of Pi-deficient plants. It can further be stated that transcriptional up-regulation of these genes is most likely causally linked to the phenotype of Pi-deficient plants.
A further novel aspect is the putative involvement of plastids in the root PSR response. Plastids are critical players in sensing environmental cues and efficient communicators of environmental conditions71. Metabolic changes in plastids can affect the plant’s overall transcript profiles and plastid-to-nucleus retrograde signalling provides a means to relay environmental signals to regulate the expression of nuclear genes72. While signalling between chloroplasts and the nucleus is generally associated with photosynthetic active cells, recent evidence indicates that signals perceived in chloroplasts might also affect root processes. A recent study showed that light-induced alternative splicing triggered by metabolic changes in chloroplasts was only observed in roots when their connection to the leaves was intact, suggesting that the induction of alternative splicing in roots requires a signal that travels form shoots to roots73. Retrograde signalling can be initiated by changes in the oxidation state of plastoquinone, which is likely to be affected by Pi deficiency as well. Three possible, not mutually exclusive scenarios could explain how the low Pi status of plastids is conveyed to the nucleus. Firstly, changes in the plastid metabolic state might trigger the production of signalling molecules that exit the organelle and are transported into the nucleus where they induce the expression of PSR genes. This scenario is supported by the observation that the hypersensitive to Pi starvation7 (hps7) mutant exhibited a more severe attenuation of root growth when compared to the wild type74. Root growth inhibition in this mutant was associated with an up-regulation of photosynthesis-related genes and subsequent accumulation of ROS in roots that may trigger the expression of growth-related genes. The H2O2 concentration decreases in response to Pi deficiency, which has been linked to morphological alterations caused by Pi deficiency75. Secondly, increased transport and sequestering of Pi in the plastid may deplete cytoplasmic Pi pools and indirectly trigger the expression of nuclear PSR genes. In an alternative scenario, reduced PS and altered states of photosynthetically active chloroplasts in the leaves might induce the production of a leaf-to-root signal that communicates the leaf’s Pi status to the roots and initiate the root PSR. In this case, alterations in root plastid gene expression are conserved and mirror the responses of leaves without having direct effects on the root PSR. Such plastid-to-nucleus signalling might also be important for other nutrient deficiencies that have a strong impact on plastid-related processes such as iron or manganese deficiency.
An unexpected finding of our investigation was the possible involvement of chromatin organization in the PSR. Our data suggest that chromatin loop dynamics are involved in the formation of root hairs in response to Pi deficiency-triggered auxin stimuli in a scenario in which increased auxin responsiveness upon Pi deficiency and active DNA demethylation may fine-tune PID and ARF expression to increase root hair density of Pi-deficient plants. DNA methylation patterns were shown to be of critical importance for a proper Pi starvation response76. Loop formation appears to be required for proper auxin signaling in response to Pi deficiency, which in turn is critical for the root hair phenotype typical of Pi-deficient plants.
We also observed down-regulation of genes involved in secondary cell wall formation as part of a reprogramming of cell wall–related carbohydrate metabolisms under conditions of Pi deficiency and an up-regulation of genes involved in glucosinolate biosynthesis. Glucosinolates are chemo-protective secondary metabolites mainly found in the order Brassicales that have repellent activity against insects and pathogens. Plant responses to pathogens are thought to be organ-specific, but only a few studies compared the resistance mechanism of different plant parts77. Both leaves and roots are capable to synthesize methionine-derived aliphatic and tryptophan-derived indolic glucosinolates78, but roots contain more indolic glucosinolates compared to leaves79. Both aliphatic and indolic glucosinolates are produced in Arabidopsis trichomes to protect the plant against pathogens80. Similar to trichomes, root hairs represent the first barrier to the environment, but knowledge about defence responses in roots is limited. Secreted secondary metabolites are critical in root resistance responses81 but glucosinolates have not been implicated with root hairs. In the present study, we observed robust up-regulation of genes involved in alipathic glucosinolate production in roots upon Pi deficiency in a context that discriminates for processes related to root hair development. This is indicative of an increased defence requirement under low Pi conditions. Glucosinolate biosynthesis has been mainly associated with above-ground defence responses; the observed up-regulation of glucosinolate biosynthesis was unanticipated and adds a novel aspect to the PSR of Arabidopsis.
In summary, the current data allow for the discovery of novel aspects of root hair development in response to Pi starvation that can guide follow-up research, to validate and extent the present findings. It should be noted that while our data have been filtered for genes involved in root hair morphogenesis, the biological processes described here may also reflect responses to Pi starvation that are not directly related to root hair formation or function. Although the transcriptional response to Pi deficiency is well explored, the identification of genes that are critical for a particular aspect of the acclamatory response to Pi shortage among hundreds or thousands of genes with altered expression is a challenging task. The current approach is suitable to identify functional modules comprising genes with relatively subtle changes in transcript abundance in response to changing Pi supply. The method described here can be used to analyse RNA-seq data for a more target-oriented dissection of transcriptional changes of high fidelity.
Methods
Co-expression analysis
Discriminative co-expression analysis was performed as described in Lan et al.26 with the MACCU software (http://maccu.sourceforge.net/) based on co-expression relationships with a Pearson’s coefficient greater than or equal to 0.90. Root hair-related microarray experiments were selected based on a positive gene list comprising genes with validated function in root development (Supplemental Table 1) and downloaded from NASCArrays (http://affymetrix.arabidopsis.info/). Visualization of the networks was performed with the Cytoscape software version 3.2.0 (http://www.cytoscape.org/).
Promoter motif analysis
The MEME algorithm available with the web tool MEME-LaB, accessible at http://wsbc.warwick. ac.uk/wsbcToolsWebpage/23,82, was used for discovery of promoter motifs. The parameters used in MEME-LaB were: promoter max length: 1000; stop at neighbouring gene for core promoter: TRUE; promoter min length: 50; use repeat masked sequences: TRUE; number of motifs: 10; min motif width: 6; max motif width; 12; include background model: Markov Chain Order 3 from RSAT pre-calculated background models (http://floresta.eead.csic.es/rsat/). The parameters used for background generation were: Markov order: 3; organism: Arabidopsis; sequence type: upstream; count on: both strands; prevent overlapping matches (noov); pseudo-frequencies: 0; output format: meme; decimals: 4. Subsequently, the identified motifs were compared with known TFBS using the web tools MEME-LaB and Tomtom25. Tomtom is part of the MEME Suite online platform (http://meme-suite.org/index.html). The resulting TFBS models were then manually curated to select the best model with relation to experimental data. In addition, the DNA-pattern software available at http://floresta.eead.csic.es/rsat/ was used for the identification of the TFBS in the promoter regions of the genes in each cluster.
GO Analysis
GO enrichment analysis of genes sets was performed using the ClueGO83 version 2.0.7 plugin tool in Cytoscape84 version 3.2.1 with the GO Biological Process category. Overrepresented Biological Process categories were identified using an (right-sided) enrichment test based on the hypergeometric distribution. To correct the P-values for multiple testing Bonferroni step-down was used.
Additional Information
How to cite this article: Salazar-Henao, J. E. et al. Discriminative gene co-expression network analysis uncovers novel modules involved in the formation of phosphate deficiency-induced root hairs in Arabidopsis. Sci. Rep. 6, 26820; doi: 10.1038/srep26820 (2016).
Supplementary Material
Acknowledgments
The authors thank T.J. Buckhout (HU Berlin) for critical reading of the manuscript. Work in the Schmidt laboratory is supported by Academia Sinica and MoST.
Footnotes
Author Contributions J.E.S.-H. contributed to the design of the study, carried out the promoter motif and GO enrichment analysis, interpreted data and contributed to the manuscript. W.-D.L. collected the datasets and carried out the coexpression analysis. W.S. conceived and designed the study, participated in the analysis and interpretation of the data and wrote the manuscript. All authors read and approved the final manuscript.
References
- Dolan L. et al. Cellular organisation of the Arabidopsis thaliana root. Development 119, 71–84 (1993). [DOI] [PubMed] [Google Scholar]
- Cederholm H. M., Iyer-Pascuzzi A. S. & Benfey P. N. Patterning the primary root in Arabidopsis. Wiley Interdiscip. Rev. Dev. Biol. 1, 675–691 (2012). [DOI] [PubMed] [Google Scholar]
- Petricka J. J., Winter C. M. & Benfey P. N. Control of Arabidopsis root development. Annu. Rev. Plant Biol. 63, 563–590 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan H., Scheres B. & Blilou I. JACKDAW controls epidermal patterning in the Arabidopsis root meristem through a non-cell-autonomous mechanism. Development 137, 1523–1529 (2010). [DOI] [PubMed] [Google Scholar]
- Kwak S. H., Shen R. & Schiefelbein J. Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 307, 1111–1113 (2005). [DOI] [PubMed] [Google Scholar]
- Lin Q. et al. GLABRA2 directly suppresses basic helix-loop-helix transcription factor genes with diverse functions in root hair development. Plant Cell 27, 2894–2906 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J., Kwak S. H., Wieckowski Y., Barron C. & Bruex A. The gene regulatory network for root epidermal cell-type pattern formation in Arabidopsis. J. Exp. Bot. 60, 1515–1521 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J., Huang L. & Zheng X. Regulation of epidermal cell fate in Arabidopsis roots: the importance of multiple feedback loops. Front. Plant Sci. 5, 47 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga-Wada R., Iwata M., Nukumizu Y. & Wada T. Analysis of IIId, IIIe and IVa group basic-helix-loop-helix proteins expressed in Arabidopsis root epidermis. Plant Sci. 181, 471–478 (2011). [DOI] [PubMed] [Google Scholar]
- Grebe M. The patterning of epidermal hairs in Arabidopsis–updated. Curr. Opin. Plant Biol. 15, 31–37 (2012). [DOI] [PubMed] [Google Scholar]
- Berger F., Hung C. Y., Dolan L. & Schiefelbein J. Control of cell division in the root epidermis of Arabidopsis thaliana. Dev. Biol. 194, 235–245 (1998). [DOI] [PubMed] [Google Scholar]
- Costa S. & Shaw P. Chromatin organization and cell fate switch respond to positional information in Arabidopsis. Nature 439, 493–496 (2006). [DOI] [PubMed] [Google Scholar]
- Lofke C., Dunser K. & Kleine-Vehn J. Epidermal patterning genes impose non-cell autonomous cell size determination and have additional roles in root meristem size control. J. Integr. Plant Biol. 55, 864–875 (2013). [DOI] [PubMed] [Google Scholar]
- Savage N. et al. Positional signaling and expression of ENHANCER OF TRY AND CPC1 are tuned to increase root hair density in response to phosphate deficiency in Arabidopsis thaliana. PloS One 8, e75452 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Bielenberg D. G., Brown K. M. & Lynch J. P. Regulation of root hair density by phosphorus availability in Arabidopsis thaliana. Plant Cell Environ. 24, 459–467 (2001). [Google Scholar]
- Müller M. & Schmidt W. Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiol 134, 409–419 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates T. R. & Lynch J. P. Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ. 19, 529–538 (1996). [Google Scholar]
- Chiou T. J. & Lin S. I. Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62, 185–206 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang X. Y., Lu S. & Liu D. A major root-associated acid phosphatase in Arabidopsis, AtPAP10, is regulated by both local and systemic signals under phosphate starvation. J. Exp. Bot. 65, 6577–6588 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruex A. et al. A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 8, e1002446 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P., Li W. & Schmidt W. Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol. Cell. Proteomics 11, 1156–1166 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W. D. et al. Coexpression-based clustering of Arabidopsis root genes predicts functional modules in early phosphate deficiency signaling. Plant Physiol. 155, 1383–1402 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P. et al. MEME-LaB: motif analysis in clusters. Bioinformatics 29, 1696–1697 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M. et al. RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 36, W119–127 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Stamatoyannopoulos J. A., Bailey T. L. & Noble W. S. Quantifying similarity between motifs. Genome Biol. 8, R24 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P., Li W. & Schmidt W. Genome-wide co-expression analysis predicts protein kinases as important regulators of phosphate deficiency-induced root hair remodeling in Arabidopsis. BMC Genomics 14, 210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi S. & Schmidt W. Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol 183, 1072–1084 (2009). [DOI] [PubMed] [Google Scholar]
- Medici A. et al. AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat. Commun. 6, 6274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Somerville C. & Anderson C. T. POLYGALACTURONASE INVOLVED IN EXPANSION1 functions in cell elongation and flower development in Arabidopsis. Plant Cell 26, 1018–1035 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boron A. K. et al. Proline-rich protein-like PRPL1 controls elongation of root hairs in Arabidopsis thaliana. J. Exp. Bot. 65, 5485–5495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. W. et al. Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 18, 2958–2970 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won S. K. et al. cis-element- and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol. 150, 1459–1473 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T. Phosphatidic acid: an emerging plant lipid second messenger. Trends Plant Sci. 6, 227–233 (2001). [DOI] [PubMed] [Google Scholar]
- Laxalt A. M. & Munnik T. Phospholipid signalling in plant defence. Curr. Opin. Plant Biol. 5, 332–338 (2002). [DOI] [PubMed] [Google Scholar]
- Nakamura Y. et al. Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. P. Natl. Acad. Sci. USA 106, 20978–20983 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y. et al. Transcriptomic and lipidomic profiles of glycerolipids during Arabidopsis flower development. New Phytol 203, 310–322 (2014). [DOI] [PubMed] [Google Scholar]
- Chandrika N. N. P., Sundaravelpandian K., Yu S. M. & Schmidt W. ALFIN-LIKE 6 is involved in root hair elongation during phosphate deficiency in Arabidopsis. New Phytol 198, 709–720 (2013). [DOI] [PubMed] [Google Scholar]
- Chen C. Y. & Schmidt W. The paralogous R3 MYB proteins CAPRICE, TRIPTYCHON and ENHANCER OF TRY AND CPC1 play pleiotropic and partly non-redundant roles in the phosphate starvation response of Arabidopsis roots. J. Exp. Bot. 66, 4821–4834 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W. Q. et al. Proteomics studies of brassinosteroid signal transduction using prefractionation and two-dimensional DIGE. Mol. Cell Proteomics 7, 728–738 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy K. T., Chen A. Y. & Nemhauser J. L. Steroids are required for epidermal cell fate establishment in Arabidopsis roots. P. Natl. Acad. Sci. USA 106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y. W. et al. Brassinosteroids control root epidermal cell fate via direct regulation of a MYB-bHLH-WD40 complex by GSK3-like kinases. eLife 3, 10.7554/eLife.02525 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J. M. et al. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA 111, 2367–2372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M. & Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R., Stepanova A., Chao Q. & Ecker J. R. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy M. et al. Improved protein-binding microarrays for the identification of DNA-binding specificities of transcription factors. Plant J 66, 700–711 (2011). [DOI] [PubMed] [Google Scholar]
- Kim M. J., Ruzicka D., Shin R. & Schachtman D. P. The Arabidopsis AP2/ERF transcription factor RAP2.11 modulates plant response to low-potassium conditions. Mol. Plant 5, 1042–1057 (2012). [DOI] [PubMed] [Google Scholar]
- Sundaravelpandian K., Chandrika N. N. P. & Schmidt W. PFT1, a transcriptional Mediator complex subunit, controls root hair differentiation through reactive oxygen species (ROS) distribution in Arabidopsis. New Phytol 197, 151–161 (2013). [DOI] [PubMed] [Google Scholar]
- Yi K., Menand B., Bell E. & Dolan L. A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat. Genet. 42, 264–267 (2010). [DOI] [PubMed] [Google Scholar]
- Datta S. et al. Root hairs: development, growth and evolution at the plant-soil interface. Plant Soil 346, 1–14 (2011). [Google Scholar]
- Cai W. H., Okuda K., Peng L. W. & Shikanai T. PROTON GRADIENT REGULATION 3 recognizes multiple targets with limited similarity and mediates translation and RNA stabilization in plastids. Plant J. 67, 318–327 (2011). [DOI] [PubMed] [Google Scholar]
- Marchive C. et al. Abnormal physiological and molecular mutant phenotypes link chloroplast polynucleotide phosphorylase to the phosphorus deprivation response in Arabidopsis. Plant Physiol. 151, 905–924 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah B. N., Karthikeyan A. S. & Raghothama K. G. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 143, 1789–1801 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson-Paulik J., Bastidas R. J., Chiou S. T., Frye R. A. & York J. D. Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc. Natl. Acad. Sci. USA 102, 12612–12617 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H., Tasaka M. & Shikanai T. PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J. 38, 152–163 (2004). [DOI] [PubMed] [Google Scholar]
- Tatematsu K., Ward S., Leyser O., Kamiya Y. & Nambara E. Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis. Plant Physiol. 138, 757–766 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H., Fritsch O. & Gasser S. M. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 26, 4113–4125 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. et al. The chromatin-remodeling factor AtINO80 plays crucial roles in genome stability maintenance and in plant development. Plant J. 82, 655–668 (2015). [DOI] [PubMed] [Google Scholar]
- Smith A. P. et al. Histone H2A.Z regulates the expression of several classes of phosphate starvation response genes but not as a transcriptional activator. Plant Physiol. 152, 217–225 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier C. R. & Cairns B. R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304 (2009). [DOI] [PubMed] [Google Scholar]
- Lee S. H. & Cho H. T. PINOID positively regulates auxin efflux in Arabidopsis root hair cells and tobacco cells. Plant Cell 18, 1604–1616 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F. et al. Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol. Cell 55, 383–396 (2014). [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno M. A. et al. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329, 1306–1311 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R., Quint A., Weijers D., Hooykaas P. & Offringa R. The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128, 4057–4067 (2001). [DOI] [PubMed] [Google Scholar]
- Parker J. S., Cavell A. C., Dolan L., Roberts K. & Grierson C. S. Genetic interactions during root hair morphogenesis in Arabidopsis. Plant Cell 12, 1961–1974 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S. et al. The glycerophosphoryl diester phosphodiesterase-like proteins SHV3 and its homologs play important roles in cell wall organization. Plant Cell Physiol. 49, 1522–1535 (2008). [DOI] [PubMed] [Google Scholar]
- Lee C., Teng Q., Zhong R. & Ye Z. H. Arabidopsis GUX proteins are glucuronyltransferases responsible for the addition of glucuronic acid side chains onto xylan. Plant Cell Physiol. 53, 1204–1216 (2012). [DOI] [PubMed] [Google Scholar]
- Bromley J. R. et al. GUX1 and GUX2 glucuronyltransferases decorate distinct domains of glucuronoxylan with different substitution patterns. Plant J. 74, 423–434 (2013). [DOI] [PubMed] [Google Scholar]
- Schuetz M. et al. Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol. 166, 798–U489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. M., Zeef L. A., Ellis J., Goodacre R. & Turner S. R. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17, 2281–2295 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J. H., Beers E. P. & Han K. H. Global comparative transcriptome analysis identifies gene network regulating secondary xylem development in Arabidopsis thaliana. Mol. Genet. Genomics 276, 517–531 (2006). [DOI] [PubMed] [Google Scholar]
- Bouvier F., Mialoundama A. & Camara B. A sentinel role for plastids In The Chloroplast (eds Sandelius A. S., Aronson H. ) 267–292 (Springer, 2009). [Google Scholar]
- Leister D. Retrograde signaling in plants: from simple to complex scenarios. Front Plant Sci 3, 135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrillo E. et al. A Chloroplast retrograde signal regulates nuclear alternative splicing. Science 344, 427–430 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J. et al. Suppression of photosynthetic gene expression in roots is required for sustained root growth under phosphate deficiency. Plant Physiol. 165, 1156–1170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacón-López A., Ibarra-Laclette E., Sánchez-Calderón L., Gutiérrez-Alanis D. & Herrera-Estrella L. Global expression pattern comparison between low phosphorus insensitive 4 and WT Arabidopsis reveals an important role of reactive oxygen species and jasmonic acid in the root tip response to phosphate starvation. Plant Signal. Behav. 6, 382–392 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong-Villalobos L. et al. Methylome analysis reveals an important role for epigenetic changes in the regulation of the Arabidopsis response to phosphate starvation. Proc. Natl. Acad. Sci. USA 112, E7293–302 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer D. & Mauch-Mani B. More beneath the surface? Root versus shoot antifungal plant defenses. Front. Plant Sci. 4, 256 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen T. G. et al. Integration of biosynthesis and long-distance transport establish organ-specific glucosinolate profiles in vegetative Arabidopsis. Plant Cell 25, 3133–3145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen B. L., Chen S., Hansen C. H., Olsen C. E. & Halkier B. A. composition and content of glucosinolates in developing Arabidopsis thaliana. Planta 214, 562–571 (2002). [DOI] [PubMed] [Google Scholar]
- Frerigmann H., Bottcher C., Baatout D. & Gigolashvili T. Glucosinolates are produced in trichomes of Arabidopsis thaliana. Front. Plant Sci. 3, 242 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanoue A., Burlat V., Schurr U. & Rose U. S. Induced root-secreted phenolic compounds as a belowground plant defense. Plant Signal. Behav. 5, 1037–1038 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L. & Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 (1994). [PubMed] [Google Scholar]
- Bindea G. et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology annotation networks. Bioinformatics 25, 1091–1093 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.